- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Duloxetine in the Bacterial Culture and Medium to Study Drug-gut Microbiome Interactions
Published: Vol 11, Iss 21, Nov 5, 2021 DOI: 10.21769/BioProtoc.4214 Views: 3421
Reviewed by: Juan Facundo Rodriguez AyalaDURAI SELLEGOUNDERAnonymous reviewer(s)
Abstract
Expanding our understanding of drug-gut bacteria interactions requires high-throughput drug measurements in complex bacterial cultures. Quantification of drugs in the cultures, media, and cell pellets is prone to strong matrix effects. We have developed a liquid chromatography–high resolution mass spectrometry (LC–HRMS) method for quantifying duloxetine from high-throughput gut-drug interaction experiments. The method is partially validated for its reproducibility, sensitivity, and accuracy, which makes it suitable for largescale drug screens. We extensively used this method to study biotransformation and bioaccumulation of duloxetine and other drugs in several species of gut bacteria.
Keywords: LC–HRMSBackground
Several recent clinical studies showed the impact of drugs on the gut microbiome. Such interactions can have a considerable impact on drug biotransformation, pharmacokinetics, efficacy, and toxicity of the administered drug (Falony et al., 2016; Scott et al., 2017). To study bacteria-drug interactions, systematic profiling of such interactions in human gut bacterial strains is a powerful approach (Maier and Typas, 2017; Zimmermann et al., 2019; Forslund et al., 2015). Such experiments usually involve large scale screening of several gut bacterial species with targeted drugs and require high-throughput quantification of drugs and their metabolites. Also, the complexity of bacterial cultures and gut microbiota media (GMM) used in such experiments generate strong background signals that interfere with drug analysis. The further study of drugs biotransformation and bioaccumulation requires their quantification in total culture medium, supernatant, and cell pellet, which is prone to strong matrix effects. We addressed these challenges by developing and validating an LC–HRMS method for the quantification of duloxetine. We studied duloxetine, a widely used antidepressant, and found it to be bioaccumulated by several gut microbial species. This LC–HRMS method serves as a validation of the results obtained from large screening of several hundreds of bacteria-drug pairs (Klünemann et al., 2021).
Generally, drug quantification is performed using triple quadrupole instruments due to their sensitivity and specificity. The reported methods for duloxetine quantification are mostly done in blood plasma samples (Senthamil Selvan et al., 2007; Waldschmitt et al., 2007; Chae et al., 2013; Zhang et al., 2017). However, during quantification of duloxetine in bacterial cultures using the reported methods, we found that the specificity of analysis was compromised by the background matrix of the bacterial culture. The matrix effect hampered the accurate quantification of duloxetine, especially from incubated cultures (for 48 h). The method we developed is accurate, specific, and sensitive for quantifying duloxetine from total culture medium, supernatant, and pellet.
The study of drug-gut interactions is an emerging field that is expanding our knowledge about this fascinating research area. Our method can be a useful tool for drug quantification in gut microbiome studies.
Materials and Reagents
Pipette tips (Rainin, METTLER TOLEDO, USA)
Tips GP-LTS-A-1,000 µl-768/8 (Rainin, catalog number: 17014338)
Tips GP-LTS-A-250 µl-/F-960/10 (Rainin, catalog number: 30389288)
GP-LTS-A-10 µl-960/10 (Rainin, catalog number: 30389284)
Eppendorf Polypropylene DNA LoBind Tubes (2.0 ml) (Thermo Fisher Scientific, catalog number: 10051232)
WebSeal 96-Well Plate, Round Well, Barcoded (Thermo Fisher Scientific, catalog number: 601 80-P206B)
Corning® 96 well Polypropylene Deep Well Plate (Sigma-Aldrich, catalog number: CLS3957)
Corning® 96 Round Well Microplate Storage Mat III (Sigma-Aldrich, catalog number: CLS3080)
Zone-FreeTM Sealing Films (Sigma-Aldrich, catalog number: Z721646)
Glassware purchased from DURAN (DWK Life Sciences, Wertheim, Germany)
Bacterial species used: Clostridium saccharolyticum; type strain, WM1; tax ID: 610130; internal database ID: NT5037. [Storage: 20% glycerol in mGAM medium, stored at -80°C]
Optima® LC–MS grade acetonitrile (Fisher Chemical, catalog number: A955-212)
Methanol (Fisher Chemical, catalog number: A456-212)
Water (Fisher Chemical, catalog number: W6-121)
LC–MS LiChropur® grade formic acid (Merck KGaA, catalog number: 5330020050)
Standards of Duloxetine (Sigma-Aldrich, Merck KGaA, catalog number: SML0474)
Internal standards Fluoxetine hydrochloride (Sigma-Aldrich, Merck KGaA, catalog number: F132)
Sulfamethazine (Sigma-Aldrich, Merck KGaA, catalog number: S5637)
Sulfamethizole (Sigma-Aldrich, Merck KGaA, catalog number:S5632)
PierceTM Triple Quadrupole Calibration Solution, Extended Mass (Thermo Fisher, catalog number: 88340)
mGAM medium: Gifu Anaerobic Medium Broth, Modified (HyServe, catalog number: 05433)
GMM (gut microbiota medium)
DMSO solvent
Extraction solvent (see Recipes)
1 µM Duloxetine stock solution preparation (see Recipes)
1 µM Fluoxetine stock solution preparation (see Recipes)
Mobile phase buffer preparations (see Recipes)
Equipment
Pipettes (Eppendorf AG, Hamburg, Germany)
Eppendorf Multipette® M4 (Eppendorf, catalog number: 4982000012)
Eppendorf Research plus pipette (100-1,000 µl)
Multi-channel 8-channel, 30-300 µl (Eppendorf, catalog number: 3122000051)
Multi-channel 8-channel, 0.5-10 µl (Eppendorf, catalog number: 3122000019)
Vanquish UHPLC–MS/MS system coupled to a Q-Exactive plus HRMS (Thermo Scientific, MA, USA; catalog number: IQLAAAGABHFAPUMZZZ)
ACQUITY UPLC HSS T3 column (100Å, 1.8 µm; 2.1 × 100 mm) (Waters, catalog number: 186003539)
S-Series Heated Ultrasonic Cleaning Water Bath (FisherbrandTM, catalog number:10162372)
Eppendorf MixMate for vortexing (Eppendorf, catalog number: 5353000510) or equivalent
HamiltonTM 1700 Series GastightTM Syringe 500 μl (Thermo Fisher Scientific, catalog number: 365JLT41)
Centrifuge 5418 R, refrigerated (Eppendorf, catalog number: 5401000013)
Vinyl Anaerobic Chambers Type C (Coy Laboratory Products, Inc., USA)
Water bath sonicator; FisherbrandTM S-Series heats ultrasonic cleaners (Thermo Fisher Scientific, catalog number: 10162372)
Software
Thermo Xcalibur software version 4.0.27 (Thermo Scientific, MA, USA) used for data acquisition (Q-exactive plus tune version 2.11) and qualitative (Qual Browser) and quantitative (Quan Browser) data analysis
Procedure
Bacterial Culture inoculation with duloxetine
Inoculate Clostridium saccharolyticum from stock culture (20% glycerol in mGAM medium, stored at -80°C) in 5 ml mGAM and allow it to grow overnight. Then passage it to 5 ml GMM (gut microbiota medium) and grow overnight to get the preculture. Use the preculture to inoculate the 96 deep well plate (total volume between 100 and 800 µl depending on the plate size). Measure the inoculation OD (should be at ~0.01).
Grow the culture plate for 48 h and measure OD (should be between 0.8 to 1).
Prepare drug containing medium (GMM) at a 2× concentration (100 µM duloxetine). Prepare the medium containing the bacterial species (Clostridium saccharolyticum) also with 2× inoculum from Step A2 (OD for 2× is 0.02). Mix the final medium according to the different sample requirements, e.g., 1× drug plus 1× bacteria (in this case, a mixture of 50 µl of 2× drug in GMM and 50 µl of 2× bacteria). The final concentration is then 1× bacteria and 1× drug.
Grow the culture plate for 48 h at 37°C in a vinyl anaerobic chambers type C, in a nitrogen atmosphere containing 12% CO2 and 1% hydrogen (anoxic).
Sample preparation for drug extraction
After 48 h of incubation, remove the culture plates from the anaerobic chamber and transfer 10 μl aliquots of ‘total’ culture (medium + bacteria) to a new 96-well plate. Spin down the remaining culture at 4,000 × g for 10 min to harvest the supernatant. Aliquot 10 μl of ‘supernatant’ in another 96-well plate.
Add 1 ml of cold (-20°C) extraction solvent (see Recipes) to 10 μl aliquots of both total and supernatant samples and vortex for 5 min (100-fold dilution) in the culture plates (deep 96-well plates). Then, further dilute 25 μl with 500 μl of extraction solvent (2,000-fold dilution) in polypropylene deep 96-well plates.
Close the plates with a lid, incubate for 15 min at -20°C, and centrifuge at 14,000 × g for 10 min at 4°C.
Remove 100 μl of extracted supernatant from the above plate and transfer to WebSeal 96-well plate for LC–MS analysis.
Standard calibration curve and Quality Control (QC) sample preparation
Prepare 1 μM stock of duloxetine and internal standards in DMSO (see Recipes).
Serially dilute duloxetine stock to prepare the following calibration using 2× dilutions at each level in extraction solvent.
Standard solutions in ‘total culture’: incubate drug concentrations of 0, 10, 20, 30, 40, 50, and 70 μM in a bacterial culture. Add 1 ml of cold extraction solvent in the total culture inoculate of each concentration level separately. Prepare these standards in the same manner as actual experimental conditions for samples (as mentioned in Step B3 of Sample preparation for extraction of drugs).
Standard solutions in ‘supernatant’: prepare supernatant standard solutions from above ‘total culture’ by centrifugation at 14,000 × g for 10 min at 4°C. Add 1 ml of cold extraction solvent in the supernatant of each concentration level separately. Prepare these standards in the same manner as the actual experimental conditions for samples (as mentioned in Step B3 of Sample preparation for extraction of drugs). Refer to Figure 1 for an overview of sample preparation.

Figure 1. Overall workflow for sample preparation for LC–HRMS analysis. For a description of total culture, refer to Procedure A.Prepare QC sample at 40 μM of duloxetine separately for ‘total culture’ and ‘supernatant’ as mentioned above in Steps C3 and C4.
LC–MS/MS analysis
Preparation of mobile phases:
For buffer solvent A and B preparations, see Recipes.
Mix each buffer solvents thoroughly by shaking and degas both the buffers in the water bath sonicator with loose caps for at least 10 min.
Set the flow rate of 0.3 ml/min.
Maintain the column at 40°C.
Set LC gradient for 10 min as follows: start at 10% of solvent B for 2 min, ramp up to 90% for 2 min, and hold for 2 min; next, allow 4 min of equilibration to the initial condition (10% of solvent B).
Vanquish UHPLC–MS/MS system preparation
Put up both mobile phase buffers on the UHPLC system and purge it with the mobile phase buffers for 5 min.
Set up the UPLC column and start the flow at a rate of 0.3 ml/min with both mobile phase buffers at a ratio of 90:10 (A:B).
Equilibrate the column for at least 15 min after the column pressure is stable.
MS parameter preparation and Q-exactive plus HRMS calibration
Use extended mass calibration solution and perform ‘custom calibration’ for the mass spec with Xcalibur tune software.
Set the tune MS parameters to the ESI positive mode as shown in Table 1:
Table 1. MS method parameters for Q-exactive plus
MS parameter value/unit Spray voltage 4 kV Sheath gas 30 Auxiliary gas 5 S-Lens 65 eV Capillary temperature 320°C Vaporization temperature 250°C MS resolution 35,000 or 70,000 AGC target 3e6 Maximum IT 100 ms Scan range 60 to 900 m/z Spectrum data type Profile
LC–MS injection sequence analysis
Inject 2 μl of each sample in the following manner:
Solvent blank: Inject extraction solvent as Blank to check the instrument background.
Medium blank: Extract GMM medium as described above in Procedure B and inject to check matrix background from medium.
Incubate bacterial culture blank (without drug): Same a 0 μM level mentioned in Procedure C to check matrix background after incubation.
Inject QC sample at least five times the (same as 40 μM standard solution from Procedure C) to stabilize the LC–MS system.
Inject six standard solutions from 10 to 70 μM prepared in supernatant (see Procedure C).
Inject QC sample.
Run ‘total culture’ samples (randomize the sequence of samples).
Inject QC sample after every ten samples.
Repeat the same sequence from Step G3 for supernatant samples.
At the end of each sequence, run QC sample followed by standard calibration curve.
Data analysis
Evaluation of raw data
Get the extracted ion chromatograms (XIC) for duloxetine, fluoxetine, sulfamethazine, and sulfamethizole for their exact masses with a 10 ppm accuracy window, as shown in Table 2. Use Xcalibur Qual browser for raw data visualization and quality check.
Table 2. Name, molecular formula, and exact mass of protonated ions (M + H) duloxetine and internal standards used to obtain respective XICs
Name Molecular formula Exact mass (M + H) Duloxetine C18H19NOS 298.1265 Fluoxetine C17H18F3NO 310.1418 Sulfamethazine C12H14N4O2S 279.0965 A representative chromatogram of duloxetine and its internal standards are shown in Figure 2.
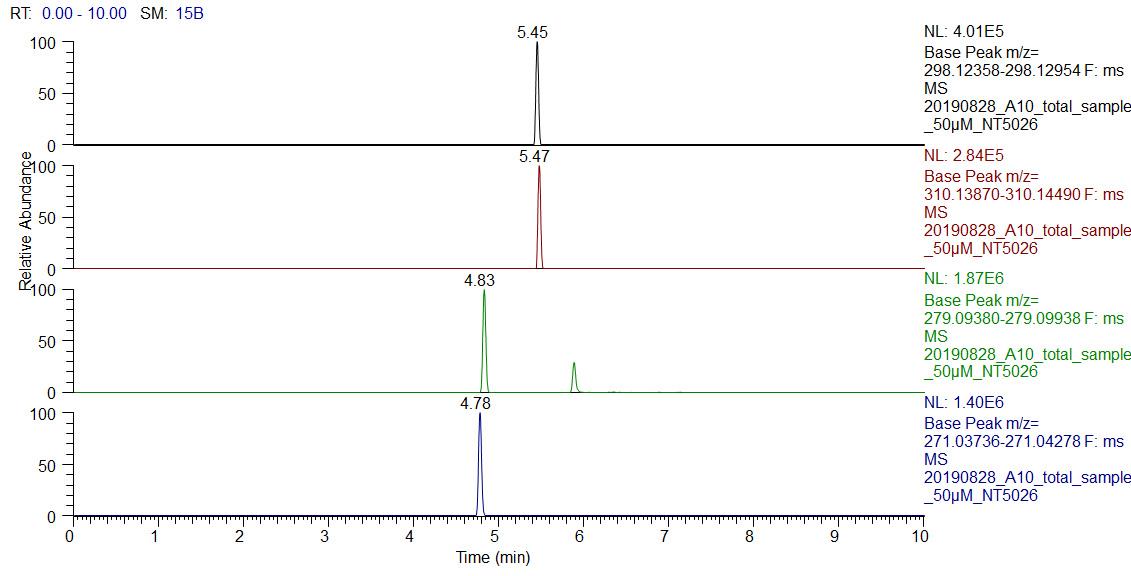
Figure 2. Representative extracted ion chromatograms of duloxetine and its internal standardsConfirm the duloxetine identity using exact mass as well as MS/MS spectra (shown in Figure 3), which can be interpreted from the chemical structure of duloxetine (shown in Figure 4). Refer to EMBL-MCF spectral library (http://curatr.mcf.embl.de) for more MS/MS data and protocols (Palmer et al., 2018).
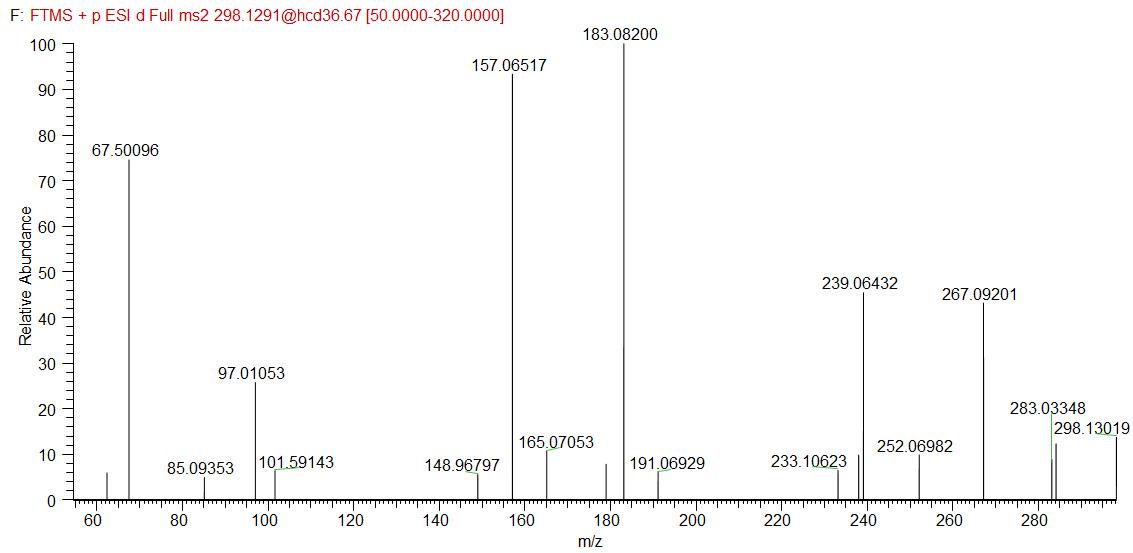
Figure 3. MS/MS spectra of duloxetine standard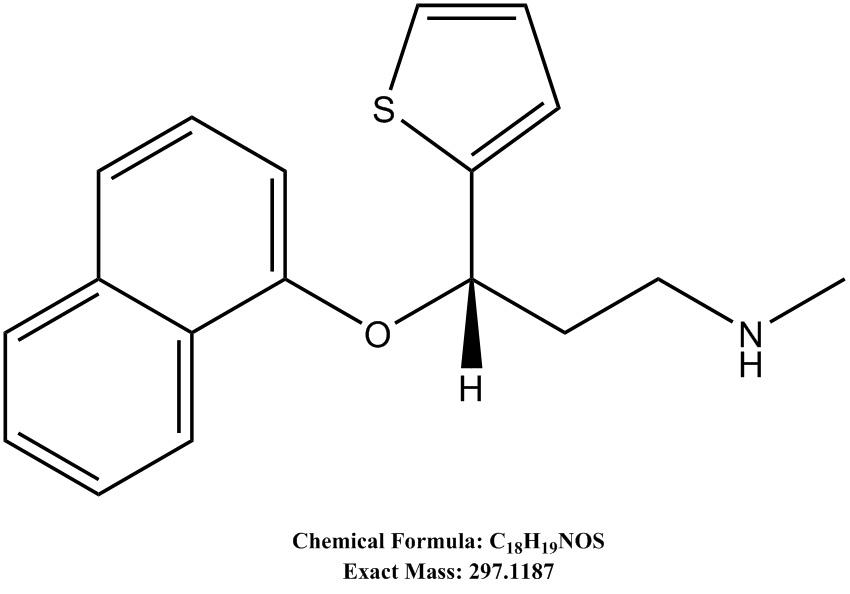
Figure 4. Chemical structure of duloxetine
Quality control evaluation with internal standards
Get the peak area from the XICs of duloxetine and its internal standards using Xcalibur Quan browser.
Calculate the concentration of QC samples injected throughout the sequence. The deviation of known and calculated concentrations should not be >10%.
Calculated the variation in peak areas internal standards across all samples in analysis sequence, which should not be >10% (shown in Figure 5).
Use peak areas of fluoxetine internal standard for normalization of respective duloxetine peaks areas. Use the slope of the standard calibration curve to calculate concentrations of duloxetine from samples.
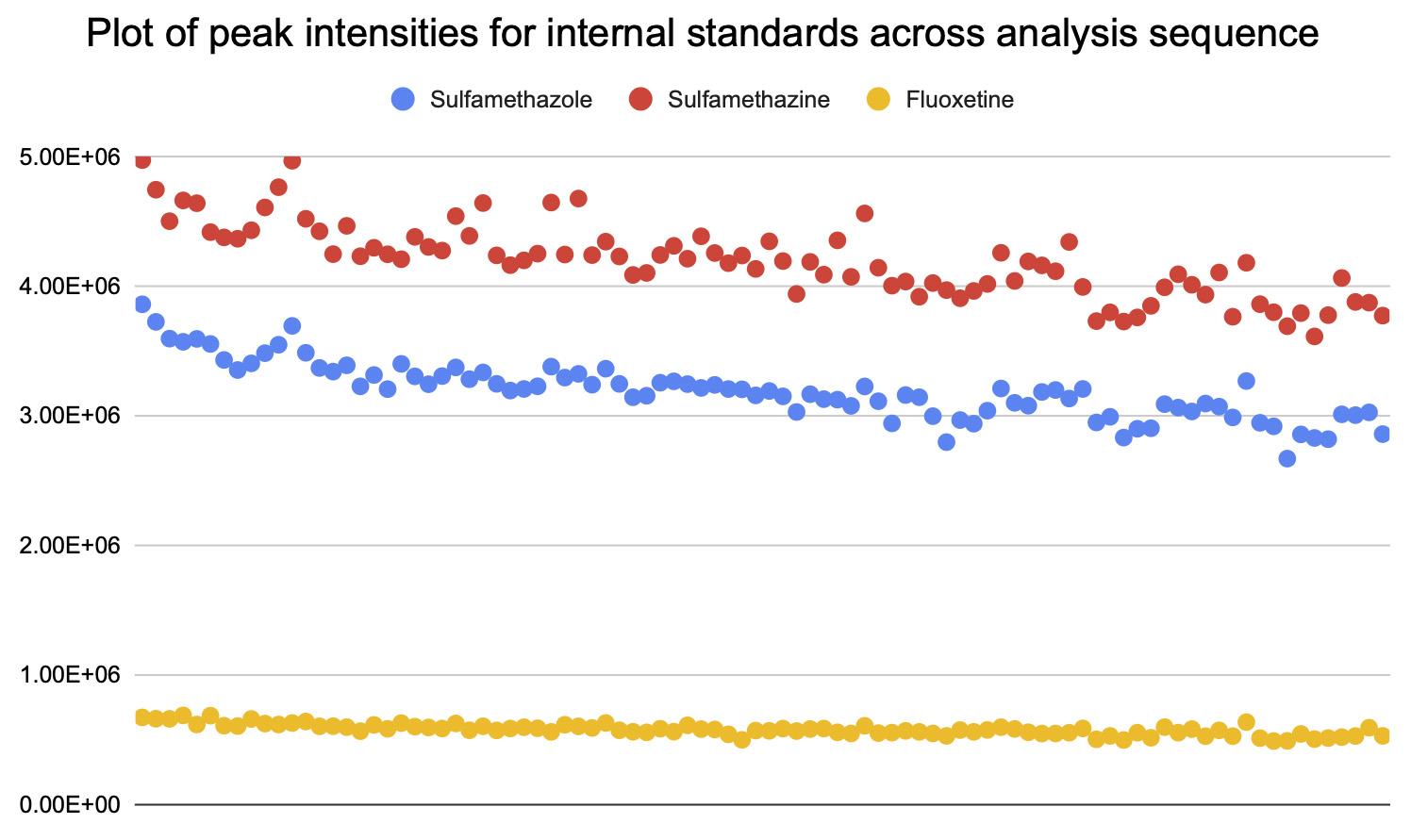
Figure 5. Plot of peak areas/intensities of internal standards from 94 bacterial culture samples. The percentage of CVs for sulfamethizole, sulfamethazine, and fluoxetine were 6.88, 6.85, and 7.54, respectively.
Recipes
Extraction solvent
Mix equal volumes of acetonitrile and methanol in a glass bottle (total volume 1,000 ml).
Prepare 1 ml of 1 µM of internal standard stock solutions for sulfamethizole (270 mg), sulfamethazine (278 mg), and fluoxetine (309 mg) in the extraction solvent (equal mix of acetonitrile and methanol).
Add an internal standard stock solution of sulfamethizole (100 µl), sulfamethazine (40 µl), and fluoxetine (2.5 µl) into 1,000 ml of extraction solvent to get final concentrations of 100 nM, 40 nM, and 2.5 nM, respectively.
1 µM Duloxetine stock solution preparation
Dissolve 297.4 ± 2 mg of duloxetine standard in 1 ml of DMSO solvent in an amber color glass vial.
1 µM Fluoxetine stock solution preparation
Dissolve 309.3 ± 2 mg of fluoxetine standard in 1 ml of DMSO solvent in an amber color glass vial.
Mobile phase buffer preparations
Preparation of buffer A: add 1 ml (0.1 %) formic acid to 1,000 ml of water.
Preparation of buffer B: add 1 ml (0.1 %) formic acid to 1,000 ml of methanol.
Acknowledgments
We thank EMBL Metabolomics and other core facilities, as well as EMBL Alxandrov team members, for their support during this project.
The sample preparation protocol was partially adapted from Zimmermann et al. (2019).
Competing interests
There are no competing interests.
References
- Chae, J. W., Baek, H. M., Kim, S. K., Kang, H. I. and Kwon, K. I. (2013). Quantitative determination of duloxetine and its metabolite in rat plasma by HPLC-MS/MS. Biomed Chromatogr 27(8): 953-955.
- Falony, G., Joossens, M., Vieira-Silva, S., Wang, J., Darzi, Y., Faust, K., Kurilshikov, A., Bonder, M. J., Valles-Colomer, M. and Vandeputte, D. (2016). Population-level analysis of gut microbiome variation. Science 352(6285): 560-564.
- Forslund, K., Hildebrand, F., Nielsen, T., Falony, G., Le Chatelier, E., Sunagawa, S., Prifti, E., Vieira-Silva, S., Gudmundsdottir, V. and Pedersen, H. K. (2015). Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528(7581): 262-266.
- Klünemann, M., Andrejev, S., Blasche, S., Mateus, A., Phapale, P., Devendran, S., Vappiani, J., Simon, B., Scott, T. A. and Kafkia, E. (2021). Bioaccumulation of therapeutic drugs by human gut bacteria. Nature 597(7877): 533-538.
- Maier, L. and Typas, A. (2017). Systematically investigating the impact of medication on the gut microbiome. Curr Opin Microbiol 39128-135.
- Palmer, A., Phapale, P., Fay, D. and Alexandrov, T. (2018). Curatr: a web application for creating, curating and sharing a mass spectral library. Bioinformatics 34(8): 1436-1438.
- Scott, T. A., Quintaneiro, L. M., Norvaisas, P., Lui, P. P., Wilson, M. P., Leung, K. Y., Herrera-Dominguez, L., Sudiwala, S., Pessia, A. and Clayton, P. T. (2017). Host-microbe co-metabolism dictates cancer drug efficacy in C. elegans. Cell 169(3): 442-456e418.
- Senthamil Selvan, P., Gowda, K. V., Mandal, U., Sam Solomon, W. D. and Pal, T. K. (2007). Determination of duloxetine in human plasma by liquid chromatography with atmospheric pressure ionization-tandem mass spectrometry and its application to pharmacokinetic study. J Chromatogr B Analyt Technol Biomed Life Sci 858(1-2): 269-275.
- Waldschmitt, C., Vogel, F., Maurer, C. and Hiemke, C. (2007). Measurement of duloxetine in blood using high-performance liquid chromatography with spectrophotometric detection and column switching. Ther Drug Monit 29(6): 767-772.
- Zhang, M., Li, H., Ji, Z., Dong, D. and Yan, S. (2017). Clinical study of duloxetine hydrochloride combined with doxazosin for the treatment of pain disorder in chronic prostatitis/chronic pelvic pain syndrome: An observational study. Medicine(Baltimore) 96(10): e6243.
- Zimmermann, M., Zimmermann-Kogadeeva, M., Wegmann, R. and Goodman, A. L. (2019). Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570(7762): 462-467.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Phapale, P. B., Blasche, S., Patil, K. R. and Alexandrov, T. (2021). Quantification of Duloxetine in the Bacterial Culture and Medium to Study Drug-gut Microbiome Interactions. Bio-protocol 11(21): e4214. DOI: 10.21769/BioProtoc.4214.
Category
Microbiology > Microbial biochemistry
Microbiology > Microbial metabolism
Biochemistry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











