- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Bacterial Infection with Listeria monocytogenes in Mice and Subsequent Analysis of Antigen-Specific CD8 T Cell Responses
Published: Vol 11, Iss 23, Dec 5, 2021 DOI: 10.21769/BioProtoc.4247 Views: 4616
Reviewed by: Giusy TornilloAmit Kumar DeyRakesh Bam

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In Vitro Co-culture of Bacterial and Mammalian Cells to Investigate Effects of Potential Probiotics on Intestinal Barrier Function
Ajitpal Purba [...] Dulantha Ulluwishewa
Jun 20, 2025 2515 Views

Artificial Metalloenzymes in Artificial Sanctuaries Through Liquid–Liquid Phase Separation
Kaixin Wang [...] Tong Wu
Oct 5, 2025 1566 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1655 Views
Abstract
Pathogens such as bacteria, viruses, fungi, or protozoa can cause acute and chronic infections in their hosts. The intracellular bacterium Listeria monocytogenes serves as a model pathogen to assess the molecular mechanisms regulating CD8 T cell activation, differentiation, and function. We set up an experimental workflow to investigate cell-intrinsic roles of the nuclear receptor NR2F6 in CD8 T cell memory formation upon Listeria monocytogenes (LmOVA) infection (Jakic et al., 2021). The current protocol details how to cultivate ovalbumin-expressing LmOVA, infect naïve C57BL/6 mice with these bacteria and determine the bacterial load in host organs. Furthermore, we describe how to evaluate antigen-specific CD8 T cell responses and discriminate between short-lived effector and memory precursor cells in vivo following LmOVA infection (Figure 1). To assess CD8 T cell-intrinsic molecular mechanisms, we integrated an adoptive cell transfer (ACT) experiment of genetically modified naïve OT-I CD8 T cells into congenic hosts before LmOVA infection.
Graphic abstract:

Figure 1. Experimental workflow depicting the steps for infection of mice with Listeria and subsequent analysis of antigen-specific CD8 memory responses. Bacteria (ovalbumin expressing Listeria monocytogenes) are thawed and grown on lysogeny broth (LB) plates overnight (ON). A single colony is picked and grown in LB medium ON. Bacteria from the exponential growth phase are then injected into a C57BL/6 mouse via tail vein injection. Colony forming units (CFU) of the bacteria can be detected in the spleen on day 3 post injection. Antigen-specific CD8 T cell immune response can be investigated during the acute phase (d3 after infection), during the peak of the adaptive immune response (d7), the clearance phase (d26), or the memory phase (d70) by flow cytometry. Created with BioRender.com.
Background
Listeria monocytogenes (Lm) is a gram-positive facultative intracellular bacterial pathogen that is the causative agent of listeriosis (Hamon et al., 2006; Khan and Badovinac, 2015). Murine listeriosis has served as a well-defined mouse infection model since the 1950s (Osebold and Sawyer, 1957). Both innate and adaptive immune cells are crucial for eliminating these bacteria (Zenewicz and Shen, 2007). Cellular responses to Lm infection are highly reproducible, and bacterial load in the host is easy to quantify. At sub-lethal doses (LD50 dose of LmOVA: ~5 × 106 CFU), Lm induces a robust immune response that is dependent on T cell-dependent bacterial clearance (Zenewicz and Shen, 2007). Thus, in addition to lymphocytic choriomeningitis virus (LCMV) infection, systemic bacterial infection with Lm has been a favored approach for the characterization of pathogen-specific CD8 T cell effector responses and memory formation for more than five decades (Mackaness, 1962; Harty and Bevan, 1995; Shen et al., 1998; Badovinac et al., 2000; Khan and Badovinac, 2015; Qiu et al., 2018; Levine et al., 2021). Lm naturally infects most mammalian hosts via the gastrointestinal tract (Pamer, 2004). However, in contrast to humans, mouse E-cadherin does not allow internalin-dependent entry into gastrointestinal epithelial cells when infected orally (Lecuit et al., 1999). Following intravenous inoculation in mice, Lm is taken up from the bloodstream by splenic and hepatic macrophages, which transport the bacteria to the T-cell zone of lymphatic organs (Merrick et al., 1997; Pamer, 2004).
Inbred mouse strains such as C57BL/6 or BALB/c respond with different MHC class I (CD8) or MHC class II (CD4) epitopes to infection by Lm. Whereas a response to the MHC-II restricted epitope listeriolysin O (LLO)190-201 is mainly observed in C57BL/6 mice, a response to the MHC-I restricted epitopes LLO91-99, and p60217-225 is dominant in BALB/c mice (Pamer et al., 1991 and 1994) (Zenewicz and Shen, 2007). Furthermore, Lm have successfully been engineered to express novel H2-Kb epitopes (Zenewicz and Shen, 2007). Other strains with attenuated pathogenicity allow the use of this bacterium in immuno-compromised mice, and the manipulation of the duration of infection by antibiotics and luminescence reporters have diversified the experimental options (Schmidt et al., 2012). One particularly useful Lm strain has been engineered to express the model antigen ovalbumin (OVA) (Pope et al., 2001; Foulds et al., 2002). This LmOVA strain is compatible with a range of animal models, such as T cell receptor (TCR) transgenic OT-I mice, and powerful immunological tools, such as OVA-tetramers for detecting antigen-specific CD8 T cells. Thus, the LmOVA strain is well suited for the investigation of transcriptional regulation, effector versus memory differentiation fates, or metabolic states of antigen-specific CD8 T cells (Pope et al., 2001; Zenewicz and Shen, 2007; Jakic et al., 2021; Harberts et al., 2021; Levine et al., 2021). Zehn et al. (2009) generated recombinant Lm strains expressing OVA protein-containing native SIINFEKL (N4) or altered OVA, where certain amino acids have been exchanged, and are termed altered peptide ligands (APL), such as A2(SAINFEKL), Y3, Q4, T4, and V4. These latter OVA variants bind H-2Kb as well as N4 but differ in their OT-I stimulation potency.
Memory formation is a hallmark of T cell-mediated immunity. However, the mechanism by which differentiation is adjusted at the molecular level, into either short-lived effector cells (SLECs) or memory precursor CD8 T cells and subsequent regulation of long-term memory, remains incompletely understood (Ahmed and Gray, 1996; Harty and Badovinac, 2008; Kaech and Cui, 2012; Chang et al., 2014; Jakic et al., 2021; Harberts et al., 2021; Levine et al., 2021). Following LmOVA infection, antigen-specific CD8 T cell responses can be investigated in vivo over time; in addition, it is possible to determine long-term persistence and re-infection responses (Jakic et al., 2021).
Here, we provide a methodological setup for LmOVA culture, infection of mice, and analysis of splenic colony-forming units (CFUs). We also describe methods that allow the investigation of blood or spleen-derived antigen-specific short-lived effector (SLECs, CD127−KLRG1+) and memory precursor (MPECs, CD127+KLRG1−) CD8 T cells, via flow cytometric analysis at different time points after LmOVA infection. Functional studies of CD8 T cell responses following antigen-specific peptide stimulation in vitro at various time-points, such as days 1-3 (early activation), day 7 (acute response), day 28 (after contraction), and day 70 (memory) are also described.
The current infection protocol is of use for the analysis of responses from innate and adaptive immune cells. Finally, the protocol is suitable for adoptive cell transfer (ACT) experiments of naïve OT-I CD8 T cells from genetically defined mouse strains into congenic hosts, with or without re-infection in vivo. This option offers the opportunity to precisely define donor CD8 T cell responses independent of a possibly altered microenvironment in germ-line deficient mouse strains.
Materials and Reagents
Listeria monocytogenes expressing ovalbumin, strain DMC 09-082 (LmOVA), carries an erythromycin resistance plasmid (Foulds et al., 2002; Ahn et al., 2016).
Notes:
Pathogen safety caution: Listeria monocytogenes is a facultatively anaerobic, gram-positive, rod-shaped coccobacillus, which causes a mild febrile illness. Disease manifestations include listeriosis in pregnancy, listeriosis of the central nervous system (CNS), febrile gastroenteritis, glandular listeriosis, local listeriosis, typhoid listeriosis, and atypical listeriosis. L. monocytogenes is commonly classified as a bio-safety class 2 organism. When working with L. monocytogenes, personal protection safety measures should be planned ahead and in accordance with official biosafety regulations. Please note that pregnant women are not advised to work with L. monocytogenes, as listeriosis can cause severe disease of the fetus or newborn baby.
All liquids should be discarded into bottles containing 10% sodium hypochlorite or an equivalent antimicrobial agent. Disposable plasticware carrying potentially infectious material should be collected and autoclaved before disposal.
Store bacteria in liquid nitrogen or at -80°C. For long-term storage, mix 500 μl bacteria in the exponential growth phase with 500 μl 20% glycerol in Lysogeny broth (LB) medium and shock freeze.
Recombinant Listeria monocytogenes strains expressing altered OVA protein termed altered peptide ligands (APL), such as A2, Y3, Q4, T4, and V4, are also available (Zehn et al., 2009).
70% ethanol
Sodium hypochlorite (Merck, catalog number: 056142500)
H2-Kb/OVAA257 PE tetramer (Baylor College of Medicine, https://www.bcm.edu/research/research-services/atc-core-labs/mhc-tetramer-production)
Antibodies (Table 1)
Table 1. Antibodies for flow cytometric analysis of CD8 responses
Antibody Clone Company Catalog number CD16/32 (Fc-block) 2.4G2 BD Biosciences 553142 CD44 FITC IM7 BD Biosciences 553133 CD45.1-PerCP A20 Biolegend 110726 CD45.2-V500 104 BD Biosciences 562129 CD45-V500 30-F11 BD Biosciences 561487 CD8-PB 53-6.7 BD Biosciences 558106 CD127-APC SB/199 Biolegend 121122 KLRG1-PE-Cy7 2F1/KLRG1 Biolegend 138416 Viability dye BD Biosciences 565388 Acridine Orange/Propidium Iodide Stain (LOGOS BIOSYSTEMS INC F23001)
Erythromycin (Sigma-Aldrich, catalog number: E5389, CAS: 114-07-8)
Glucose (Sigma-Aldrich, catalog number: G7021, CAS: 50-99-7)
Phosphate buffered saline (PBS) (Sigma, catalog number: P5493-1L)
Naïve CD8a+ T Cell Isolation Kit, mouse (Miltenyi, catalog number: 130-104-075)
Heparin, Gilvasan Pharma 5000 I.E./ml (GILVASAN 5000 I.E./ml)
OneComp eBeadsTM Compensation Beads (Thermo Fisher Scientific, catalog number: 01-1111-41)
Paraformaldehyde (PFA)
LB-Agar (Lennox) (Carl Roth, catalog number: X965.1)
LB-Agar (Lennox), granulated, 2.5 kg (Carl Roth, catalog number: 6671.3)
ROTI® Histofix 4%, 500 ml, plastic (Carl Roth, catalog number: P087.4)
Ammonium chloride (Carl Roth, catalog number: K298.1)
Potassium bicarbonate (Sigma-Aldrich, catalog number: 60339)
EDTA dihydrate (Carl Roth, catalog number: X986.2)
Lysogeny broth (LB) medium (see Recipes)
LB agar plates with erythromycin (see Recipes)
Erythromycin stock (see Recipes)
Glucose additive for cultivation of Listeria monocytogenes stock (see Recipes)
FACS buffer (see Recipes)
Erythrocyte lysis buffer (see Recipes)
Fixation buffer (see Recipes)
Buffer C (see Recipes)
Adult age and sex-matched C57BL/6 mice
Animal procedures must be in accordance with national authorities.
Disposable plastic materials
2 ml microcentrifuge tubes (Sarstedt, catalog number: 72.691)
FACS tubes 12 mm × 75 mm Universal (VWR, catalog number: 212-0294)
29 G insulin syringes 0.5 ml (VWR, catalog number: BDAM324824)
5 ml syringes (BD Biosciences, catalog number: 300185)
30 G needles 0.30 × 12 mm (Braun, catalog number: 1942110120)
15 ml Falcon® tubes (BD Biosciences, catalog number: 352096)
50 ml Falcon® tubes (BD Biosciences, catalog number: 352070)
1,000 μl, 200 μl and 20 μl pipette tips (Sarstedt, catalog numbers: 70.762.211, 70.760.211, 70.1114.210)
(Optional) Disposable inoculation loop
NuncTM cryogenic tubes (Thermo Fischer Scientific, catalog number: 36656)
Spectrophotometer 1.6 ml semi-micro-cuvette (Greiner Bio-One, catalog number: 613101)
100 µm and 70 µm cell strainers (Corning, catalog numbers: 352360 and 352350)
Pre-separation filters 30 µm (Miltenyi, catalog number:130-041-407)
Falcon® 96 well round-bottom plate (Corning, catalog number:352077)
Luna Cell Counting Slides (Logos biosystems, catalog number: L12002)
Bacterial spreader, Heathrow Scientific #HD8151 (optional: glass spreader and a Bunsen burner can be used)
Inoculation loop (Fisher Scientific, catalog number: 11576863)
500 ml bottle top vacuum filter (0.22 μm pore 33.2cm2 CA membrane, Corning, catalog number: 430513)
Petri dish 100 × 15 mm (Corning, catalog number: 351029)
Equipment
Precision Balance (Kern KEPCB60000)
LS-columns (Miltenyi, catalog number: 130-042-401)
Forceps and scissors
37°C flask-shaker (IKA KS4000i control or equivalent)
Spectrophotometer (IMPLEN OD600 DiluPhotometerTM OD600-10 or equivalent)
Calculator
Bio-safety level 1 and 2 cabinets with an Ultraviolet lamp (Thermo ScientificTM HerasafeTM KS, Class II Biological Safety Cabinet)
37°C incubator (Memmert CO2 Incubator ICO)
BD FACSCantoTM II or equivalent (BD Biosciences BD FACSCantoTM II Flow Cytometry System)
Precision scale (Kern, item no. KEPCB60000)
Vortex mixer (MERCK StuartTM Scientific SA8 vortex mixer Z648531)
Centrifuge equivalent to ROTINA 420/420 R (HettichTM Swing-out Rotor)
Centrifuge equivalent to Eppendorf Centrifuge 5418/5418R 5418R
LUNATM Automated Cell Counter (Logos Biosystems L10001-LG)
1000 µl, 200 μl and 20 µl pipettes (Gilson #FA10006M, #FA10005M, # FA10003M)
Miltenyi MidiMACS Starting Kit (Miltenyi Biotec, catalog number: 130-042-501)
Vacuum pump (Millipore MPWP6122050)
Animal restrainer (MERCK, Z756903 Mouse restrainer with dorsal access)
Software
GraphPad Prism 9.01 (https://www.graphpad.com/scientific-software/prism/)
Microsoft Excel (https://www.microsoft.com)
FlowJo 10.7.2 (https://www.flowjo.com/solutions/flowjo)
Procedure
Preparation of LmOVA
Notes:
All steps should be performed in a biosafety cabinet class II. Sterilize all surfaces of the cabinet with UV light and 70% ethanol before and after the procedures.
Wear personal protective equipment, including a lab coat, gloves, and safety glasses.
Place a vial with frozen bacteria on ice in a biosafety level 2 cabinet. With a sterile 200 μl pipette tip or a disposable inoculation loop, take a smear of the frozen bacteria and spread it onto an LB plate with erythromycin (see scheme below for spreading movements). Change the tip or inoculation loop, drag it through the first streak, and spread it in the next 1/4 of the plate. Repeat this process with the secondary streak. With each new streak, drag a new tip or loop across the latest streak before starting the spread for efficiency. It is advisable to spread bacteria from 2-3 different vials onto separate LB plates to ensure colony growth. Place the plate up-side-down at 37°C (Figure 2).
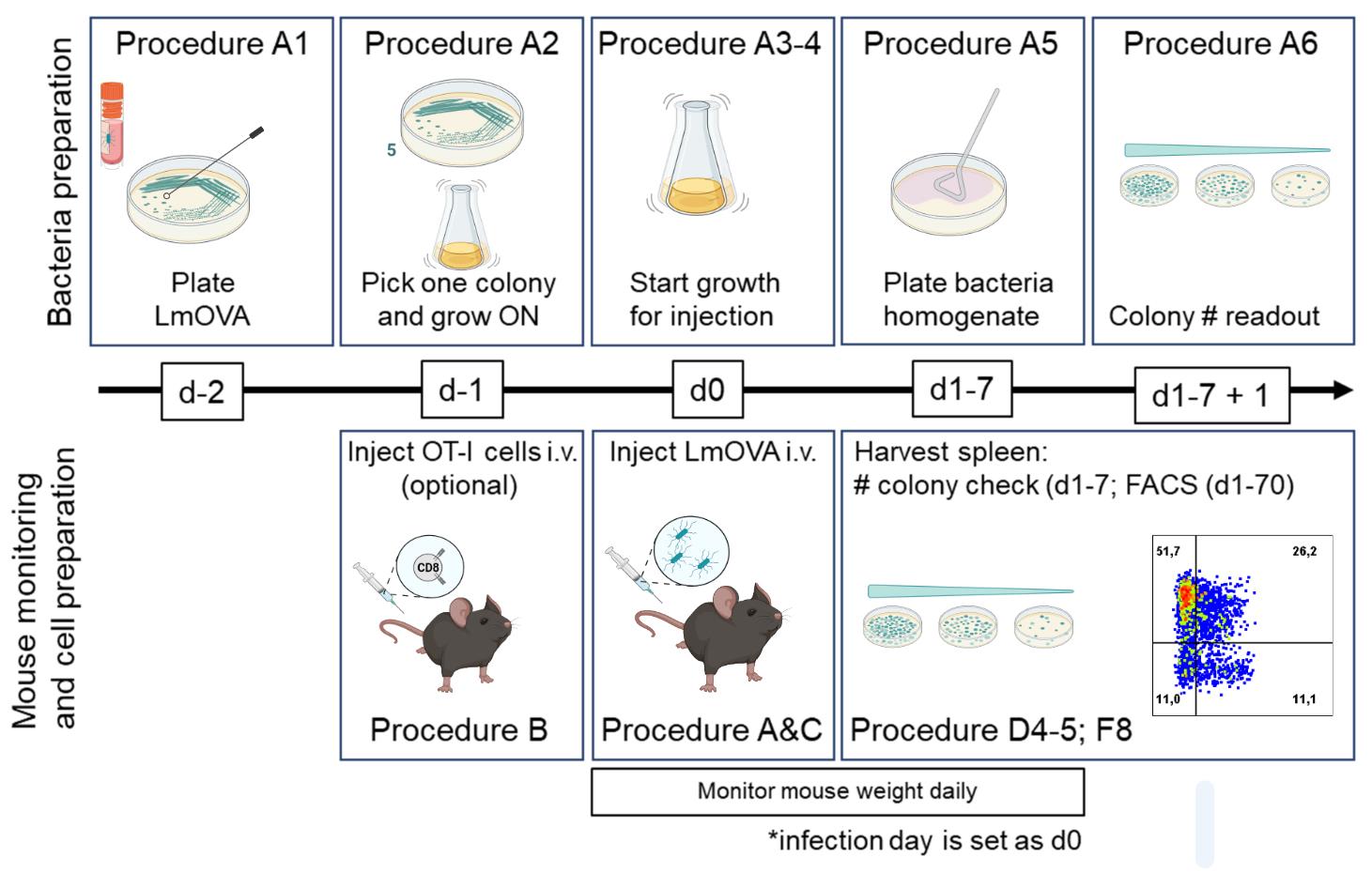
Figure 2. Experimental outline. Listeria growth and dilution, procedures A1 and A3-4. Adoptive transfer and infection of mice with LmOVA, procedures B, C, D-F. Created with BioRender.com.After 24 h, single colonies of bacteria should be visible (if not, leave the bacteria for another 24 h). Pick one colony with a sterile 200 μl pipette tip, and stir it in 100 ml of LB medium + erythromycin at 37°C in a sterile Erlenmeyer flask (the volume of the culture should not exceed 20% of the total volume). Cover the lid of the flask with aluminum or parafilm, and place it in a flask-shaker (225 rpm) at 37°C overnight (Figure 2).
The following morning, transfer 100 μl of the overnight growth culture into a sterile Erlenmeyer flask with fresh pre-warmed 100 ml of LB medium + erythromycin and place in a 37°C flask-shaker (225 rpm). Save at least 10 ml of the fresh LB-medium + erythromycin to use as a blank when determining bacterial growth with a spectrophotometer. Determine corrected OD600 every 2 h by pipetting 1 ml of the fresh LB-medium + erythromycin into a 1.6 ml semi-micro-cuvette and setting it as blank, followed by pipetting 1 ml of the bacterial growth in a new 1 ml cuvette and measuring OD600. When corrected OD600 corresponds to an OD where bacteria are in exponential growth (for details, see below), growth can be stopped by placing the bacteria on ice, and bacteria can be prepared for injection into the mouse (Figure 2).
Notes:
The OD600 over a whole day should be determined in a pilot experiment to identify the OD600 where bacteria are optimally growing in an exponential phase. This can differ depending on the spectrophotometer, LB-medium, temperature, etc. Exponential bacterial growth is at corrected OD600 0.5-1.0, and bacteria typically reach corrected OD600 = 0.5 after 5 h (Jones and D’Orazio, 2013; Ahn et al., 2016).
It is essential to use the same batch of LB-medium + erythromycin for bacterial growth and blank.
When taking out the flask with bacteria from the 37°C shaker, work quickly and put bacteria back immediately if optimal corrected OD600 has not been reached.
Once the optimal bacterial density is reached, the concentration of the bacteria can be calculated using the following formula: [c] = corrected OD600 × 11 × 108/ml.
Perform serial dilutions in sterile PBS using 2 ml Eppendorf tubes, e.g., if the initial bacterial concentration is 6.6 × 108/ml, prepare serial dilutions by adding 757.6 μl (corresponding to 5 × 108 bacteria) of bacterial solution to 242.4 μl of PBS, followed by four serial dilutions using 100 μl into 900 μl of PBS, to get the final concentration of 5 × 104 bacteria/ml (Figure 3).
Note: Initial volume will depend on the concentration of bacteria and needs to be recalculated each time. Make sure to mix properly by pipetting up and down, and prepare excess material as backup material. For mouse tail vein injection, dilute bacteria to 5 × 104 bacteria/ml in order to inject 104 CFU per mouse in a total volume of 200 μl (Procedure C below).
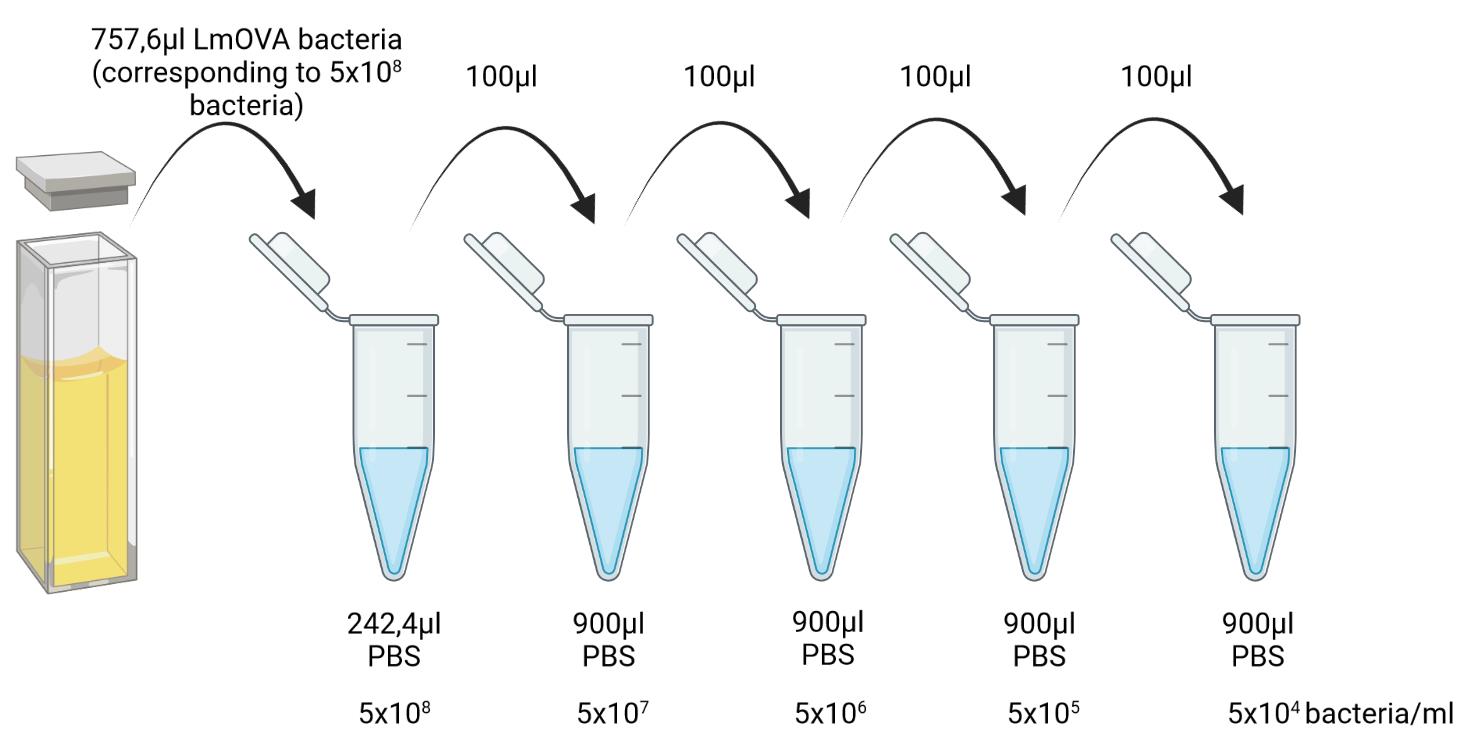
Figure 3. Bacterial serial dilutions. One specific example of serial dilutions of bacteria is shown. Created with BioRender.com.After infecting the mice (see Procedure C), use the final dilution of the left-over bacteria to control for the concentration of bacteria in the injected material. Dilute the bacteria serially 1:10, 1:100, 1:1,000, and 1:10,000 in PBS and plate 100 μl of each dilution in technical duplicates by spreading onto LB + erythromycin plates. Grow up-side-down for 24 h at 37°C (Figure 2).
After 24 h, count the colonies. The number of colony-forming units (CFU) should correspond to 5 × 104 bacteria/ml in the original mixture (Figure 2); for further details, see Table 2.
Notes:
Plates with >300 colonies or <30 colonies should not be used to calculate cells numbers due to inaccuracy.
Mix the serial dilutions of the bacteria thoroughly before plating.
Glycerol stocks for long-term storage can be prepared by mixing 500 μl bacteria in the exponential growth phase with 500 μl 20% glycerol in Lysogeny broth (LB) from an overnight culture in a cryogenic vial and stored at -80°C or in liquid nitrogen.
(Optional) Adoptive transfer of OT-I CD8 T cells
Notes:
This step is optional in case one wishes to study the dynamics of antigen-specific CD8 T cell responses. The adoptive cell transfer should be done one day before or on the day of the infection.
Keep cells and reagents on ice unless otherwise indicated.
Sacrifice one OT-I transgenic (CD45.2) mouse using CO2, isoflurane, or ketamine-xylazine according to the applicable rule for work with experimental mice.
Collect lymphoid organs, such as spleen and lymph nodes, from the mouse into cold PBS containing 2% FCS (FACS buffer, see Recipes) in 15 ml tubes on ice. This donor mouse should be sex-and-age-matched to the congenic (CD45.1) recipient.
Bring the organs to a biosafety cabinet class I. Insert a 100 µm cell strainer into a 50 ml tube and place on ice. Place the lymphoid organs onto the cell strainer and carefully mash in circular motions with the blunt end of a 5 ml syringe. Gently rinse the strainer into the 50 ml tube with the FACS buffer using a 1 ml pipette. Repeat the procedure until no tissue clumps are left in the strainer.
Remove the cell strainer and screw the cap onto the 50 ml tube. Centrifuge at 1,200 rpm (equivalent to 293 rcf) for 7 min at 4°C. Open the tube in the biosafety cabinet and decant the supernatant.
Add 3 ml of 1× erythrocyte lysis buffer (see Recipes) and mix by pipetting up and down. Incubate for 3 min at RT. Top up with 6 ml of FACS buffer and centrifuge at 1,200 rpm for 7 min at 4°C.
Decant the supernatant and resuspend the pellet in 1 ml of Buffer C (see Recipes). The pellet should now have a whitish color.
Mix the cell suspension thoroughly by pipetting up and down with a 200 µl pipette. Add 4-9 ml of Buffer C. Count cells, e.g., using a Luna cell counter: In a U-bottom well of a 96-well plate mix 1.5 µl of Acridine Orange/Propidium Iodide Stain + 13.5 µl of cell suspension. Load 10 µl on the slide to assess the fraction of live cells. Alternatively, any cell counter or a hemocytometer and trypan blue can be used.
Remove 1 × 106 cells and save in FACS buffer at 4°C for flow cytometry analysis at a later time point (see below in Procedure E).
Isolate naïve CD8 T cells using the Naïve CD8a+ T Cell Isolation Kit, as per the manufacturer’s instructions.
For every 1 × 107 cells, resuspend the pellet in 40 µl of Buffer C + 10 µl of biotin-antibody cocktail in a 15 ml tube. Mix well and incubate for 10 min at 4°C.
For 1 × 107 cells, add 20 of µl Buffer C + 20 µl of anti-biotin microbeads + 10 µl of anti-CD44 microbeads. Mix well and incubate for another 15 min at 4°C.
Add 10 ml of Buffer C and centrifuge at 287 × g for 7 min at 4°C.
Decant the supernatant and resuspend the pellet in 3 ml of Buffer C.
Place a pre-cooled (4°C) LS column in the magnetic field of the Miltenyi MidiMACS separator.
Rinse the column with 3 ml of ice-cold buffer C. Discard the flow-through.
Place a 15 ml tube on ice and position it below the outlet of the column. Load the 3 ml of cell suspension onto the column and collect the flow-through in a 15 ml tube. Rinse each column once with 3 ml of Buffer C. The total volume of the collected flow-through is 6 ml.
Count cells (see Step B7).
Remove 1 × 106 cells and save in FACS buffer at 4°C for flow cytometry analysis at a later time-point (see below in Procedure E).
Centrifuge at 287 × g for 7 min at 4 °C. Dilute cells in sterile PBS to reach the number of cells that need to be transferred per mouse.
For experiments with 24 h engraftment prior to Lm infection and day 1 post-infection read-out, inject 3 × 106 cells per mouse in 100 μl of PBS (room-temperature, RT); for d3 post-infection read-out, inject 1 × 106 cells per mouse in 100 μl of PBS (RT); and for d7 or later post-infection read-out, inject 2 × 104 cells per mouse in 100 μl of PBS (RT). Resuspend cells prior to injection into the tail vein.
Note: Engraftment can be assessed 24 h after adoptive cell transfer (when LmOVA is injected) by collecting 50 μl of peripheral blood according to the guidelines of the relevant animal experiment ethical permit and proceeding with flow cytometry analysis as described below (see below in Procedure E).
Injection of LmOVA and monitoring of animals
Perform the infection of mice in a bio-safety level 2 (BSL-2) animal facility. Animals need to be housed in individually ventilated cages (IVC).
Note: After infection, all used cages, including litter, need to be autoclaved.
Measure individual mouse baseline body weight using a lockable box and a precision scale that can be disinfected.
Note: Ideally, mice should get used to the daily weighting process 3-5 days prior to the experiment in order to avoid weight loss due to stress responses. It is also advisable to weigh the mice at the same time each day in order to avoid fluctuations due to food intake.
Put an individual mouse into the restrainer.
Note: To better visualize the vein and facilitate injection, the animals can be placed below an infrared lamp for 5-10 min, being careful not to overheat the animals.
A video demonstrating this procedure can be found at the following link: Intravenous Injection in the Mouse: https://researchanimaltraining.com/articles/intravenous-injection-in-the-mouse/.
Intravenously inject well mixed LmOVA, as prepared in Procedure A, into mice via the tail vein using a 29G insulin syringe. Inject 200 μl of a 5 × 104 bacteria/ml dilution per mouse for a total of 104 bacteria (Figure 2). Discard the needle and syringe in a biohazard sharps container for later autoclaving.
Note: Infection can be done either in healthy wild-type mice or in mice that had received OT-I cells the day before by adoptive cell transfer.
To determine the actual CFU that was injected, follow procedure A5 (see above).
Monitor the weight and general wellbeing of the animals every day for the first 7 days after bacterial infection. An initial weight drop is expected on days 1-3; later on, a recovery in weight can be observed. Monitor the consistency of the feces, which should be softer and lighter in color compared to normal conditions.
Note: Check the general appearance of the mice carefully, corresponding to your experimental animal license. Severe pain, suffering, and distress must be prevented.
Preparing spleen homogenates for plating
Notes:
Lm can be detected in the spleen for up to 7 days after infection in C57BL/6 wild-type mice. In animals that had received OT-I cells, the bacteria are cleared faster and are rarely detected on day 7 (unpublished observation). The bacterial load can be assessed during the acute phase (24 h and 3 days after infection) and/or during peak immune response (7 days after infection).
A fraction of the spleen can be used to evaluate immune cells via flow cytometry. In this case, after recording the spleen weight, the preparation of single-cell suspensions should be generated following steps 1-8 in procedure B, and subsequently, only 50% of the cell suspension will be lysed in 0.14%Tergitol/PBS for plating onto LB Agar plates. The other 50% can be used for flow cytometric analysis as described in procedure E.
All work should be performed in a biosafety cabinet class II and with adequate protective gear.
Sacrifice animals as described in Procedure B1 and collect spleens in 15 ml conical tubes containing 5 ml of cold FACS buffer; store the tubes on ice.
Bring the spleen to a biosafety cabinet class II. Record the weight of the spleen. If CD8 T cell responses are to be analyzed in parallel by flow cytometry, proceed with the weighing of 50% of the spleen.
Mash the spleen (or 50% of the organ) in a sterile 100 mm × 15 mm Petri dish in 5 ml of 0.14%Tergitol/PBS. Tergitol causes immediate cell lysis and releases intracellular bacteria.
Prepare serial 10-fold dilutions of the cell lysate in PBS ranging from 1:10 to 1:1,000,000 and plate 100 μl of each dilution in technical duplicates by spreading onto LB + erythromycin LB Agar plates. Grow upside-down at 37°C for 24 h (Figure 2).
After 24-48 h, count the colonies. Plates with CFU between 30-300 should be used to determine the bacterial load (Figure 2). An example calculation is provided in Table 2:
Table 2. Example calculation of CFUs
CFU = colony forming units, Ga CFU = Grand CFU, V = Volume, mg = milligram, hom. = homogenate. Average CFU is multiplied by 10 to get concentration per ml (100 μl × 10 = 1 ml).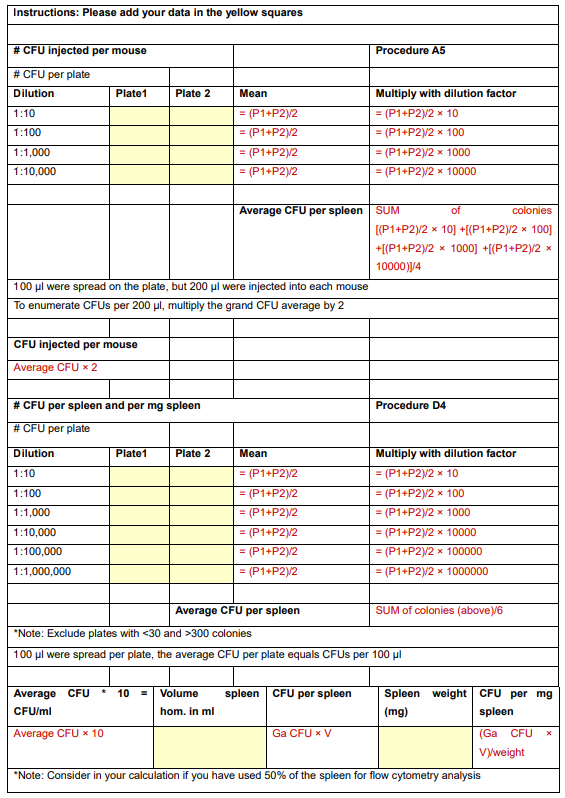
Preparing peripheral blood mononuclear cells (PBMCs) for flow cytometry analysis
Notes:
The same animal can be analyzed at various time points after LmOVA infection, such as early (d3), peak (d7), memory (d70), or after re-infection.
PBMCs can also be collected one day after ACT to assess the engraftment of OT-I cells.
Collect 50 μl of peripheral blood (approx. 2-3 drops) from either the mandibular or tail vein into an Eppendorf tube with 10 μl of heparin.
Transfer the tube into a biosafety cabinet class II.
Transfer 50 μl of the blood/heparin mixture into 10 ml of homemade erythrocyte lysis buffer (see Recipes) in a 50 ml tube and incubate for 5 min at RT. Then add 20 ml of FACS buffer.
Centrifuge at 287 × g for 7 min at 4°C and decant the supernatant. Resuspend the pellet in 1 ml of FACS buffer and count the cells as described above (Procedure B7).
Transfer 0.5 × 106-1 × 106 cells per FACS tube and centrifuge at 1,200 rpm (equivalent to 293 rcf) for 7 min at 4°C and decant the supernatant.
Add 10 μl of Fc-Block (1:25 in FACS buffer), resuspend, and incubate for 5 min at RT or 4°C.
Add 50 μl of the antibody mixture (diluted as described below) and stain for 30 min at 4°C, protecting from light.
Note: Staining with SIINFEKL-tetramer should be performed at RT.
Add 1 ml of FACS buffer, centrifuge at 1,200 rpm (equivalent to 293 rcf) for 7 min at 4°C, and decant the supernatant.
Fix the cells with 1% PFA/PBS for 20 min at RT.
Wash the cells with 1 ml of FACS buffer and centrifuge at 1,200 rpm (equivalent to 293 rcf) for 7 min at 4°C and decant the supernatant.
Resuspend the pellet in 200 μl of FACS buffer and acquire on a flow cytometer. To set the compensation, cells or OneComp eBeadsTM can be used.
Preparing single-cell suspensions from spleens for flow cytometry analysis
Prepare single-cell suspensions as described in Procedure B, Steps 1-8.
Centrifuge the cells at 287 × g for 7 min at 4°C and decant the supernatant.
Add 10 μl of Fc-Block (1:25 in FACS buffer) per tube, resuspend, and incubate for 5 min.
Add 50 μl of the antibody mixture (diluted as described below) per tube and stain for 30 min at 4°C, protect from light.
Note: Staining with SIINFEKL-tetramer should be performed at RT.
Add 1 ml of FACS buffer, centrifuge at 287 × g for 7 min at 4°C, and decant the supernatant.
Fix the cells with 1% PFA/PBS for 20 min at RT.
Wash with 1 ml of FACS buffer, centrifuge at 287 × g for 7 min at 4°C, and decant the supernatant.
Resuspend the pellet in 200 μl of FACS buffer and acquire on a flow cytometer (Figures 2 and 4).
Table 3. Antibody-mixture panels for flow cytometry analyses.
The ACT engraftment panel is used for the blood samples described in procedure B12 (optional). The other two panels are used for detecting antigen-specific CD8 T cells in the spleen and blood, with the following gating schema: CD45+CD8+ pre-gated short-lived effector cells (SLECs: CD127−KLRG1+) and memory-precursor cells (MPECs: CD127+KLRG1−) are shown.
Panel for ACT engraftment check Panel for ACT mice Panel for non-ACT mice Dilution SIINFEKL tetramer-PE SIINFEKL tetramer-PE 1:100 CD45.1-PerCP CD45.1-PerCP CD44-FITC 1:200 KLRG1-PE-Cy7 KLRG1-PE-Cy7 1:200 CD127-APC CD127-APC 1:200 CD8-PB CD8-PB CD8-PB 1:200 CD45.2-V500 CD45.2-V500 CD45-V500 1:200
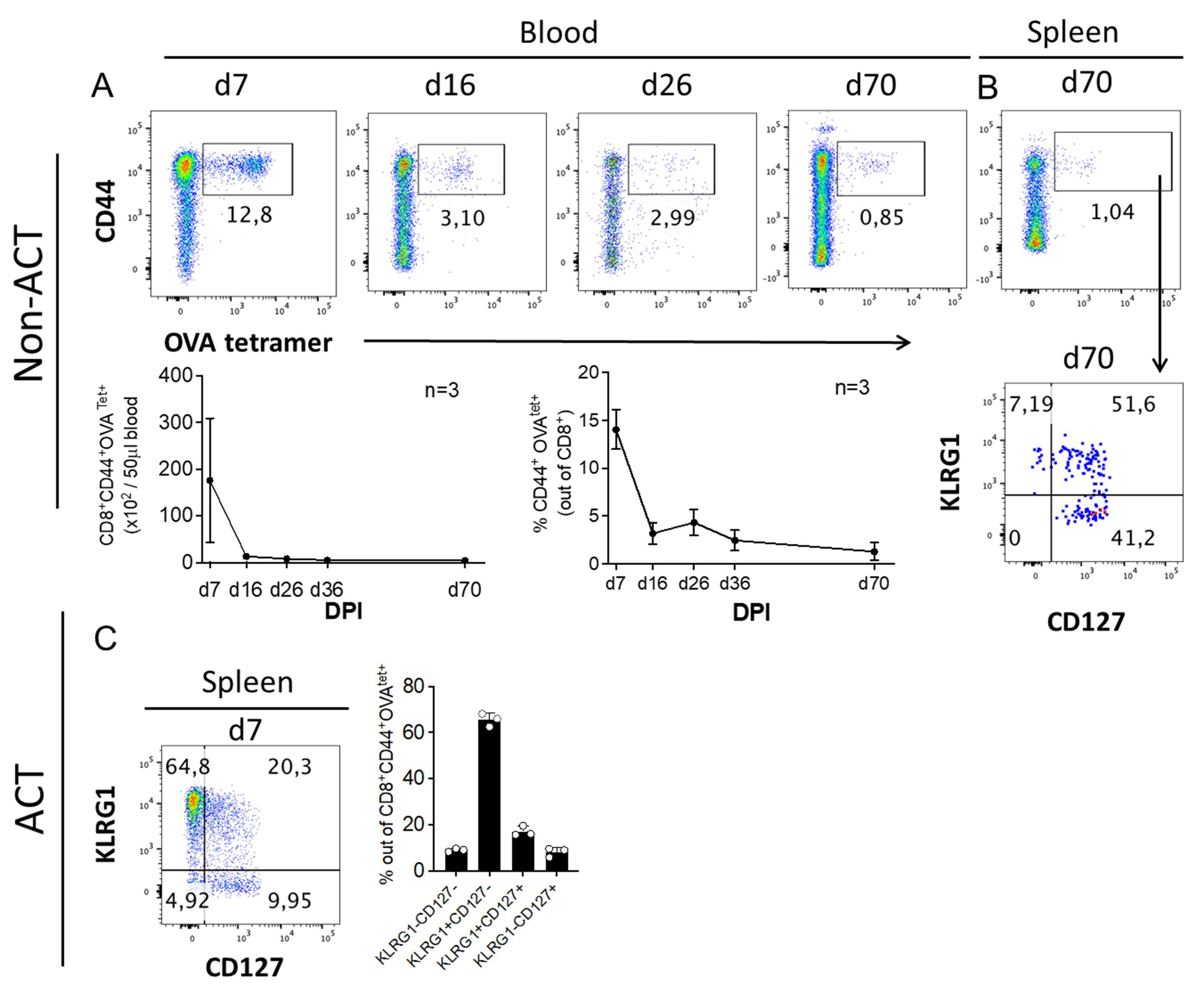
Figure 4. Example flow cytometry data analysis of antigen-specific CD8 T cells. (A) C57BL/6 mice were infected with 1 × 104 CFU LmOVA. The frequency of antigen-specific (CD44+OVAtet+) CD8 T cells were monitored in the blood for up to 70 days post-infection (d.p.i). Representative dot plots and quantification of CD44+OVAtet+ cells within CD8+ T cells are shown for the indicated time points post-infection. (B) Representative dot plots of CD44+OVAtet+ CD8+ T cells within SLECs (CD127−KLRG1+) and MPECs (CD127+KLRG1−) in the spleen on 70 d.p.i. (C) Representative dot plot and quantification of splenic SLEC (CD127−KLRG1+) and MPEC (CD127+KLRG1−) OT-I CD8 T cells at d7 after adoptive transfer of OT-I T cells and infection 1 day later, pre-gated on CD45.2+CD45.1−CD8+CD44+. Data shown represent at least two independent experiments, n = 3-7. Each dot in a bar graph represents data from one individual mouse. Results are shown as mean ± SD, reprinted with permission from Nature Publishing Group (Jakic et al., 2021).
Data analysis
Determining bacterial load in procedures A5 and D4:
Colony-forming units (CFUs) from spleens, see Supplementary Figure 1B in Jakic et al. (2021) .
Determining mouse weight loss:
The weight is calculated relative to day 0 (before infection), see Supplementary Figure 1B in Jakic et al. (2021) .
Determining the antigen-specific immune response:
Analyze flow cytometry data using, e.g., FlowJo software. Calculate relative frequencies of CD45.2 OT-I cells (ACT experiments) or antigen-specific (OVAtet+) CD8 T cells. Gate on lymphocytes after doublet exclusion (FSC-A vs. FSC-H), for MPEC (KLRG1-CD127+) and/or SLEC (KLRG1+CD127-) evaluation see Figures 1E, 2B, 4C, 6A, 6D, and Supplementary Figures 5B, 5E, 7A (Jakic et al., 2021). Numerical data can be exported to Excel to calculate absolute cell numbers in the spleen and in a defined volume of blood.
Perform statistical analysis and generate graphs, e.g., in GraphPad Prism. Normality of the data should be assessed via, e.g., Shapiro–Wilk test. When normally distributed, statistical analysis using unpaired Student’s t-test for samples with equal variance (F test), two-way ANOVA, or mixed-effects model analysis (REML) can be performed. When not normally distributed, the Mann-Whitney U test can be performed. Data are ideally presented as mean±SD.
Note: For adoptive transfer experiments, proceed only with mice that show successful engraftment, i.e., OT-I cells are detected in the peripheral blood 24h after engraftment.
Recipes
LB medium
Dissolve 20 g of LB agar powder according to Lennox (Carl Roth #X965.1) per liter of distilled water.
Sterilize by autoclaving.
Prior to use, add glucose at a final concentration of 500 mg/ml and erythromycin at a final concentration of 5 µg/ml.
Store at 4°C for up to one month.
LB agar plates
Dissolve 17.5 g LB agar powder according to Lennox (Carl Roth #6671.3) in 500 ml of distilled water (pH adjusted to 7).
Sterilize by autoclaving.
Cool liquid agar to <50°C prior to adding erythromycin at a final concentration of 5 µg/ml.
Pour agar into 10 cm Petri dishes in a safety cabinet.
Allow full polymerization, keep sterile, and store at 4°C.
Erythromycin (Stock: 5 mg/ml; final concentration: 5 μg/ml)
Dissolve 50 mg of erythromycin in ethanol and bring to a total volume of 10 ml.
Store aliquots of 1 ml (in 1.5 ml reaction tubes) at 4°C.
Add 1 ml of erythromycin stock solution to 1 L of LB medium/agar.
Glucose additive for cultivation of Listeria monocytogenes (Stock: 500 mg/ml; final concentration: 5 mg/ml)
Dissolve 25 g glucose in water and bring to a final volume of 50 ml.
Sterilize by filtration using a 0.22 μm filter and store at 4°C.
Add 10 ml of glucose stock solution to 1 L of LB medium/agar.
FACS buffer
Mix 20 ml Fetal calf serum (FCS) with 1 L sterile PBS.
Store at 4°C for up to one month.
Erythrocyte lysis buffer (10×) (Cold Spring Harbor Protocols, 2006)
For storage at 4°C for up to one month, prepare with distilled H2O:
Chemical Formula Molar mass (MW) End molarity Add to 250 ml H2O Ammonium chloride NH4Cl 53.491 g/ml 1.5 M 20.0591 g Potassium bicarbonate KHCO3 100.115 g/mol 100 mM 2.5029 g EDTA dihydrate C10H14N2Na2O8·2H2O 372.24 g/ml 1 mM 93.06 mg Filter through a 0.22 µm-filter using a vacuum pump. Store at 4°C and prepare a fresh 1× dilution by diluting directly in distilled water prior to use.
Fixation buffer
Dilute 4% formaldehyde stock solution (Carl Roth #P087.4) with PBS to generate a 1% solution.
Use the solution immediately to fix the cells (as described in procedure E above).
Store stock solution in an air-vented chemical cabinet, protected from light.
Buffer C
PBS pH 7.2
BSA, 0.5% (w/v)
2 mM EDTA
Mix all the components by stirring. Sterile filter with a 0.22 µm filter using a vacuum pump. Store at 4°C for up to one month.
Acknowledgments
This work was supported by the FWF Austrian Science Fund with the following grant (N-HK.: P28694-B30). This protocol describes experiments published in Jakic et al. (2021).
Competing interests
The authors declare no conflict of financial or non-financial interest.
Ethics
Animal procedures were approved by the Austrian Federal Ministry of Education, Science and Research (BMWFW-66.011/0064-WF/V/3b/2016; BMWFW-66.011/0112-WF/V/3b/2017).
References
- Ahmed, R. and Gray, D. (1996). Immunological memory and protective immunity: understanding their relation. Science 272(5258): 54-60.
- Ahn, J. J., Selvanantham, T., Zhang, M. A., Mallevaey, T. and Dunn, S. E. (2016). Experimental Infection with Listeria monocytogenes as a Model for Studying Host Interferon-γ Responses. J Vis Exp(117): 54554.
- Badovinac, V. P., Tvinnereim, A. R. and Harty, J. T. (2000). Regulation of antigen-specific CD8+ T cell homeostasis by perforin and interferon-γ. Science 290(5495): 1354-1358.
- Chang, J. T., Wherry, E. J. and Goldrath, A. W. (2014). Molecular regulation of effector and memory T cell differentiation. Nat Immunol 15(12): 1104-1115.
- Cold Spring Harbor Protocols. (2006). Red Blood Cell Lysis Buffer. doi:10.1101/pdb.rec390.
- Foulds, K. E., Zenewicz, L. A., Shedlock, D. J., Jiang, J., Troy, A. E. and Shen, H. (2002). Cutting edge: CD4 and CD8 T cells are intrinsically different in their proliferative responses. J Immunol 168(4): 1528-1532.
- Hamon, M., Bierne, H. and Cossart, P. (2006). Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol 4(6): 423-434.
- Harberts, A., Schmidt, C., Schmid, J., Reimers, D., Koch-Nolte, F., Mittrucker, H. W. and Raczkowski, F. (2021). Interferon regulatory factor 4 controls effector functions of CD8+ memory T cells. Proc Natl Acad Sci U S A 118(16): e2014553118.
- Harty, J. T. and Badovinac, V. P. (2008). Shaping and reshaping CD8+ T-cell memory. Nat Rev Immunol 8(2): 107-119.
- Harty, J. T. and Bevan, M. J. (1995). Specific immunity to Listeria monocytogenes in the absence of IFNγ. Immunity 3(1): 109-117.
- Jakic, B., Olson, W. J., Siegmund, K., Klepsch, V., Kimpel, J., Labi, V., Zehn, D., Baier, G. and Hermann-Kleiter, N. (2021). Loss of the orphan nuclear receptor NR2F6 enhances CD8+ T-cell memory via IFN-γ. Cell Death Dis 12(2): 187.
- Jones, G. S. and D'Orazio, S. E. F. (2013). Listeria monocytogenes: cultivation and laboratory maintenance. Curr Protoc Microbiol 31: 9B 2 1-9B 2 7.
- Kaech, S. M. and Cui, W. (2012). Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 12(11): 749-761.
- Khan, S. H. and Badovinac, V. P. (2015). Listeria monocytogenes: a model pathogen to study antigen-specific memory CD8 T cell responses. Semin Immunopathol 37(3): 301-310.
- Lecuit, M., Dramsi, S., Gottardi, C., Fedor-Chaiken, M., Gumbiner, B., and Cossart, P. (1999). A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. Embo J 18: 3956-3963.
- Levine, L. S., Hiam-Galvez, K. J., Marquez, D. M., Tenvooren, I., Madden, M. Z., Contreras, D. C., Dahunsi, D. O., Irish, J. M., Oluwole, O. O., Rathmell, J. C. et al. (2021). Single-cell analysis by mass cytometry reveals metabolic states of early-activated CD8+ T cells during the primary immune response. Immunity 54(4): 829-844 e825.
- Mackaness, G. B. (1962). Cellular resistance to infection. J Exp Med 116: 381-406.
- Merrick, J. C., Edelson, B. T., Bhardwaj, V., Swanson, P. E. and Unanue, E. R. (1997). Lymphocyte apoptosis during early phase of Listeria infection in mice. Am J Pathol 151(3): 785-792.
- Osebold, J. W. and Sawyer, M. T. (1957). Immunization studies on listeriosis in mice. J Immunol 78(4): 262-268.
- Pamer, E. G. (1994). Direct sequence identification and kinetic analysis of an MHC class I-restricted Listeria monocytogenes CTL epitope. J Immunol 152(2): 686-694.
- Pamer, E. G. (2004). Immune responses to Listeria monocytogenes. Nat Rev Immunol 4(10): 812-823.
- Pamer, E. G., Harty, J. T. and Bevan, M. J. (1991). Precise prediction of a dominant class I MHC-restricted epitope of Listeria monocytogenes. Nature 353(6347): 852-855.
- Pope, C., Kim, S. K., Marzo, A., Masopust, D., Williams, K., Jiang, J., Shen, H. and Lefrancois, L. (2001). Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. J Immunol 166(5): 3402-3409.
- Qiu, Z., Khairallah, C. and Sheridan, B. S. (2018). Listeria monocytogenes: A Model Pathogen Continues to Refine Our Knowledge of the CD8 T Cell Response. Pathogens 7(2):55.
- Schmidt, N. W., Khanolkar, A., Hancox, L., Heusel, J. W. and Harty, J. T. (2012). Perforin plays an unexpected role in regulating T-cell contraction during prolonged Listeria monocytogenes infection. Eur J Immunol 42(3): 629-640.
- Shen, H., Miller, J. F., Fan, X., Kolwyck, D., Ahmed, R. and Harty, J. T. (1998). Compartmentalization of bacterial antigens: differential effects on priming of CD8 T cells and protective immunity. Cell 92(4): 535-545.
- Zehn, D., Lee, S. Y. and Bevan, M. J. (2009). Complete but curtailed T-cell response to very low-affinity antigen. Nature 458(7235): 211-214.
- Zenewicz, L. A. and Shen, H. (2007). Innate and adaptive immune responses to Listeria monocytogenes: a short overview. Microbes Infect 9(10): 1208-1215.
- Zehn, D., Lee, S. Y., and Bevan, M. J. (2009). Complete but curtailed T-cell response to very low-affinity antigen. Nature 458: 211-214.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Jakic, B., Kimpel, J., Olson, W. J., Labi, V. and Hermann-Kleiter, N. (2021). Bacterial Infection with Listeria monocytogenes in Mice and Subsequent Analysis of Antigen-Specific CD8 T Cell Responses. Bio-protocol 11(23): e4247. DOI: 10.21769/BioProtoc.4247.
Category
Immunology > Immune cell function
Microbiology > Microbe-host interactions > Bacterium
Cell Biology > Cell isolation and culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link