- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Optimization of Extracellular Flux Assay to Measure Respiration of Anchorage-independent Tumor Cell Spheroids
Published: Vol 12, Iss 4, Feb 20, 2022 DOI: 10.21769/BioProtoc.4321 Views: 4575
Reviewed by: Masahiro MoritaKimberly Dunham-SnaryAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Assessment of Cellular Redox State Using NAD(P)H Fluorescence Intensity and Lifetime
Thomas S. Blacker [...] Gyorgy Szabadkai
Jan 20, 2017 14864 Views

FACS-based Glucose Uptake Assay of Mouse Embryonic Fibroblasts and Breast Cancer Cells Using 2-NBDG Probe
Shengli Dong and Suresh K Alahari
Apr 20, 2018 11539 Views

Measurement of Oxygen Consumption Rate (OCR) and Extracellular Acidification Rate (ECAR) in Culture Cells for Assessment of the Energy Metabolism
Birte Plitzko and Sandra Loesgen
May 20, 2018 76077 Views
Abstract
Three-dimensional (3D) cell culture models are widely used in tumor studies to more accurately reflect cell-cell interactions and tumor growth conditions in vivo. 3D anchorage-independent spheroids derived by culturing cells in ultra-low attachment (ULA) conditions is particularly relevant to ovarian cancer, as such cell clusters are often observed in malignant ascites of late-stage ovarian cancer patients. We and others have found that cells derived from anchorage-independent spheroids vary widely in gene expression profiles, proliferative state, and metabolism compared to cells maintained under attached culture conditions. This includes changes in mitochondrial function, which is most commonly assessed in cultured live cells by measuring oxygen consumption in extracellular flux assays. To measure mitochondrial function in anchorage-independent multicellular aggregates, we have adapted the Agilent Seahorse extracellular flux assay to optimize measurements of oxygen consumption and extracellular acidification of ovarian cancer cell spheroids generated by culture in ULA plates. This protocol includes: (i) Methods for culturing tumor cells as uniform anchorage-independent spheroids; (ii) Optimization for the transfer of spheroids to the Agilent Seahorse cell culture plates; (iii) Adaptations of the mitochondrial and glycolysis stress tests for spheroid extracellular flux analysis; and (iv) Suggestions for optimization of cell numbers, spheroid size, and normalization of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) values. Using this method, we have found that ovarian cancer cells cultured as anchorage-independent spheroids display altered mitochondrial function compared to monolayer cultures attached to plastic dishes. This method allows for the assessment of mitochondrial function in a more relevant patho/physiological culture condition and can be adapted to evaluate mitochondrial function of various cell types that are able to aggregate into multicellular clusters in anchorage-independence.
Graphic abstract:
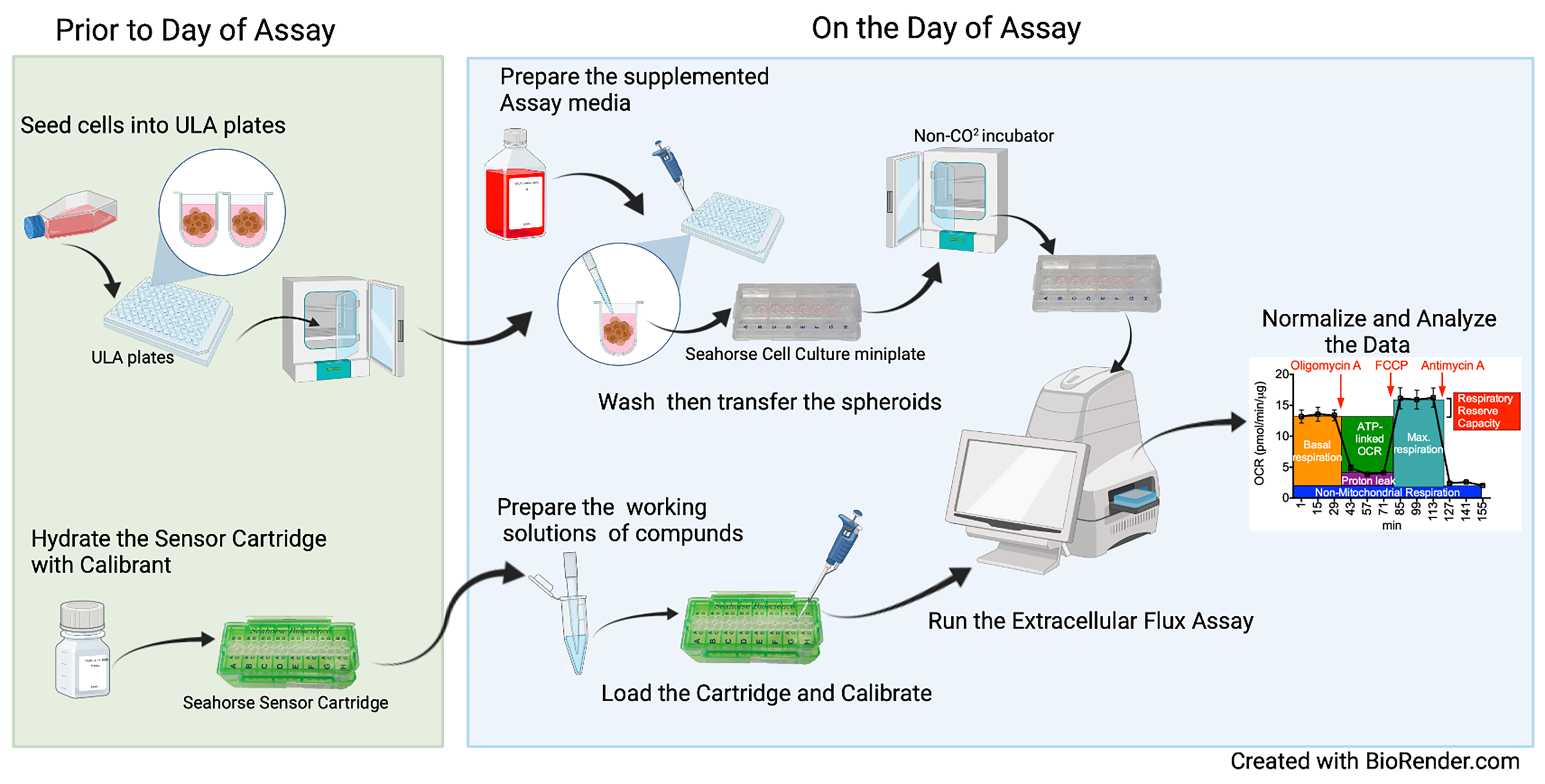
Workflow of the Extracellular Flux Assay to Measure Respiration of Anchorage-independent Tumor Cell Spheroids.
Background
Epithelial ovarian cancer remains the deadliest gynecological malignancy with an estimated five-year survival rate below 29% for patients with advanced stage disease, which is when most patients are first diagnosed (Torre et al., 2018). At these later stages, ovarian cancer cells have metastasized to distant sites across the peritoneum. This spread of ovarian cancer across the peritoneal cavity is facilitated by the passive dissemination of tumor cells directly into the peritoneal ascites fluid. In the malignant ascites, tumor cells can be found as both single cells and as multicellular clusters, also commonly referred to as “spheroids”. Formation of spheroid aggregates, which are enriched in cancer stem cells (Liao et al., 2014; Kenda Suster and Virant-Klun, 2019), are key to the progression of ovarian cancer, as these ensure maintenance of intercellular survival signaling among cells detached from the primary tumor (Tan et al., 2006; Al Habyan et al. 2018). In addition, cancer stem-like cells in spheroids display metabolic flexibility. They shift their metabolism based on nutrient availability and metabolic demands, potentially allowing tumor cells to survive in ascites and invade the neighboring peritoneal organs (Anderson et al.., 2013; Kim et al.., 2020; Ghoneum et al., 2020). To advance our understanding of ovarian cancer metastatic progression and associated changes in tumor metabolism, it is important to develop methods that allow us to study the metabolic changes in anchorage-independent tumor spheroids, which mimic tumor cell clusters found in malignant ascites of patients.
The following protocol utilizes ultra-low attachment (ULA) plates to generate multi-cellular tumor cell spheroid clusters in order to study their metabolic phenotype using the Seahorse XFp (Agilent) extracellular flux assay platform. Spontaneous clustering of tumor cells in anchorage independence is facilitated by the lack of ability to attach to the hydrophilic neutrally charged hydrogel coating of ULA plates. Compared to cells cultured in conventional attached monolayer conditions on plastic surfaces, spheroids more closely reflect the metastatic tumor cells found in anchorage-independent multicellular clusters in malignant ascites. This makes spheroids a good in vitro model to better simulate the in vivo behavior of tumor cells and to study their unique characteristics related to cell survival, drug sensitivity, hypoxia, and tumor metabolism. During optimization of this protocol, we determined that uniform spheroid formation is integral to minimizing variability in extracellular flux measurements. The use of ULA platforms with small well sizes, such as 96 or 384 wells per plate, permits the generation of uniformly sized spheroids by controlling the seeding density. The use of bioprinting in combination with magnetic nanoparticles has been investigated as an alternate method for metabolic studies of 3D tumor-fibroblast co-cultures (Noel et al., 2017). However, for cell types that spontaneously cluster into spheroids in response to anchorage independence, the ULA culture platform represents a relatively straightforward approach to generate uniform multicellular clusters, without the need for further manipulation.
The Seahorse extracellular flux analyzers specifically measure oxygen consumption (OCR) and extracellular acidification rates (ECAR), allowing for the quantification of mitochondrial respiration and an indirect indication of glycolysis, respectively. Several past studies have interrogated mitochondrial function in 3D cultures using single well assays and measured the response to single or a limited number of metabolic stress compounds, with the need to independently determine the rates of oxygen consumption and extracellular acidification (Rodríguez-Enríquez et al., 2008; Schafer et al., 2009; Mandujano-Tinoco et al., 2013; Bloch et al., 2014; Marin-Hernandez et al., 2017). We have optimized the transfer of spheroids from 96 well ULA plates to the Seahorse cell culture plates and the use of this platform for assessment of spheroid bioenergetics, allowing for multi-well analysis and sequential addition of pharmacological agents to manipulate OCR/ECAR. Below are described the use of the mitochondrial stress test and the glycolysis stress test, together with how spheroid culturing and transfer is best achieved. Using substrate limited media and pharmacological approaches, the assay is easily modified to gain further insight into the specific metabolic activity of spheroids. Although not discussed in detail below, iterations of the extracellular flux assay include the palmitate oxidation stress test, which is a modified assay that determines the capacity of cells to oxidize palmitate in the absence or limitation of other exogenous carbon substrates. We refer the reader to the Seahorse Agilent website for alternate assays to assess substrate utilizations. The protocol was developed using the Agilent Seahorse XFp Metabolic Analyzer, and can be applied to the Seahorse XF96 format, as these two platforms utilize cell culture plates with similar surface areas and media volumes. It can also be adapted for the larger surface area of the XF24 platform wells. Hence, the method explained in this paper can be used as a master template for performing not only the mentioned mitochondrial and glycolysis stress tests, but also altered extracellular flux assays with 3D anchorage-independent tumor cell spheroids.
The method outlined below was originally designed for the study of ovarian cancer spheroids cultured in anchorage-independent conditions, which mimic tumor cell clusters commonly found in malignant ascites of ovarian cancer patient (Kim et al., 2020). It can be adapted to other cancer cell lines, primary cells, stem cell cultures, or cell types where 3D cell culture represents a more patho/physiologically relevant culture condition.
Materials and Reagents
96 Well round bottom ultra-low attachment (ULA) plates (Corning, catalog number: 7007)
0.2 µm sterile syringe filter (polyethersulfone membrane, VWR, catalog number: 28145-501)
T-75 tissue culture flask, vent cap, sterile (Celltreat, catalog number: 229341)
0.2 µm sterile vacuum filtration system with polyethersulfone membrane for cell culture media (VWR, catalog number: 10040-436)
Tumor cells:
ES-2 (ATCC, catalog number: CRL-1978)
NIHOVCAR3 (ATCC, catalog number: HTB-161)
OVCA433 cells (generously provided by Dr. Susan Murphy, Duke University)
Note: Experiments should be limited to cells in culture for less than 10 passages.
Cell culture media:
ES-2 cells: McCoy’s 5A + L-glutamine (Corning, catalog number: 10-050-CV), supplemented with 10% FBS
OVCAR3 cells: RPMI 1640 + L-glutamine (Gibco, catalog number: 11875119), supplemented with 20% FBS and 0.01 mg/mL bovine insulin
OVCA433 cells: RPMI 1640 + L-glutamine (Corning, catalog number: 10-040-CV) supplemented with 10% FBS
Note: Media are sterile filtered using the following 0.2 µm vacuum Filters. If desired, Penicillin/Streptomycin may be added to the culture media, depending on cell lines used.
Fetal Bovine Serum (FBS) (VWR, catalog number: 97068-085)
Phosphate buffered saline (PBS, sterile; Corning, catalog number: 21-040-CV)
0.25% Trypsin-EDTA (sterile) (Gibco, catalog number: 25200114)
Seahorse XFp FluxPaks includes Seahorse XFp cell culture miniplates (8 wells), XFp sensor cartridges (8 wells) and XF calibrant solution (Agilent, catalog number: 103022-100)
Seahorse XF base assay media (DMEM, Agilent, catalog number: 103575-100; RPMI, Agilent, catalog number: 103576-100), supplemented on the day of assay as described below, pH 7.4
XF 100 mM Pyruvate Solution (Agilent, catalog number: 103578-100); alternatively sterile 100 mM Pyruvate solution may be sought from other vendors or made in house.
XF 200 mM Glutamine Solution (Agilent, catalog number: 103579-100), alternatively sterile 200 mM Glutamine solution may be sought from other vendors or made in house.
Sterile DEPC-treated ddH2O (Quality Biological, catalog number: 351-068-131); alternatively sterile autoclaved ddH2O can be used.
PierceTM BCA Protein Assay (Thermo Scientific, catalog number: 1859078)
Sodium chloride
Tris-HCl pH 8.0 (Tris Base; Sigma, catalog number: TRIS-RO)
NP40/IgepalCA-630 (Sigma, catalog number: 13021)
Sodium deoxycholate (Sigma, catalog number: D6750)
Sodium phosphate dibasic (Sigma, catalog number: S9763)
Sodium phosphate monobasic (anhydrous; Sigma, catalog number: S3139)
Sodium phosphate buffer
Sodium dodecyl sulfate (SDS; Sigma, catalog number: L3771)
Ethylenedinitrilotetraacetic acid (EDTA; Sigma, catalog number: E9884)
Sodium fluoride (NaF; Sigma, catalog number: 201154)
Protease inhibitor cocktail (Thermo Scientific, catalog number: 87785)
D-glucose (Sigma, catalog number: G8270)
2-Deoxy-D-glucose, 2-DG (Sigma, catalog number: D8375)
Oligomycin A (Sigma, catalog number: 75351) (see Recipes, Table 3)
FCCP (Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; Sigma, catalog number: C2920) (see Recipes, Table 3)
Antimycin A (Sigma, catalog number: A8674) (see Recipes, Table 3)
Rotenone (Sigma, catalog number: 45656) (see Recipes, Table 3)
RIPA lysis buffer (see Recipes)
Equipment
Agilent Seahorse XFp extracellular flux analyzer (Agilent, model: S7802A)
Humidified cell culture incubator (37°C, 5% CO2)
Non-CO2 Humidified cell culture incubator (37°C)
Tissue culture tabletop centrifuge compatible for 15 mL Falcon tubes (Eppendorf, model: Centrifuge 5702)
Tissue culture light microscope with 4× objective and camera
Laminar flow biosafety cabinet (Thermo Scientific, model: 1300 Series A2)
Software
Seahorse Wave Desktop 2.6 Software (Agilent, https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolic-analysis/xf-software/seahorse-wave-desktop-software-740897)
Procedure
Spheroid Culture
Grow tumor cells to ~70% confluence in a T-75 cell culture flask with normal supplemented growth media optimized for the cell lines of choice.
One to three days prior to the Seahorse extracellular flux assay, wash cells with 5 mL of warm PBS and detach using 2 mL of 0.25% trypsin/EDTA by incubating at 37°C for 10 min for ES-2 and OVCAR3 cells, and 15 min for OVCA433 cells. The time for trypsinization should be optimized for the cell lines used.
Deactivate trypsin/EDTA with 6 mL of fully supplemented growth media, collect cells, and centrifuge at 500 × g for 5 min. Remove trypsin/media and wash cells with 5 mL of warm PBS. Repeat centrifugation and remove PBS.
Resuspend cells in fully supplemented growth media (5 mL). In a 1.5 mL tube, add trypan blue at a 1:1 ratio to 0.2 mL of resuspended cells, and count viable cells via the trypan blue exclusion method, using a hemocytometer or automated cell counter. Derive final cell numbers by considering the trypan blue dilution factor. Alternate cell counting methods may be used. A single T-75 flask of ovarian cancer cells will typically yield ~0.8 × 106cells/mL.
Dilute cells to 5 × 103 cells/mL and seed into a 96-well round bottom ULA plate, by pipetting 1,000 cells per well in 200 µL of growth media.
Notes:
The use of round bottom ULA plates is recommended, as this is the most effective method for producing uniform-sized single spheroids per well. This ensures accurate transfer of predetermined size and numbers of spheroids to the Seahorse assay plate (Figure 1).
The need for 1,000 cells per well was determined for ovarian cancer cell lines ES-2, OVCAR3, and OVCA433 to achieve uniform spheroid formation in 96 well ULA plates. Cell numbers per spheroid may require optimization depending on the cell lines used. It should be kept in mind that increasing cell numbers may lead to larger spheroids with hypoxic cores, potentially influencing mitochondrial bioenergetics. Moreover, adequate penetrance of compounds used in the Seahorse stress assays needs to be considered.
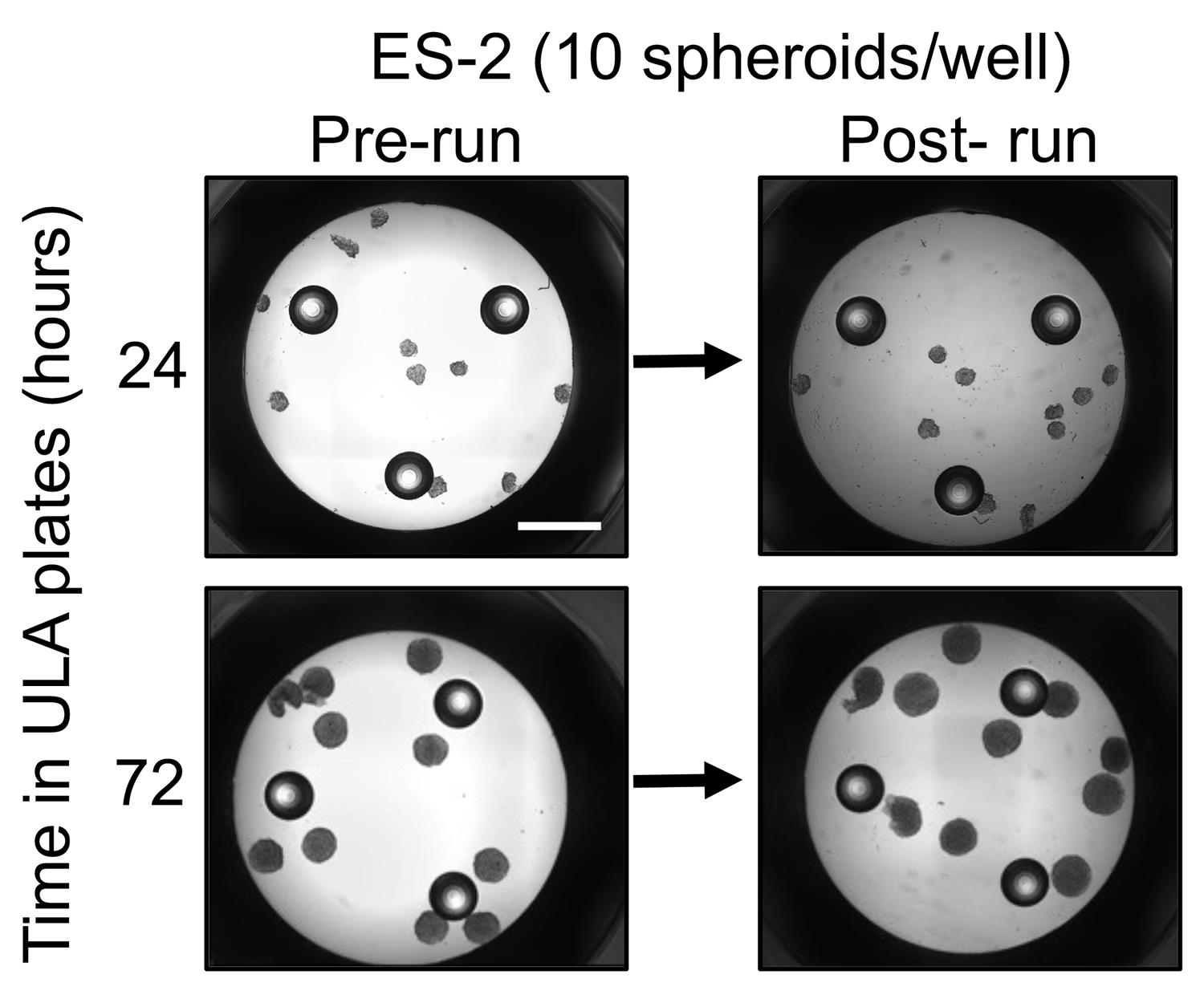
Figure 1. Image of ES-2 cell spheroids pre and post extracellular flux assay run. Spheroids remain intact after the completion of the assay. Scale bar = 1mm.
Incubate cells in ULA plates for 24-72 h under normal cell culture conditions in a humidified cell culture incubator (37°C, 5% CO2). The timing in ULA conditions is dependent on the experimental design and cell lines used.
If comparing bioenergetics of spheroids to attached cells, follow steps A1–A4, and seed cells directly into Seahorse XFp wells. The number of cells seeded per well is dependent on proliferation rate differences between attached and spheroid cultured cells, therefore, the same number of cells should be plated in attached conditions relative to the number of spheroids per Seahorse well x cells/spheroid, for comparison.
Preparation of Seahorse Reagents and assay setup
Note: The following should be prepared a day before the Seahorse extracellular Flux Assay.
Hydrate the XFp sensor cartridge, by adding 200 µL of sterile ddH2O to each well and 400 µL into the outer moats surrounding the wells of the utility plate. Place sensor cartridge into a 37°C humidified non-CO2 incubator overnight. (For more information on preparing the cartridge refer to: https://www.agilent.com/en/product/cell-analysis/how-to-run-an-assay#chapter2_3, https://www.agilent.com/cs/library/usermanuals/public/Hydrating_an_XFp_Sensor_Cartridge.pdf).
Turn on the Agilent Seahorse XFp Analyzer to ensure the temperature of the machine is at 37°C on the day of the assay.
Open the wave desktop software and navigate to the template tab.
Select the XFp cell Mito Stress test template. Under the tab “group definitions” in “assay navigation”, assign the experimental groups and proceed to design your plate map with your groups in the next tab. Lastly, under the “protocol” tab, set up the spheroid mitochondrial stress test assay with the following parameters:
Three repeats: mix for 3 min, and measure for 3 min.
Inject Oligomycin A in Port A.
Three repeats: mix for 3 min, and measure for 3 min.
Inject FCCP in Port B.
Three repeats: mix for 3 min, and measure for 3 min.
Inject Rotenone/Antimycin A.
Three repeats: mix for 3 min, and measure for 3 min.
Select the XFp cell Glycolysis Stress test template. Define your experimental groups and design your plate maps in the assay navigation. Set up the protocol for the spheroid glycolysis stress test assay with the following run times:
Three repeats: mix for 3 min, and measure for 3 min.
Inject Glucose in Port A.
Three repeats: mix for 3 min, and measure for 3 min.
Inject Oligomycin in Port B.
Three repeats: mix for 3 min, and measure for 3 min.
Inject 2-Deoxy-D-Glucose in Port C.
Three repeats: mix for 3 min, and measure for 3 min.
Note: The above assay run protocols may need to be adjusted for individual experimental setups. If a response plateau is not reached after the indicated number of reads following addition of drug, the number of reads and measure times should be increased to ensure that maximal effects of compounds are recorded. For dense and larger spheroids, five repeats of mix and measure after addition of a drug is recommended.
Save the templates as new protocols and transfer to the Agilent Seahorse XFp Analyzer via the network or a USB drive.
Note: For more information on experiment design and assay setup in Wave please refer to: https://www.agilent.com/en/product/cell-analysis/how-to-run-an-assay - chapter3_4, https://www.agilent.com/cs/library/usermanuals/public/S7894-10000_Rev_C_Wave_2_6_User_Guide.pdf).
Transfer of Spheroids into Seahorse Cell Culture Plates
On the day of the assay, prepare the Seahorse XF assay media:
For the mitochondrial stress test, supplement 10 mL of Seahorse XF base media (DMEM or RPMI, depending on cell line) with 1 mM sodium pyruvate, 2 mM glutamine, and 10 mM glucose. A volume of 10 mL of assay media is sufficient to run one XFp miniplate and to make compound dilutions. If multiple plates are assayed, or the assay is performed in a XF96 platform, the amount of assay media prepared should be scaled up accordingly.
For the glycolysis stress test, supplement 10 mL of Seahorse XF base medium with 2 mM glutamine.
Note: Seahorse XF assay media lacks sodium bicarbonate to maintain low buffering capacity. For the same reason, it is recommended to avoid the use of serum in the assay media. If cells are sensitive to a lack of FBS during the assay run, addition of 1% of FBS can be tested.
Warm the assay medium to 37°C prior to adjusting the pH to 7.4, using HCl or NaOH. Filter sterilize using a 0.2 µm syringe filter.
Replace growth media with Seahorse XF assay media and move spheroids from round bottom ULA well plates into the XFp cell culture plate, approximately 1–2 h prior to the assay run, using the following steps (C4–C10):
Replace growth media in each ULA plate well with Seahorse assay media. Carefully remove 180 µL of growth media (from 96-well ULA plate) using a P200 pipette, by tilting the plate and angling the pipette tip towards the upper back of the well, avoiding aspiration of spheroids, which will be present in the center of the round bottom ULA well. Each ULA plate well should contain one spheroid. Inspect wells under the microscope for any loss of spheroids due to the aspiration procedure.
Carefully add 200 µL of Seahorse Assay media to each ULA well by slowly pipetting media against the side of the well.
Remove 180 µL of Seahorse assay media using a P200 pipette, as in step C4. A multichannel pipette can also be used to perform the washing steps (C3–C5).
Repeat steps C3–C5 to ensure that minimal residual sodium bicarbonate from growth medium remains.
Note: An additional wash step (C4–C5) can be added if deemed necessary.
Remove each spheroid from a ULA well by carefully aspirating it in the remaining liquid (~40 µL) using a P1000 pipette, and transfer it to a well of the Seahorse XFp cell culture miniplate (wells B–G). Observe the ULA well and the XFp miniplate wells under the microscope to ensure successful transfer.
Repeat step 8 to transfer the desired number of spheroids per XFp cell culture plate well. The optimal number of spheroids per XFp well should be determined empirically (Figure 2). Let the spheroids settle to bottom and adjust the media volume in the wells to a total of 180 µL.
Add 180 µL of Seahorse assay media to wells A & H of the Seahorse XFp cell culture miniplate. Do not add spheroids to these wells, as they are used as a blank reference in the assay.
Inspect wells using a microscope and take images with a low magnification objective prior to the assay run (Figure 1). This is to ensure successful transfer of the spheroids in each well, and adjust the number of spheroids if needed, in case of a failed transfer.
Place the XFp cell culture miniplate containing spheroids in Seahorse assay media into a humidified 37°C non-CO2 incubator for 60 min prior to the assay, to de-gas the plate. This ensures diffusion of CO2 from the cells and media, prior to the measurements of O2 and H+ in the assay.
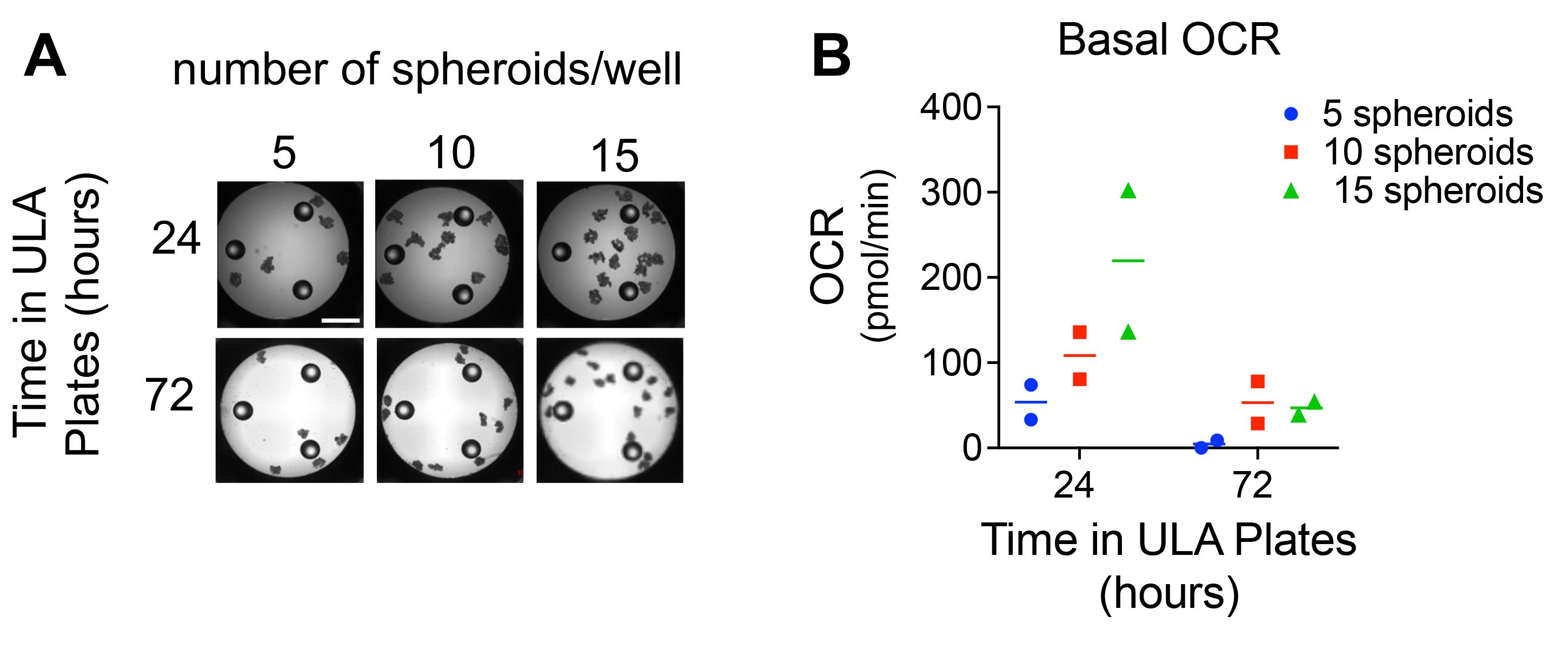
Figure 2. Optimization of spheroid number and ULA culture time. A. Images of OVCAR3 spheroids after transfer into the XFp minicell culture plate wells. Scale bar=1 mm. B. Basal OCR measurements were obtained from the indicated number of spheroids per well, using the Seahorse XFp extracellular flux analyzer. Prior to the assay, spheroids were cultured in ULA plates for either 24 or 72 h (n=2, with mean OCR shown).
Seahorse XFp mitochondrial stress test
On the day of the assay, remove the H2O from the hydrated XFp sensor cartridge and replace with 200 µL of prewarmed XF Calibrant. Leave in 37°C humidified non-CO2 incubator until the assay run.
Prepare the mitochondrial stress test reagents under sterile conditions (refer to the Recipes section for details). Prepare 300 µL of 10× reagents by diluting the stock solutions with pre-warmed XF assay media to the desired concentrations (see Table 1). The optimal concentrations for Oligomycin and FCCP can be adjusted based on empirical determination for each cell line. It is recommended to determine the optimal concentrations that yield maximal responses by testing a range of final well concentration between 0.125–2 µM of FCCP and between 0.5–2 µM of Oligomycin A.
Consult the following method to carry out oligomycin A and FCCP titration assays: https://www.agilent.com/cs/library/usermanuals/public/New_Cell_Line_Characterization.pdf.
Table 1. Preparation of compound solutions for Mitochondrial Stress test.
Port Designation Final Well Conc.
(µM)
Stock Solution Conc.
(µM)
Stock Solution Volume (µL) Assay Media Volume (µL) 10× Port Conc.
(µM)
Volume added to port (µL) Port A:
Oligomycin
1.0 50 60 240 10 20 1.5 90 210 15 20 Port B:
FCCP
0.5 50 30 270 5 22 1.0 60 240 10 22 2.0 120 180 20 22 Port C:
Rotenone /Antimycin A
0.5 25 60 240 5 25 Load the 10× concentrated reagents into the drug injection ports of the hydrated sensor cartridge, at the volumes indicated in Table 1. Care should be taken while pipetting, to ensure equal loading and avoid the creation of air bubbles, which can interfere with the injection of compounds (Figure 3A).
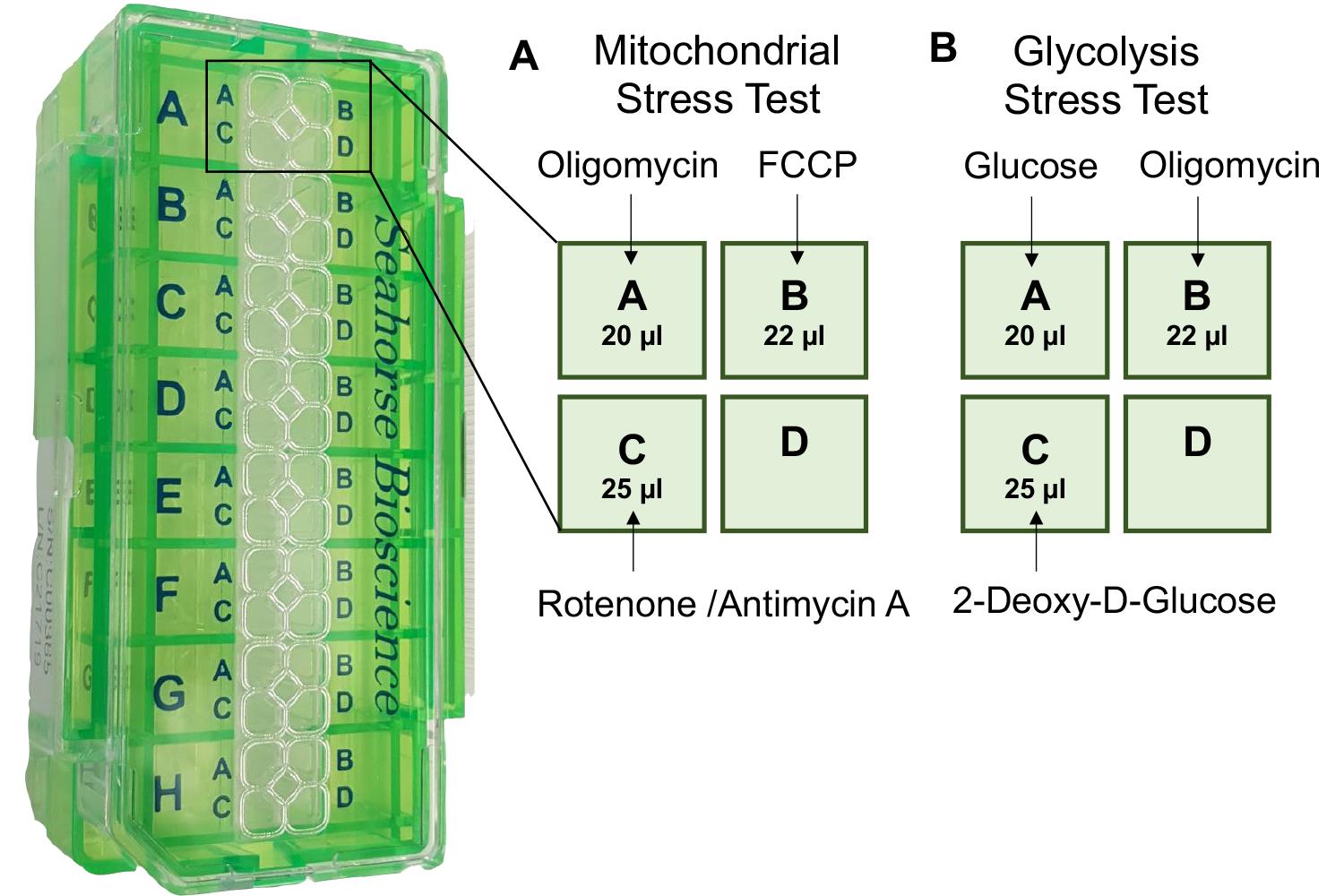
Figure 3. Loading the cartridge ports with test compounds. A. Mitochondrial stress test. B. Glycolysis stress test.For further details on loading the compounds into the cartridge please see: https://www.agilent.com/en/product/cell-analysis/how-to-run-an-assay#chapter3_4, https://www.agilent.com/cs/library/usermanuals/public/Loading%20Cartridge%20XFp.pdf.
Touch the start button on the XFp Analyzer home view to see the available list of assay templates. Run the Agilent Seahorse XFp cell mitochondrial stress test by selecting the protocol created in step B4. After assigning the experimental groups, click the protocol page and “Start Assay”.
Place the utility plate with the hydrated sensor cartridge and loaded ports containing the mitochondrial stress test reagents on the instrument tray. Ensure that the cartridge lid has been removed and place the plate in the correct orientation. Click continue to initiate machine calibration (~20 min).
Following calibration, remove the utility plate and replace with the Seahorse XFp cell culture miniplate containing the spheroids. Ensure that the cell culture plate lid has been removed and place the plate in the correct orientation. Press “continue” to start the assay.
Seahorse XFp glycolysis stress test
On the day of the assay, remove the diH2O from the hydrated XFp sensor cartridge and replace with 200 µL of XF Calibrant. Leave in 37°C humidified non-CO2 incubator until the assay run.
Prepare the glycolysis stress test reagents under sterile conditions (refer to the Recipes section for details). Prepare 300 µL of 10× reagents by diluting the stock solutions with pre-warmed XF assay media to the desired concentrations (see Table 2). The optimal concentration for Oligomycin can be adjusted based on empirical determination for each cell line.
Table 2. Preparation of compound solutions for glycolysis stress test.
Port Designation Final Well Conc.
Stock
Conc.
Stock Solution Volume (µL) Assay Media Volume (µL) 10× Port Conc. Volume added to port (µL) Port A:
Glucose
10 mM 100 mM 300 0 100 mM 20 Port B:
Oligomycin A
1.0 µM 50 µM 60 240 10 µM 22 1.5 µM 90 210 15 µM 22 2.0 µM 120 180 20 µM 22 Port C:
2-Deoxy-D-Glucose
50 mM 500 mM 300 0 500 mM 25 Load the 10× concentrated reagents into the drug injection ports of the hydrated sensor cartridge, at the volumes indicated in Table 2. Care should be taken while pipetting, to ensure equal loading and avoid the creation of air bubbles (Figure 3B).
Touch the start button on the XFp Analyzer home view to see the available list of assay templates. Run the Agilent Seahorse XFp cell glycolysis stress test, by selecting the protocol created in step B5. After assigning the experimental groups, click the Protocol page and “Start Assay”.
Proceed by following steps D5–6.
Normalization
Following the completion of the assay, remove the cell culture miniplate and image the spheroids as in step C11, to ensure spheroid integrity and even distribution of spheroids in the assay well post run (Figure 1).
Prior to data analysis, choose a method to normalize the results. Examples of data normalization include:
Number of spheroids per well. Prior to the assay, equal numbers of spheroids are plated into assay plate wells. This can be achieved by use of 96 well ULA plates to culture spheroids, as these tend to generate a single spheroid per well. Hence, the exact number of desired spheroids/assay well can be accurately transferred into the assay well plate. Post assay, the wells are imaged to count the number of spheroids again, to ensure that no loss or break-up of spheroids occurred during the assay. OCR and ECAR values are then divided by the number of spheroids per assay well and expressed per spheroid (e.g., OCR: pmol/min/spheroid). This method is appropriate when manipulation of experimental parameters is not expected to change spheroid morphology or size, and can be useful when wells vary in spheroid numbers.
Number of total cells per well. This normalization takes into account the total number of cells per spheroid and the total number of spheroids per well. This normalization per cell is an appropriate method when comparing OCR and ECAR values between cells cultured as spheroids and those cultured in attached conditions. The total number of cells is calculated by multiplying the initial seeding density of a single spheroid with the final number of spheroids per XFp well. For comparison to attached conditions, the same total number of cells/well are seeded directly into the Seahorse assay plate. The OCR and ECAR measurement are then expressed per cell (e.g., OCR: pmol/min/cell). This normalization should be carried out to determine if cell density per spheroid affects bioenergetics. If desired, cell counts can be determined again post assay. Spheroids are collected from the assay wells, transferred to 1.5 mL Eppendorf tubes, centrifuged (500 × g for 5 min), and resuspended in 0.25% trypsin/EDTA. Following 5 min incubation at 37°C, and gentle resuspension after 1:1 addition of trypan blue solution, cells are counted using a hemocytometer, or other preferred cell counting method.
Protein concentration per well. Spheroids are collected from the assay wells post assay, transferred to 1.5 mL Eppendorf tubes, centrifuged (500 × g for 5 min), media is aspirated, and cells are resuspended in 50 μL of lysis buffer, followed by protein quantification using a preferred method, such as the Bicinchoninic Acid (BCA) protein assay. The total protein per well is used to normalize the OCR and ECAR measurements across the wells (e.g., OCR: pmol/min/mg protein). For samples with different spheroid sizes, this should be used as the method of normalization.
Note: It is recommended to save the cells post-assay to allow for normalization of protein content, in addition to spheroid and cell numbers.
Export the assay result from the Agilent Seahorse XFp Analyzer. Add values for normalization and analyze the data using the Agilent Seahorse Wave software.
Data analysis
Use the normalized results to either generate the mitochondrial stress test report or the glycolysis stress test report via the Wave software, or export the results out of the software and proceed to use the read measurements to extract the following data:
Mitochondrial Stress test:
Determine which of the repeat measurements following compound addition represents the maximal response to each compound in the mitochondrial stress test.
Basal Mitochondrial OCR: Subtract the non-mitochondrial OCR (remaining OCR after Antimycin A/rotenone addition) from the basal OCR reading prior to injections.
ATP-linked OCR: Subtract the OCR obtained after ATP synthase inhibition (the lowest OCR reading after Oligomycin injection) from the basal OCR measurement (baseline measurement prior to injections).
Maximal Respiration: Subtract the non-mitochondrial OCR (remaining OCR after Antimycin A/rotenone addition) from the highest OCR measurement following injection of the mitochondrial uncoupler FCCP.
Respiratory reserve: subtract the basal (baseline measurement prior to the first injection) OCR measurement from the measurement that gave maximum OCR after FCCP addition.
Note: The response time to FCCP is variable in spheroids and dependent on cell line. The maximal OCR reading should be used to derive the respiratory reserve calculation. For each cell line, the timing of maximal response should be determined based on consistency of response to FCCP in experimental replicates.
H+ Proton Leak: Subtract the last OCR measurement (post Rotenone/Antimycin A injection) from the OCR after ATP synthase inhibition (the lowest OCR post Oligomycin injection).
OCR/ECAR ratios indicate the relative contribution of mitochondrial respiration to glycolysis. This can be represented as a ratio of Basal OCR measurement / Basal ECAR measurement, or plotted as in Figure 4.
Glycolysis Stress test:
Determine which of the repeat measurements following compound addition represents the maximal response to each compound and use these for the following calculations.
Basal Glycolysis: Subtract the ECAR measurement after inhibiting glycolysis with 2-Deoxy-D-glucose from the highest ECAR following glucose addition.
Maximal ECAR: The highest ECAR measurement following Oligomycin A injection.
Glycolytic Capacity: Subtract the ECAR measurement after inhibiting glycolysis with 2-Deoxy-D-glucose from the maximal ECAR following Oligomycin A addition.
Glycolytic Reserve: Subtract the maximal ECAR following glucose addition from the maximal ECAR upon addition of Oligomycin A.
Use preferred analysis and graphing software to analyze statistics and generate graphs of data.
Additional resources provide information on calculation of the above (Hill et al., 2012; Dier et al., 2014; Divakaruni et al., 2014; Plitzko and Loesgen, 2018).
Representative Results
Figure 2 above illustrates the need to optimize spheroid numbers per well and ULA culture times. To obtain single uniform spheroids, 1,000 OVCAR3 cells were seeded per well in 96 round bottom ULA plates and cultured in anchorage-independence for 24 or 72 h prior to transfer to the Seahorse cell culture assay plate. Spheroids were imaged pre-run to confirm the successful transfer and even distribution of intact spheroids (Figure 2A). Basal Oxygen consumption rates are shown for increasing numbers of spheroids per well (5, 10, and 15, Figure 2B). Increasing the number of spheroids per Seahorse well is reflected by increases in OCR levels. While this suggests that normalization against spheroid count can allow for comparison of groups with randomized number of initial spheroids seeded, it is recommended that equal numbers of spheroids are compared between experimental groups. For each cell line used, investigators should optimize the time spent in anchorage independence (culture in ULA prior to assay), initial cell seeding density (cells per spheroid), and number of spheroids/well in the extracellular flux assay.
Time in anchorage independence can affect mitochondrial respiratory function of spheroids and is dependent on cell lines. For example, OVCAR3 cells become less energetic over time in anchorage-independence (24 h vs. 72 h), decreasing both ECAR and OCR. Conversely, ES-2 cells become more energetic over time in anchorage independence (Figure 4A and 4B). This can be a consequence of cells continuing to proliferate in anchorage-independent conditions, as previously observed for ES-2 cells (Figures 1 and 2A; Dier et al., 2014). Conversely, OVCAR3 cells maintain a similar spheroid size over time in ULA plates, indicative of a quiescent state. When comparing the ECAR/OCR ratios at different time points, ES-2 cells reduce ECAR relative to OCR over time in ULA plates, suggesting a potential shift from reliance on glycolysis to oxidative phosphorylation (Figure 4C). In comparison, the ECAR/OCR ratio remains constant for OVCAR3 cells at both 24 and 72 h in anchorage independence. This example illustrates that cells differ in behavior in anchorage independence, and that normalization needs to be tailored to the experimental design. In this case, normalization to cell number or protein content post-assay would be recommended as an additional step to account for changes in proliferation.
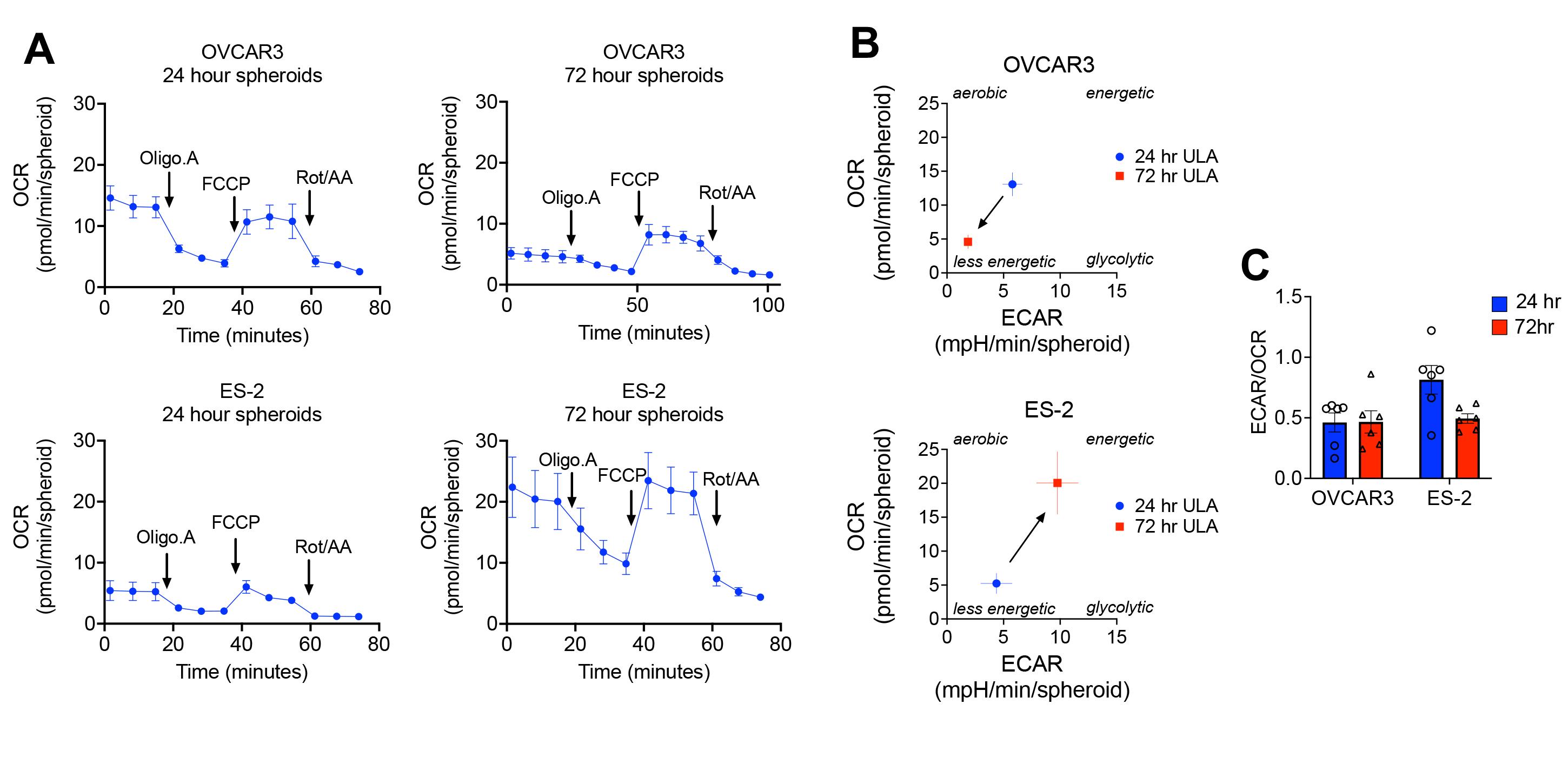
Figure 4. Bioenergetics profiles of different cell lines and dependency on time in ULA culture conditions. A. OCR measurements are shown over the course of the mitochondrial stress test assay (n=6). In the mitochondrial stress test, 1.0 µM Oligomycin and 1.0 µM FCCP were used. B. Basal OCR and ECAR values indicate that bioenergetic changes differ between cell lines, as cells are maintained for different times in anchorage-independent culture conditions. C. ECAR/OCR ratios provide information on the relative changes in oxidative phosphorylation in anchorage independence.
To compare the bioenergetic profile of tumor spheroids to the same cells cultured in attached conditions, cell lines were cultured for 24 h in 96 well ULA plates (1,000 cells per well) prior to performing the mitochondrial stress test assay (Figure 5). For analysis, 10 spheroids/well were used, and these were compared to cells in attached conditions, which were seeded at a density of 10,000 cells/well (an equivalent number of cells compared to 10 spheroids/well made of 1,000 cells each) into the seahorse cell culture miniplate 24 h prior to the assay. On the day of the assay, spheroids were transferred to the remaining empty wells in the same mini cell culture plate. The mitochondrial stress test assay demonstrates that cells cultured as spheroids can have distinct bioenergetic profiles compared to their attached counterparts (Figures 5A–4B), and their metabolic response to ULA culture again differs between ES-2 and OVCAR3 cells.
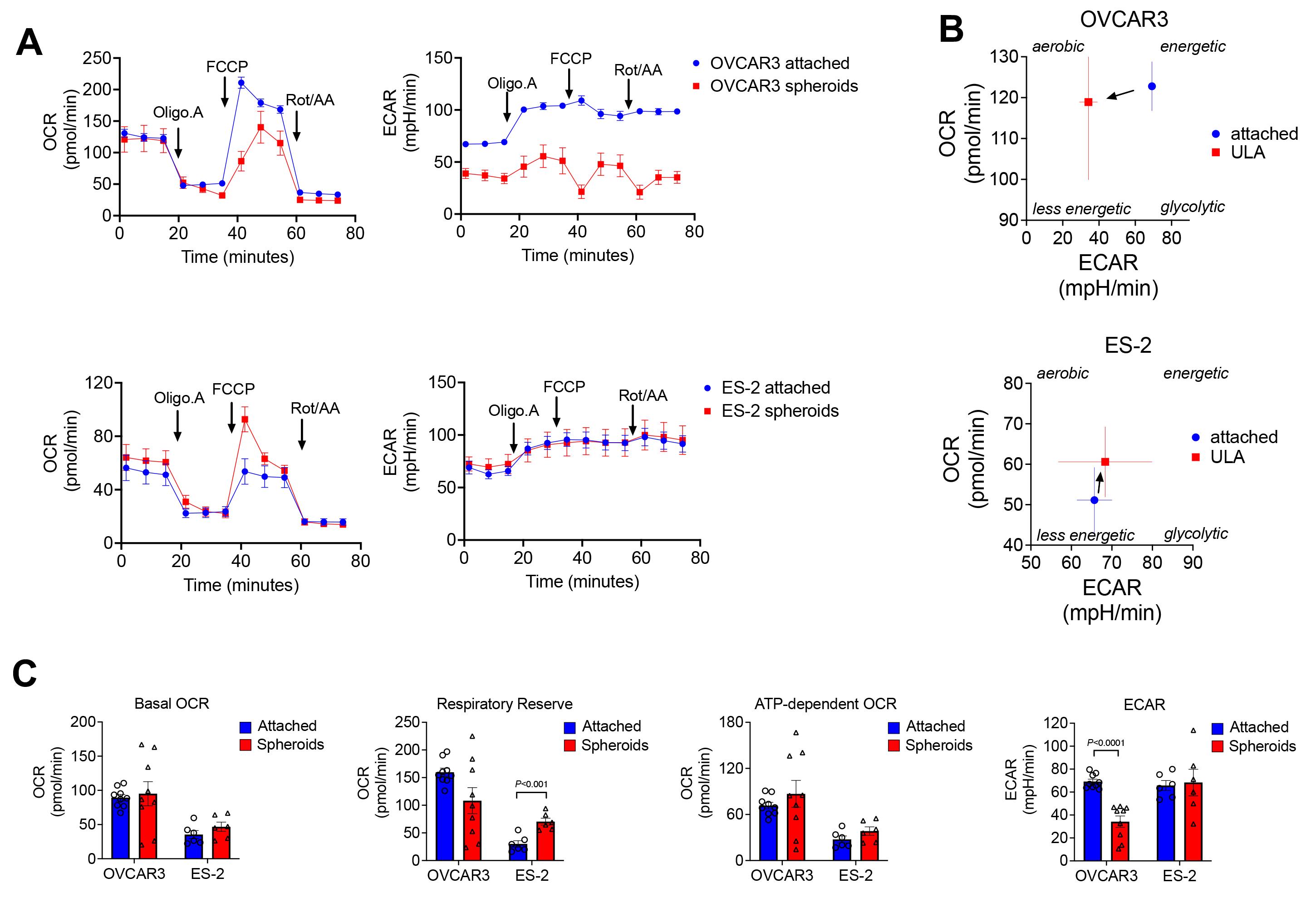
Figure 5. Changes in bioenergetics between attached and anchorage-independent conditions. A. OCR and ECAR were measured during the mitochondrial stress test Assay (1.0 µM Oligomycin and 1.0 µM FCCP; n=9 OVCAR3; n=6 ES-2). B. Plots of basal OCR and ECAR measurements demonstrate the bioenergetic phenotypes of OVCAR3 and ES-2 cells cultured in attached and ULA conditions. C. Example of mitochondrial stress test analysis as described in “Data analysis” above (multiple unpaired t-tests).
Of note is the decreased in ECAR in OVCAR3 spheroids compared to attached cells, and the increase in respiratory reserve in ES-2 cell spheroids, suggesting that different mechanisms contribute to a shift from glycolysis to reliance on mitochondrial respiration in spheroids between the two cell lines (Figure 5C). Additionally, we have utilized the extracellular flux assay to compare the glycolytic activity of attached and anchorage-independent spheroids by performing the glycolysis stress test on the high-grade serous cell line OVCA433. For examples of data generated using the glycolysis stress, we refer the reader to our previous publication (Kim et al., 2020). Briefly, the glycolysis stress test similarly demonstrated that cells in spheroids primarily utilize glucose for respiration, while the same cells cultured in attached conditions utilized glucose primarily for glycolysis (Kim et al., 2020).
Notes
Depending on cell line and spheroid size, oxygen consumption can be detected with as little as five spheroids per well. However, due to variable distribution of spheroids on the plate, low spheroid numbers yielded more variability in OCR readings, and this may be due to the distribution of spheroids too far from the detection probe. To avoid variability in OCR readings, plating spheroids into the center of each well between the three well posts is recommended.
The XFp platform has limitations related to small number of assay wells per plate. We recommend running three technical replicates per plate, if a control and experimental group are to be compared. If two experimental groups and a control are to be compared, two technical replicates can be run, or each experimental group compared to the control in separate plates. For larger sets of experimental groups, it is recommended that the assay be scaled up to the XF96 format.
For adaptation of this method to the larger surface area of the XF24 platform wells, we recommend optimizing spheroid numbers and scaling the test compound volumes based on the manufacture’s protocols. Seahorse capture screens (Agilent, catalog number: 101122-100) are available for the XF24 platform culture plates, and this platform may be an alternative for loosely aggregated cells and maintaining spheroid integrity during the assay.
The Seahorse XF cell mito stress test kit (Agilent, catalog number: 103010-100) and seahorse XF cell glycolysis stress test kit (Agilent, catalog number: 103017-100) are alternative sources of the compounds used in the mitochondrial and glycolysis stress assays.
Recipes
Preparing Stock Concentrations of Compounds
Use DMSO to solubilize the compounds.
Table 3. Mitochondrial Stress Test Compound Stock Solutions.
Stock concentration (mM) Compound (mg) DMSO (mL) *Oligomycin A 10 5 0.632 *FCCP 10 10 3.934 *Antimycin A 10 25 4.69 **Rotenone 1 10 25.3 *Oligomycin A, FCCP, and AA are soluble in DMSO. Make aliquots and store at -20°C.
**Rotenone is soluble in chloroform at a concentration of 50 mg/mL, or in DMSO at 0.5 mg/mL. Due to the volatile nature of chloroform, we chose to dilute in DMSO for long-term storage. However, rotenone can be prepared in chloroform if desired. Make aliquots and store at -80°C.
Use the appropriate assay media to dilute DMSO stocks to 10–50 μM 10× working stock solutions used in the Seahorse XFp assay protocol. Note: Mix Antimycin A and Rotenone in 1:1 ratio to obtain final concentration of 25 μM.
Additional compound preparations for Glycolysis Stress Test:
D-Glucose: Prepare 1 M and 100 mM stock solutions in sterile diH2O.
2-Deoxy-D-Glucose: Prepare a 500 mM stock solution in sterile diH2O.
RIPA lysis buffer
Prepare RIPA buffer by adding the following components at indicated concntrations to a final volume of 500 mL:
50 mM Tris-HCl pH 8.0 (to make 1 M stock Tris-HCL pH 8.0 stock solution, dissolve 121.1 g of Tris base in 800 mL of H2O, and adjust the pH to 8.0 by adding concentrated HCl, before adjusting the volume to 1 L)
150 mM NaCl
1% NP40/Igepal
1% sodium deoxycholate
10 mM sodium phosphate pH 7.2 (to make 0.1 M phosphate buffer stock, first prepare 0.5 M sodium phosphate dibasic and phosphate monobasic stock solutions in a final volume of 500 mL of ddH2O, dilute each stock to 0.1 M, and adjust the 0.1 M sodium phosphate dibasic solution to pH 7.2, by adding 0.1 M sodium phosphate monobasic solution)
0.1% SDS
2 mM EDTA
50 mM NaF
Store at 4°C. Prior to use, aliquot 10 mL into a Falcon tube, add 1 Protease inhibitor cocktail tablet, and dissolve.
Acknowledgments
This work was supported by NIH grants R01CA242021 (N.H.) and R01CA230628 (N.H. & M.K.). This protocol was derived from our previous publication: Kim YS et al., Context-dependent activation of SIRT3 is necessary for anchorage-independent survival and metastasis of ovarian cancer cells. Oncogene. 2020 Feb;39(8):1619-1633 (Kim et al., 2020).
Competing interests
The Agilent Seahorse XFp extracellular flux analyzer (S7802A) was awarded to Nadine Hempel through an equipment grant from Agilent.
References
- Al Habyan, S., Kalos, C., Szymborski, J. and McCaffrey, L. (2018). Multicellular detachment generates metastatic spheroids during intra-abdominal dissemination in epithelial ovarian cancer. Oncogene 37(37): 5127-5135.
- Anderson, A. S., Roberts, P. C., Frisard, M. I., McMillan, R. P., Brown, T. J., Lawless, M. H., Hulver, M. W. and Schmelz, E. M. (2013). Metabolic changes during ovarian cancer progression as targets for sphingosine treatment. Exp Cell Res 319(10): 1431-1442.
- Bloch, K., Smith, H., van Hamel Parsons, V., Gavaghan, D., Kelly, C., Fletcher, A., Maini, P. and Callaghan, R. (2014). Metabolic alterations during the growth of tumour spheroids. Cell Biochem Biophys 68(3): 615-628.
- Dier, U., Shin, D. H., Hemachandra, L. P., Uusitalo, L. M. and Hempel, N. (2014). Bioenergetic analysis of ovarian cancer cell lines: profiling of histological subtypes and identification of a mitochondria-defective cell line. PLoS One 9(5): e98479.
- Divakaruni, A. S., Paradyse, A., Ferrick, D. A., Murphy, A. N. and Jastroch, M. (2014). Analysis and interpretation of microplate-based oxygen consumption and pH data. Methods Enzymol 547: 309-354.
- Ghoneum, A., Gonzalez, D., Abdulfattah, A. Y. and Said, N. (2020). Metabolic Plasticity in Ovarian Cancer Stem Cells. Cancers (Basel) 12(5): 1267.
- Hill, B. G., Benavides, G. A., Lancaster, J. R., Jr., Ballinger, S., Dell'Italia, L., Jianhua, Z. and Darley-Usmar, V. M. (2012). Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem 393(12): 1485-1512.
- Kenda Suster, N. and Virant-Klun, I. (2019). Presence and role of stem cells in ovarian cancer. World J Stem Cells 11(7): 383-397.
- Kim, Y. S., Gupta Vallur, P., Jones, V. M., Worley, B. L., Shimko, S., Shin, D. H., Crawford, L. C., Chen, C. W., Aird, K. M., Abraham, T., et al. (2020). Context-dependent activation of SIRT3 is necessary for anchorage-independent survival and metastasis of ovarian cancer cells. Oncogene 39(8): 1619-1633.
- Liao, J., Qian, F., Tchabo, N., Mhawech-Fauceglia, P., Beck, A., Qian, Z., Wang, X., Huss, W. J., Lele, S. B., Morrison, C. D. and Odunsi, K. (2014). Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism. PLoS One 9(1): e84941.
- Mandujano-Tinoco, E. A., Gallardo-Perez, J. C., Marin-Hernandez, A., Moreno-Sanchez, R. and Rodriguez-Enriquez, S. (2013). Anti-mitochondrial therapy in human breast cancer multi-cellular spheroids. Biochim Biophys Acta 1833(3): 541-551.
- Marin-Hernandez, A., Gallardo-Perez, J. C., Hernandez-Resendiz, I., Del Mazo-Monsalvo, I., Robledo-Cadena, D. X., Moreno-Sanchez, R. and Rodriguez-Enriquez, S. (2017). Hypoglycemia Enhances Epithelial-Mesenchymal Transition and Invasiveness, and Restrains the Warburg Phenotype, in Hypoxic HeLa Cell Cultures and Microspheroids. J Cell Physiol 232(6): 1346-1359.
- Noel, P., Munoz, R., Rogers, G. W., Neilson, A., Von Hoff, D. D. and Han, H. (2017). Preparation and Metabolic Assay of 3-dimensional Spheroid Co-cultures of Pancreatic Cancer Cells and Fibroblasts. J Vis Exp(126): 56081.
- Plitzko, B. and Loesgen, S. (2018). Measurement of Oxygen Consumption Rate (OCR) and Extracellular Acidification Rate (ECAR) in Culture Cells for Assessment of the Energy Metabolism. Bio-protocol 8(10): e2850.
- Rodríguez-Enríquez, S., Gallardo-Perez, J. C., Aviles-Salas, A., Marin-Hernandez, A., Carreno-Fuentes, L., Maldonado-Lagunas, V. and Moreno-Sanchez, R. (2008). Energy metabolism transition in multi-cellular human tumor spheroids. J Cell Physiol 216(1): 189-197.
- Schafer, Z. T., Grassian, A. R., Song, L., Jiang, Z., Gerhart-Hines, Z., Irie, H. Y., Gao, S., Puigserver, P. and Brugge, J. S. (2009). Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461(7260): 109-113.
- Tan, D. S., Agarwal, R. and Kaye, S. B. (2006). Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol 7(11): 925-934.
- Torre, L. A., Trabert, B., DeSantis, C. E., Miller, K. D., Samimi, G., Runowicz, C. D., Gaudet, M. M., Jemal, A. and Siegel, R. L. (2018). Ovarian cancer statistics, 2018. CA Cancer J Clin 68(4): 284-296.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Javed, Z., Worley, B. L., Stump, C., Shimko, S. S., Crawford, L. C., Mythreye, K. and Hempel, N. (2022). Optimization of Extracellular Flux Assay to Measure Respiration of Anchorage-independent Tumor Cell Spheroids. Bio-protocol 12(4): e4321. DOI: 10.21769/BioProtoc.4321.
Category
Cancer Biology > Cellular energetics > Cell biology assays > Metabolism
Cell Biology > Cell metabolism > Respirometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link