- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Bacterial Infection and Hypersensitive Response Assays in Arabidopsis-Pseudomonas syringae Pathosystem
Published: Vol 11, Iss 24, Dec 20, 2021 DOI: 10.21769/BioProtoc.4268 Views: 5311
Reviewed by: Andrea PuharGang YuShweta Panchal

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantitative Estimation of Auxin, Siderophore, and Hydrogen Cyanide Production in Halo and Drought-Tolerant Bacterial Isolates for Cucumber Growth
Zeinab Fotoohiyan and Ali Salehi Sardoei
Oct 5, 2025 1342 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1655 Views

Reproducible Emu-Based Workflow for High-Fidelity Soil and Plant Microbiome Profiling on HPC Clusters
Henrique M. Dias [...] Christopher Graham
Jan 20, 2026 404 Views
Abstract
Arabidopsis thaliana-Pseudomonas syringae pathosystem has been used as an important model system for studying plant-microbe interactions, leading to many milestones and breakthroughs in the understanding of plant immune system and pathogenesis mechanisms. Bacterial infection and plant disease assessment are key experiments in the studies of plant-pathogen interactions. The hypersensitive response (HR), which is characterized by rapid cell death and tissue collapse after inoculation with a high dose of bacteria, is a hallmark response of plant effector-triggered immunity (ETI), one layer of plant immunity triggered by recognition of pathogen-derived effector proteins. Here, we present a detailed protocol for bacterial disease and hypersensitive response assays applicable to studies of Pseudomonas syringae interaction with various plant species such as Arabidopsis, Nicotiana benthamiana, and tomato.
Background
Pseudomonas syringae is a Gram-negative phytopathogenic bacterial species that causes diseases on a broad host range, namely bacterial speck in tomato and canker disease in pepper and kiwifruit (Lewis Ivey and Miller, 2000; Basim et al., 2004; Mazzaglia et al., 2012; Xin and He, 2013). Over the last two decades, Pseudomonas syringae has also been an important model pathogen for studying bacterial ecology, pathogenesis mechanisms, and plant immune system (Xin et al., 2018). Due to its importance in basic biology research, as wells as in outbreaks of economically-important diseases, it was selected as the number one of the top 10 plant pathogenic bacteria in molecular plant pathology (Mansfield et al., 2012).
P. syringae bacteria are generally used as foliar pathogens in laboratories, although in nature they cause diseases in various organs. P. syringae enters plant leaf tissue through wounds or open stomata during natural infections, and uptakes nutrients in the apoplastic space of the leaves for multiplication (Xin and He, 2013). Bacterial disease assays are powerful tools in plant pathology studies. Two inoculation approaches, surface inoculation (i.e., by dipping or spray) and infiltration (i.e., by a needle-less syringe or vacuum), are commonly used in laboratories (Katagiri et al., 2002). Here we present step-by-step procedures for bacterial disease assays by syringe infiltration, which bypasses pathogen entry through stomata and plant “stomatal defense”, and is broadly used in studying plant “apoplast defense”. In addition, we also describe detailed procedures of the hypersensitive response assay, in which recognition of pathogen effectors by plant immune receptors triggers fast tissue cell death, and the rate of cell death can be used as a readout of the strength of plant immunity. Although this protocol is presented using the Arabidopsis-P. syringae pathosystem, it can be easily adapted to different pathosystems, such as Nicotiana benthamiana-P. syringae and tomato-P. syringae with slight modifications on bacterial inoculum.
Materials and Reagents
Eppendorf tubes (1.5ml and 2 ml, Thermo Fisher Scientific, catalog number: 509-GRD-Q and 508-GRD-Q)
0.22 μm Millex-GP Syringe Filter (Merck, catalog number: SLGPR33RB)
96-well plate (200 μl, Round Bottom, Beyotime, catalog number: FPT016)
Paper towels
Pipette tips (Thermo Fisher Scientific QSP, catalog number:112NXL-Q)
1 ml needleless syringe (LABSTAR, catalog number: BX150)
Arabidopsis thaliana accession Col-0, fec (Gimenez- Ibanez et al., 2009) and rps2 (Mindrinos et al., 1994)
Note: Arabidopsis Col-0 plant contains the RPS2 gene, which mediates the recognition of effector protein AvrRpt2 and induces plant ETI resistance to Pst DC3000(avrRpt2). fls2 efr cerk1 (fec) triple mutant, which is mutated in three major pattern-recognition receptor genes; rps2 mutant is mutated in the RPS2 gene, encoding the receptor recognizing AvRpt2.
Pseudomonas syringae pv. tomato (Pst) DC3000 and Pst DC3000(avrRpt2) (Mudgett et al., 1999)
Sodium hypochlorite (Sinopharm Chemical Reagent, catalog number: 80010428)
Sterilized water (e.g., Milli-Q)
Mixed soil, which contains substrate (PINDSTRUP), vermiculite (Size: 1-3 mm) and perlite (Size: 3-5 mm), the ratio of these materials is 3:9:1 in mixed soil.
Tryptone (OXOID, catalog number: LP0042B)
Yeast Extract Powder (OXOID, catalog number: LP0021B)
Potassium dihydrogen phosphate (KH2PO4) (Sinopharm Chemical Reagent, catalog number: 10017608)
Sodium chloride (NaCl) (Sinopharm Chemical Reagent, catalog number: 10019318)
Magnesium sulfate (MgSO4) (Sinopharm Chemical Reagent, catalog number: 20025117)
Agar powder (Shanghai DingGuo Biotech, catalog number: DH010-1.1)
Rifampicin (Yeasen Biotechnology, catalog number: 60234ES08)
Spectinomycin (Sangon Biotech, catalog number: A600901-0005)
75% ethanol (Sinopharm Chemical Reagent, catalog number: 80176965)
Luria-Marine (LM) solid medium (see Recipes)
Rifampicin stock stock (50 g/L, 1,000×) (see Recipes)
Spectinomycin stock stock (50 g/L, 1,000×) (see Recipes)
Equipment
Ultra-low temperature freezer (-75°C freezer, New Brunswick Scientific)
Tray (size: 310 g), transparent plastic dome, pot (size: 8 cm) and mesh (pore size: mesh 18 = 880 μm)
Arabidopsis growth chamber (Percival and JIUPU)
Pipette (1 ml, Rainin, model: L-1000PL for export)
Centrifuge (Eppendorf, model: 5425R)
Spectrophotometer (Thermo Fisher Scientific, model: NanoDrop ONEC)
Steel ball (5 mm in diameter; SSCB, catalog number: KH000268)
Millex-GP Syringe Filter Unit (Merck, catalog number: SLGPR33RB)
Camera (Canon, model: EOS 80D)
Vortex Oscillator (Scientific Industries, model: Vortex-Genie 2)
Tweezer
Beaker (Thermo Scientific, catalog number: 1201-1234)
Cork borer (Sigma-Aldrich, catalog number: Z165220-1SET, 7.5 mm in diameter)
TissueLyser (Shanghai Jingxin Industry, model: Tissuelyser-48)
Stereoscope (Leica, model: MDG41)
Autoclave
Temperature and Humidity Data Logger (Easylog, model: EL-21CFR-2-LCD)
Software
Microsoft Excel
GraphPad Prism 8
Procedure
Growing Arabidopsis plants in soil
Sterilize Arabidopsis seeds with 5% sodium hypochlorite for 7-10 min, and then wash the seeds with sterilized water 5 times; place the sterilized seeds in the dark at 4°C for two days.
Note: The cold treatment will synchronize germination.
Place the mixed soil in an ultra-low temperature freezer (below -20°C) overnight.
Note: Autoclaving soil is usually harmful for seed germination and plant growth in our hands. We therefore used a freezing treatment to kill insect eggs and larvae in the soil to prevent insect infestation during plant growth, without causing any visible effect on seed germination.
Place the soil into small plastic pots, cover the pots with mesh, and fix the mesh with a rubber band (Figure 1A).
Use a pipette to sow sterilized seeds (from A1, about 200-300 seeds/ml) in the soil (about 20-30 seeds per pot, 4-8 seeds at each corner).
Grow plants in environmentally-controlled growth chambers, with relative humidity set at 60%, temperature at 22°C, light intensity at 100 μE⋅m-2⋅s-1 with a 12 h light/12 h dark photoperiod.
Remove excess seedlings after one week, and keep 4 seedlings per pot.
Water the plants with tap water every 2-3 days. Four- to five-week-old plants (Figure 1B) were used for our experiments.
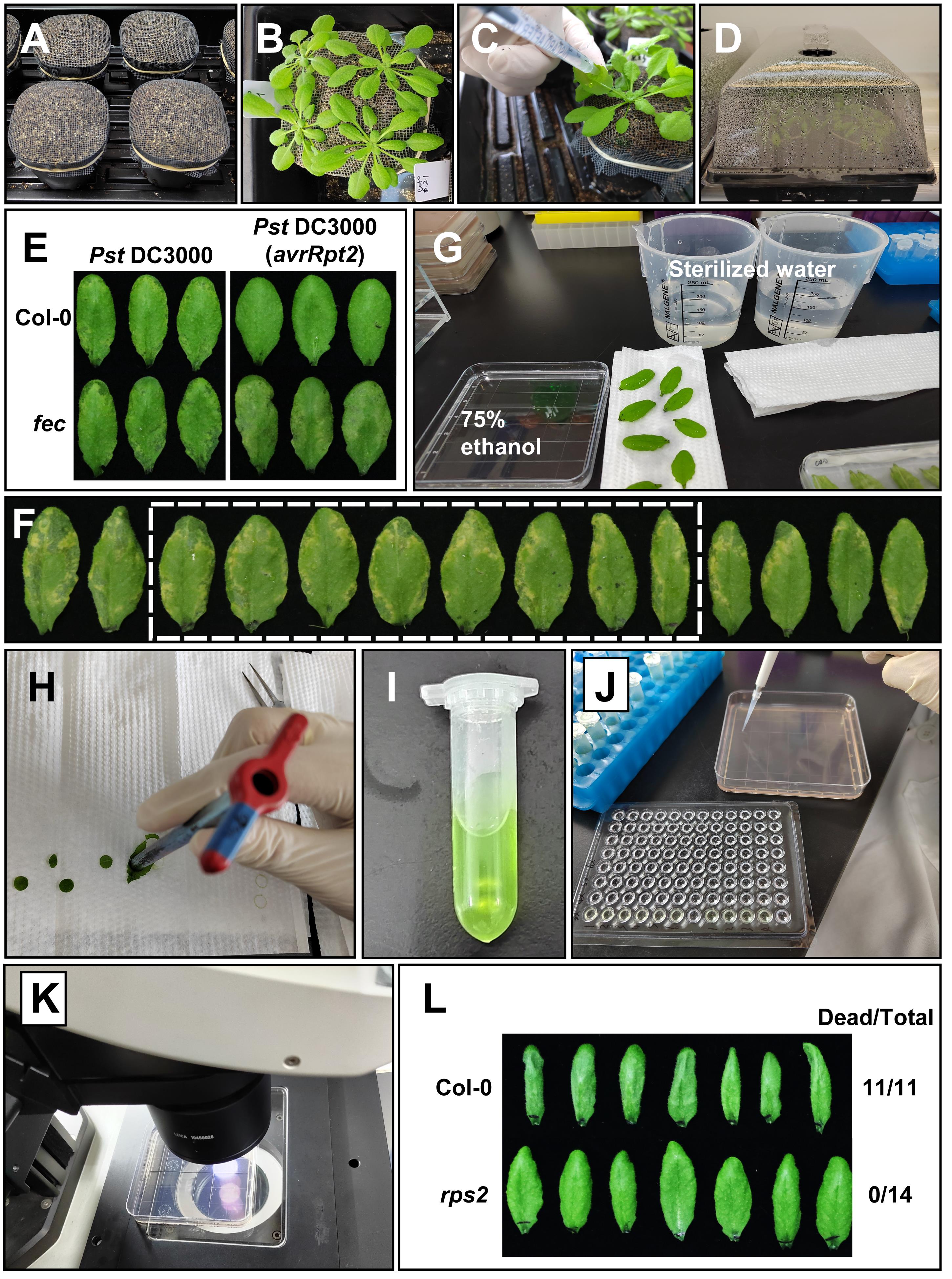
Figure 1. Experimental procedures of bacterial infection and hypersensitive response assays. A. Preparation of the mixed soil in pots. B. Appearance of 4-week-old Arabidopsis plants grown in environmentally-controlled growth chambers. C. Inoculation of leaves with bacterial solutions using a 1 ml needleless syringe. D. Inoculated plants are covered with a dome to keep high humidity, for the disease to develop in the greenhouse. E. Photograph of inoculated leaves 3 days after infiltration. F. Disease phenotype 3 days after infiltration in Col-0 plants, the white dotted box represents selected representative samples. G. Sterilization and rinsing of the sampled leaves. H. Sampling of leaf discs using a cork borer (7.5 mm in diameter). I. Ground leaf solution. J. Dilute the extracted solutions in different dilution ratios, and then take 10 μl from each dilution and place on LM agar plates. K. Count the colonies with a stereoscope. L. Photographs of tissue collapse phenotype for HR assay. Pst DC3000 (avrRpt2) bacteria were infiltrated at OD600 of 0.2 and images were taken about 7 h post infiltration (hpi).
Preparing Pst strains for inoculation
Streak out the Pst strains from -75°C freezer onto Luria-Marine (LM) solid medium containing antibiotics, and allow to grow in a 30°C incubator for 2 days.
Culture the bacterial strains in 4-6 ml LM liquid medium supplemented with the appropriate antibiotics [50 mg/L rifampicin for Pst DC3000; 50 mg/L spectinomycin and 50 mg/L rifampicin for Pst DC3000(avrRpt2), which contains the pDSK600-avrRpt2 construct with spectinomycin resistance], shaking at 200 rpm and 30°C for 12-16 h.
Note: The bacterial culture should reach mid-log growth phase (OD600 = 0.6-1.0).
Transfer 1.5 ml bacterial culture to a 2 ml Eppendorf tube, and collect by centrifugation at 2,500 × g for 5 min.
Remove the supernatant and resuspend the pellet with 2 ml sterilized water to wash.
Centrifuge the bacterial solution at 2,500 × g for 5 min, and remove the supernatant, and then resuspend the pellet with 1 ml sterilized water.
Adjust the bacterial solution to a cell density of OD600 = 0.2 (~1 × 108 cfu/ml) with sterilized water, measured with a spectrophotometer.
For bacterial disease assay
Inoculation of Arabidopsis with Pst strains
Dilute the bacterial solution from B6 with sterilized water to a cell density from OD600 = 0.001 (~5 × 105 cfu/ml) to 0.002 (~1 × 106 cfu/ml).
Inoculate 3 marked leaves (from the abaxial side of the leaves) per plant with adjusted bacterial solution using a 1 ml needleless syringe (Figure 1C and Video 1). We estimate that approximately 100-200 μl are necessary to fully infiltrate one adult leaf. We usually inoculate 4 plants for each strain, and inoculate different plants with different strains in each pot.
Video 1. Syringe infiltration demonstration.Wipe off the solution on the surface of the infiltrated leaves with a paper towel.
Keep inoculated plants under ambient humidity for about 1 h to allow evaporation of excess water from the leaf.
Cover the tray with a transparent plastic dome to keep high humidity until sampling, and place plants back in the growth chamber for the disease to develop (Figure 1D).
Recording the disease symptoms and counting the number of bacteria
Harvest samples after 2-4 days (varies among experiments, due to different plant genotypes/bacterial strains), remove all inoculated leaves from the plants, and take a photo to record the chlorosis and necrosis symptoms (Figure 1E).
Select 6-8 leaves that are representative of the symptoms (e.g., the middle level; Figure 1F), and place them in a 75% ethanol solution for ~30 s to kill the bacteria on the leaf surface.
Place the leaves on the paper towel to quickly remove excess ethanol, and then rinse the leaves with sterilized water twice (Figure 1G).
Dry the leaves with the paper towel, take two leaf discs from each leaf using a cork borer (7.5 mm in diameter) and four discs from two different leaves as one biological repeat, place the leaf discs into a 2 ml Eppendorf tube containing 200 μl of sterilized water and one or two steel balls (5 mm in diameter); collect three to four repeats from each treatment (Figure 1H).
Grind the leaf discs by TissueLyser at 30 Hz, for 1 min.
Quickly spin the extracted solutions (5,000 × g, 10 s) to move the solution from the tube caps to the inside of the tube; open the tube and add 800 μl of sterilized water to the tube, briefly vortex, and mix well by Vortex Oscillator (Figure 1I).
Serially dilute the bacterial solutions with sterilized water (i.e., by 10×, 100×, 1,000×, etc.), and then take 10 μl from each dilution and place on LM agar plates supplemented with rifampicin (at 50 mg/L). Perform two technical replicates for each sample (Figure 1J), and air dry the plates at room temperature.
Note: As 10 μl from 1 ml of the extracted solution were placed on a LM agar plate, this is the equivalent of a 100-fold dilution of the bacteria from 4 leaf discs. If we take 10 μl from 1 ml of the extracted solution to 90 μl of sterilized water and then take 10 μl to place on the LM agar plate, this is equivalent to another 10-fold dilution. Serial 10-fold dilutions are done for each sample by repeating this process. We usually dilute the extracted solution to 10-4, 10-5 and 10-6 for Pst DC3000, and dilute to 10-2, 10-3 and 10-4 for Pst DC3000 (avrRpt2).
Place the air-dried LM agar plates in an incubator at 30°C for colonies to grow.
Count the colonies with a stereoscope 24 h after incubation (Figure 1K). It can also be counted by eye if the colonies are well separated and grow to large sizes (e.g., after more than 24 h incubation).
For the hypersensitive response assay
Inoculation of Arabidopsis plants with Pst strains
Inoculate 3-4 marked leaves (from the abaxial side of the leaves) per plant with Pst DC3000(avrRpt2) strain at a cell density of OD600 = 0.2 (~108 cfu/ml) using a 1 ml needleless syringe (Figure 1C). Inoculate 4-6 plants for each genotype.
Wipe off the solution on the surface of the leaves with a paper towel, let the leaves dry, and place the plants back in growth conditions without cover and under ambient humidity. Check tissue collapse starting from 4-5 h after infiltration.
Note: Different effector proteins lead to different ETI response intensities and different rates of HR, so different strains may require distinct observation time for HR.
Harvest leaves, count the number of leaves showing cell death, and take a photo (Figure 1L).
Note: When cell death occurs, ions will leak out from the cell, and the electrolyte leakage can be measured using a Electrolytic conductivity meter over a time course after infiltration, providing a more quantitative way of assessing HR (protocols described in Hatsugai and Katagiri, 2018).
Data analysis
Analysis of the data from disease assay:
Enter the number of bacteria from dilutions (we usually choose the dilutions with 10-100 colonies for calculation) with corresponding dilution factor in Microsoft Excel.
Average the two replicates of each sample. Multiply this average by a multitude of dilution factor to get the total number of colonies (colony forming units, CFU) in the sample. For example, if we get an average of 56 and the dilution factor is 10-5, then the total number of colonies is 56 × 105 CFU.
Calculate the total leaf area. The area of one leaf disc of 7.5 mm diameter is equal to π multiplied by 0.375 squared, so the area is 0.441786 cm2; there are four leaf discs in one sample, so the total leaf area is 1.767 cm2.
Divide the total number of bacteria of each sample by the leaf area to get the number of colonies per unit area. This value represents the disease susceptibility of the plant, and the higher the value, the more susceptible the plant is. For example, the number of colonies per unit area of the above sample from step1 is 56 × 105/1.767 = 3169213 CFU/cm2.
Take the logarithm of base 10 of the number of colonies per unit area of each sample. For example, the above value from step 4 is Log10 3169213 = 6.5 Log10 (CFU/cm2).
Enter all values from all samples in GraphPad, and calculate the average and standard deviation (SD) of 3 biological replicates, draw the figure and determine statistical significance using two-way ANOVA with Tukey’s test by GraphPad (Figure 2).
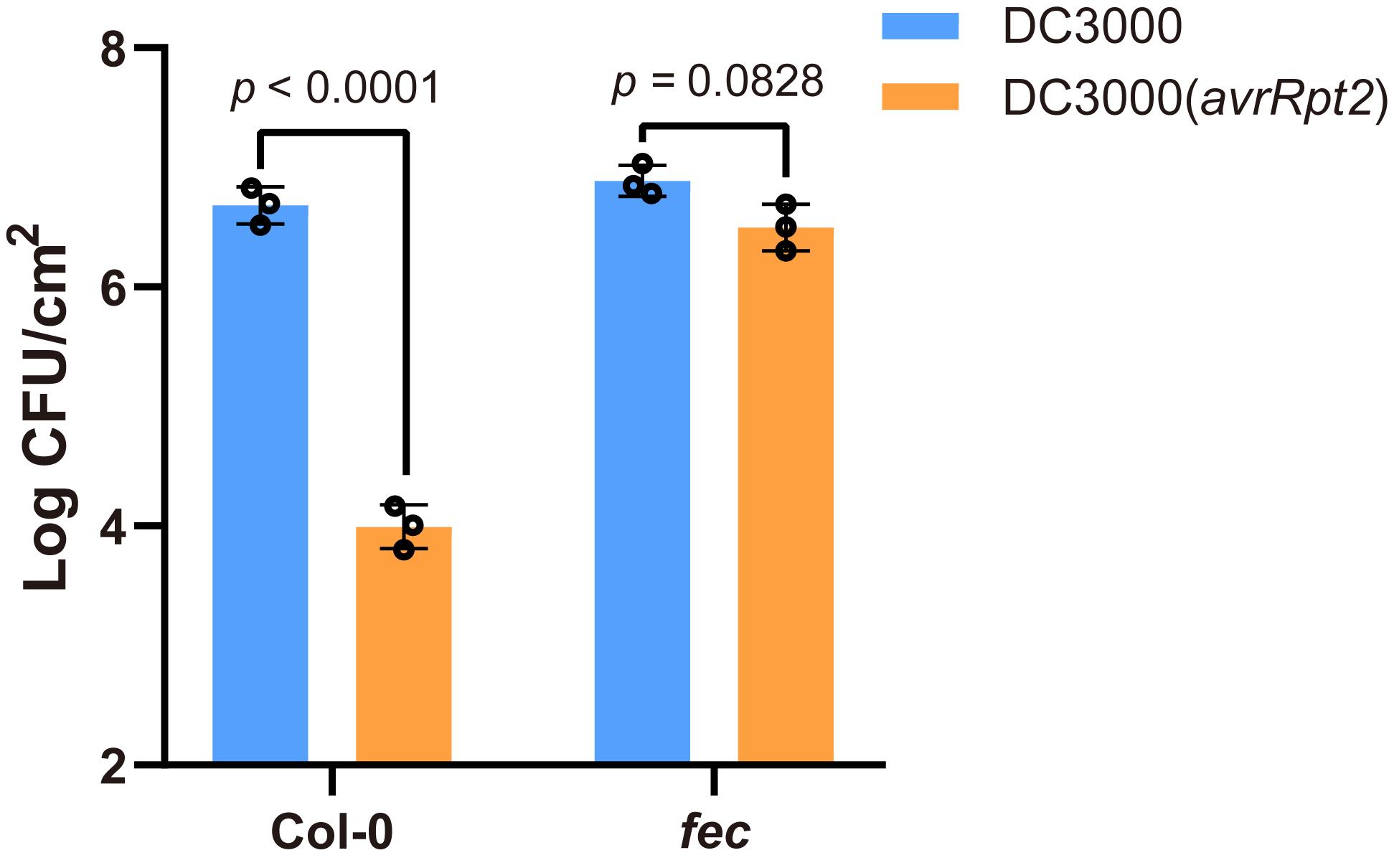
Figure 2. Example graph of disease infection assay. The above graph was generated from one biological replicate of disease assay from Yuan et al. (2021) . Pst DC3000 (avrRpt2) bacteria were infiltrated into Arabidopsis leaves at OD600 of 0.002 and populations were determined at 3 dpi (mean ± S.D.; n = 3 biologically independent samples). Data were analyzed using two-way ANOVA with Tukey’s test.
Notes
We found that plant health status is one of the key determinants of disease phenotypes. Stressed or sub-optimal plants often give inconsistent or even opposite results. Abiotic conditions (such as light intensity/photoperiod, temperature, and water/soil wetness) in the chamber influence the health and basal defense level of plants. It’s important to use healthy plants that are grown under optimized conditions, and have a minimized or low level of basal defense for pathogen-related assays. We usually use 10 or 12 h light, keep the watering frequency/amount (not watering too much each time and avoid leaving standing water in the tray), and use plants at appropriate ages (usually around 4 weeks, as older plants sometimes tend to get anthocyanin accumulation and higher basal defense). Plants that look dark green or purple in the center of the rosettes are usually not used in our assays. Plants that are infected by insects or fungi also can not be used.
Leaf age may affect hormonal level and plant immunity. Leaves of a similar age should be used in these assays. We usually use 3 leaves that are middle-aged and fully expanded from each plant. Bacterial dose and sampling time (e.g., day 2 instead of day 3 or 4) can be adjusted in experiments, depending on plant condition, and bacterial strains used.
Environmental factors such as light, temperature and humidity affect disease development and severity level. We found disease development to be very sensitive to air humidity, so we usually cover the inoculated plants with a transparent plastic dome (fully covered) to keep relatively high air humidity inside (Figure 1D, above 90% relative humidity) for 3-4 days.
For hypersensitive response assays, wounding and leaf-age seem to have a major effect on cell death rate. We observed that mechanical wounding during syringe infiltration dramatically accelerates tissue collapse. Thus, care should be taken during injection with the syringe. We also found that younger leaves are usually associated with faster cell death, so leaves at similar age should be chosen for a fair comparison, or to observe a less obvious HR phenotype.
Recipes
Luria-Marine (LM) medium
Tryptone 10 g/L
Yeast Extract Powder 6 g/L
KH2PO4 1.5 g/L
NaCl 0.6 g/L
MgSO4 0.35 g/L
Dissolve the ingredients in distilled water, adjust the pH to 7 with 10 N sodium hydroxide (NaOH) solution, and autoclave the solution.
For LM solid medium, add 15 g/l agar powder in liquid medium before autoclaving.
Rifampicin stock solution (1,000×)
Dissolve 2.5 g of the rifampicin powder in 50 ml dimethyl sulfoxide (DMSO) to make a 50 g/L stock solution, mix well by vortex, aliquot the solution into 1.5 ml Eppendorf tubes, and store at -20°C.
Spectinomycin stock solution (1,000×)
Dissolve 2.5 g of the spectinomycin powder in 50 ml sterilized water to make 50 g/L stock solution, mix well by vortex, sterilize the solution by filtering through a 0.22 μm Millex-GP Syringe Filter, aliquot into 1.5 ml Eppendorf tubes, and store at -20°C.
Acknowledgments
This work was supported by the Chinese Academy of Sciences, Center for Excellence in Molecular Plant Sciences/Institute of Plant Physiology and Ecology, National Key Laboratory of Plant Molecular Genetics and Chinese Academy of Sciences Strategic Priority Research Program (Type-B; project number: XDB27040211). M.Y. was supported by National Postdoctoral Program for Innovative Talents (project number: BX2021313). We would like to thank Dr. Jun Fan at China Agricultural University for the advice on freezing treatment of soil for insect control.
Competing interests
The authors declare no competing interests.
References
- Basim, H., Basim, E., Yilmaz, S., Dickstein, E. R. and Jones, J. B. (2004). An Outbreak of Bacterial Speck Caused by Pseudomonas syringae pv. tomato on Tomato Transplants Grown in Commercial Seedling Companies Located in the Western Mediterranean Region of Turkey. Plant Dis 88(9): 1050.
- Gimenez-Ibanez, S.,Ntoukakis, V. and Rathjen, J. P. (2009). The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis.Plant Signal Behav 4(6): 539-41.
- Hatsugai, N. and Katagiri, F. (2018). Quantification of Plant Cell Death by Electrolyte Leakage Assay. Bio-protocol 8(5): e2758.
- Katagiri, F., Thilmony, R. and He, S. Y. (2002). The Arabidopsis thaliana-Pseudomonassyringae interaction. Arabidopsis Book 1: e0039.
- Lewis Ivey, M. L. and Miller, S. A. (2000). First Report of Bacterial Canker of Pepper in Ohio. Plant Dis 84(7): 810.
- Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., Dow, M., Verdier, V., Beer, S. V., Machado, M. A., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13(6): 614-629.
- Mazzaglia, A., Studholme, D. J., Taratufolo, M. C., Cai, R., Almeida, N. F., Goodman, T., Guttman, D. S., Vinatzer, B. A. and Balestra, G. M. (2012). Pseudomonas syringae pv. actinidiae (PSA) isolates from recent bacterial canker of kiwifruit outbreaks belong to the same genetic lineage. PLoS One 7(5): e36518.
- Mindrinos, M., Katagiri, F., Yu, G. L. and Ausubel, F. M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78(6):1089-99.
- Mudgett, M. B. and Staskawicz, B. J.(1999). Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol Microbiol 32(5): 927-41.
- Xin, X. F. and He, S. Y. (2013). Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51: 473-498.
- Xin, X. F., Kvitko, B. and He, S. Y. (2018). Pseudomonas syringae: what it takes to be a pathogen. Nat Rev Microbiol 16(5): 316-328.
- Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., Cai, B. and Zhou, J. M. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Natrue 592(7852): 105-109.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yuan, M. and Xin, X. F. (2021). Bacterial Infection and Hypersensitive Response Assays in Arabidopsis-Pseudomonas syringae Pathosystem. Bio-protocol 11(24): e4268. DOI: 10.21769/BioProtoc.4268.
Category
Plant Science > Plant immunity > Host-microbe interactions
Microbiology > Pathogen detection
Biological Sciences > Microbiology
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










