- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
High-throughput 3D Spheroid Formation and Effective Cardiomyocyte Differentiation from Human iPS Cells Using the Microfabric Vessels EZSPHERETM
Published: Vol 11, Iss 21, Nov 5, 2021 DOI: 10.21769/BioProtoc.4203 Views: 3799
Reviewed by: Alessandro DidonnaFereshteh AzediAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Human iPSC-Derived Neuron and Oligodendrocyte Co-culture as a Small-Molecule Screening Assay for Myelination
Stefanie Elke Chie [...] Maria Consolata Miletta
May 5, 2025 3414 Views

Isolation and Culture of Ferret Airway Stem Cells
Ziying Yan [...] Feng Yuan
Jul 20, 2025 2411 Views

Optimization of Adipogenic Differentiation Protocol for Murine and Human Cell Culture Models
Junwan Fan [...] Wenyan He
Jan 20, 2026 214 Views
Abstract
High-throughput 3D spheroid formation from human induced pluripotent stem cells (hiPSCs) can be easily performed using the unique microfabric vessels EZSPHERE, resulting in effective and large scale generation of differentiated cells such as cardiomyocytes or neurons. Such hiPSC-derived cardiomyocytes (hiPSC-CMs) or neurons are very useful in the fields of regenerative medicine or cell-based drug safety tests. Previous studies indicated that 3D spheroids arising from hiPSCs are effectively differentiated into high quality hiPSC-CMs by controlling Wnt signals through utilization of the microfabric vessels EZSPHERE. Here, we describe a simple and highly efficient protocol for generating a large number of uniformly sized hiPSC spheroids and inducing them for cardiac differentiation using the EZSPHERE. This method comprises the collection and dissociation of the spheroids from cardiac differentiation medium, in the middle stage of the whole cardiac differentiation process, and re-seeding the obtained single cells into the EZSPHERE to re-aggregate them into uniform hiPSC-CM spheroids with controlled size. This re-aggregation process promotes non-canonical Wnt signal-related cardiac development and improves the purity and maturity of the hiPSC-CMs generated.
Graphic abstract:
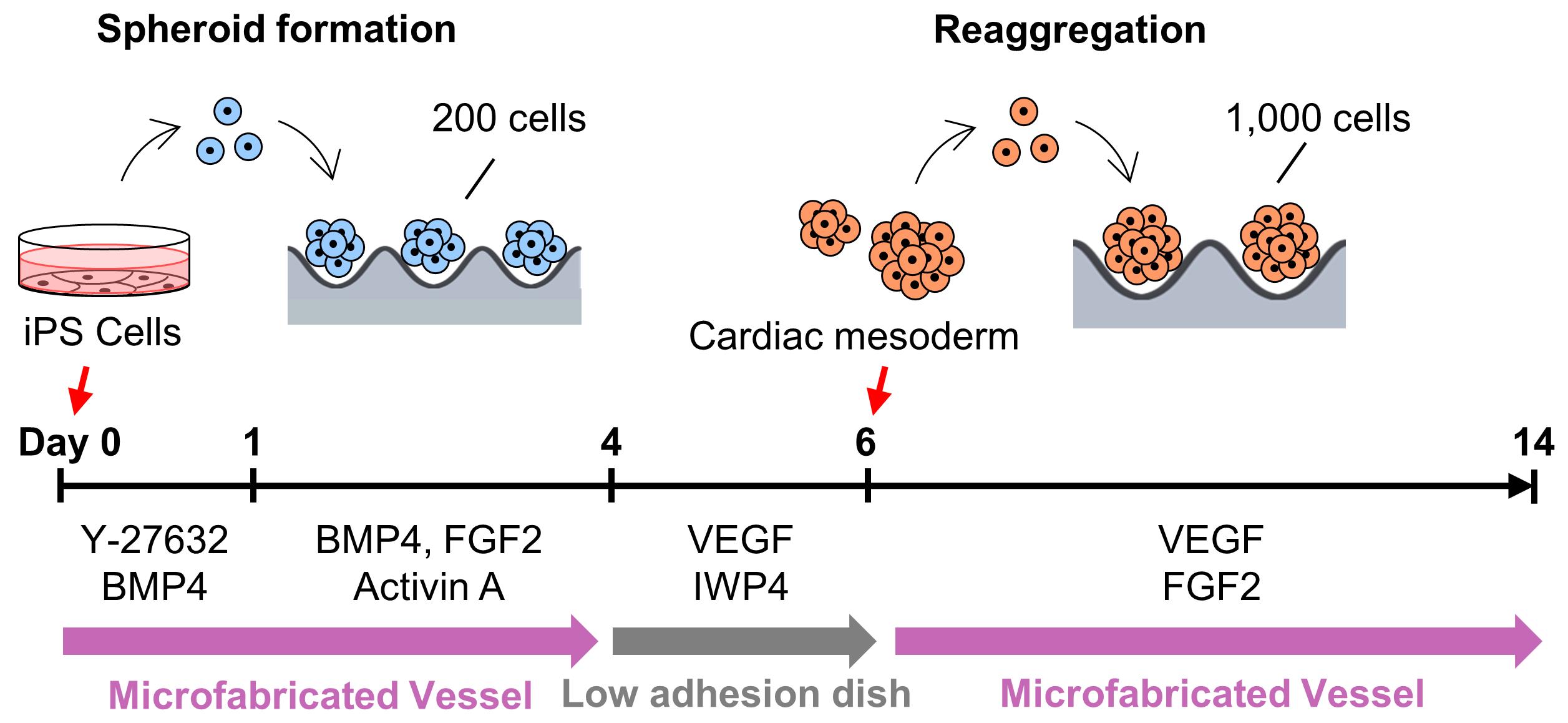
Overview of cardiac differentiation from iPSCs by spheroid formation and reaggregation using EZSPHERE.
Background
Human induced pluripotent stem cell-derived Cardiomyocytes (hiPSC-CMs) are expected to become a cell source for applications in regenerative medicine of heart diseases and drug discovery. Heart diseases remain the leading cause of death worldwide (WHO, 2018). In non-human primates, transplanted hiPSC-CMs improve cardiac contraction function and are sufficient to regenerate the infarcted heart (Shiba et al., 2016). Previous studies, represented by the CiPA Project (Blinova et al., 2017) and the JiCSA Project (Yamazaki et al., 2018), have shown the utility of hiPSC-CMs for prediction of the risk of cardiotoxicity, like Torsades de Pointes. It was also shown that maturity of the generated hiPSC-CMs alters the response to drugs (da Rocha et al., 2017). Thus, the development of a high-throughput and robust protocol for generating high-quality hiPSC-CMs is urgent for the realization of regenerative medicine and drug discovery.
hiPSCs could be directly differentiated into cardiomyocytes in either 2D adhesion culture or 3D cell aggregates/spheroid culture methods. In particular, the spheroid culture method is easier to scale-up and better at promoting maturation of the generated hiPSC-CMs, compared with the 2D culture method (Pal et al., 2013). In the cardiac differentiation process, regulation of canonical and non-canonical Wnt at the right time strongly promotes pluripotent stem cell differentiation into cardiomyocytes, not other mesodermal cell types (Loh et al., 2016). In addition, expression levels of the Wnt signal-related genes Wnt5A and Wnt11 are affected by spheroid size (Hwang et al., 2009, Chen et al., 2015). Therefore, we sought to develop a highly efficient and robust cardiac differentiation protocol by achieving high-throughput spheroid formation with simultaneous active spheroid size control.
In this study, we used the uniquely shaped microfabric dishes, “EZSPHERE,” designed for high-throughput production of uniform spheroids on one culture dish, with its high-density micro-wells (Sato et al., 2016). Initially, a basic spheroid culture protocol, optimized for cardiac differentiation (Yang et al., 2008, Bauwens et al., 2014, Kempf et al., 2014), was combined with the EZSPHERE dishes. For spheroid formation from the hiPSCs, Type 900 EZSPHERE dishes with a diameter of 100 mm were used. Each of them had approximately 14,000 individual micro-wells with regular size (400-500 μm in diameter and approximately 100 μm in depth). On calculation, approximately 14,000 spheroids could be formed in each EZSPHERE dish, and their initial seeding cell number per spheroid ranged 200-220. As per standard protocol, activin A, BMP-4, and FGF2 were added to the spheroids in the EZSPHERE to induce differentiation into Primitive Streak on day 1. On day 4, the spheroids were collected, washed, and transferred onto a flat bottom low cell adhesion dish, followed by treatment with Wnt inhibitor. On day 6, the size uniformity of the spheroids was lost, but 62% of the cell population had differentiated into PDGFR-α/KDR double-positive cardiac mesoderm-like cells (Kattman et al., 2011; Birket et al., 2015). To control spheroid size once more, the differentiated spheroids were collected and dissociated into single cells. Then, the single cells obtained were re-aggregated by re-seeding onto the dishes of Type 903 EZSPHERE (with a diameter of 35 mm), which have approximately 1,000 individual micro-wells with larger size (approximately 800 μm diameter and 400 μm depth) in each dish. The re-aggregated spheroids increased the number of cardiac Troroponin T (CTNT) positive cells faster than that of the non-reaggregated spheroids. In addition, the cardiac maturity (estimated by the gene expression level of MYL2 per MYL7) was much higher in the re-aggregated spheroids compared with that of the spheroids that did not undergo the re-aggregation process. Performing the re-aggregation process also raised the expression levels of the non-canonical Wnt signals Wnt5A and Wnt11. In mice, Wnt5A and Wnt11 are essential for the generation of second heart field progenitor cells (Cohen et al., 2012). Wnt11 is also known for its role in generating myocardial electrical gradient patterns through the regulation of non-canonical Wnt/Ca2+ signaling during zebrafish heart development (Panakova et al., 2010). To reveal the detail of its molecular mechanism, further study is required. However, these data strongly suggest that the cardiomyocyte differentiation process performed on EZSPHERE, including a re-aggregation step for the induced cardiac mesoderm/progenitor, is effective in producing cardiomyocytes with high purity and maturation levels.
Materials and Reagents
EZSPHERE Dish 100 mm Type 900 (AGC Techno Glass, IWAKI, catalog number: 4020-900)
EZSPHERE Dish 35 mm Type 903 (AGC Techno Glass, IWAKI, catalog number: 4000-903)
EZ-BindShut II 100 mm Dish (AGC Techno Glass, IWAKI, catalog number: 4020-800LP)
Cell culture T25 Flask (AGC Techno Glass, IWAKI, catalog number: 3100-025)
5 ml Pipet (AGC Techno Glass, IWAKI, catalog number: 7103-005)
Cell scraper (AGC Techno Glass, IWAKI, catalog number: 9010-320)
70 μm cell strainer (Corning, Falcon, catalog number: 352350)
Pasteur pipettes (AGC Techno Glass, IWAKI, catalog number: IK-PAS-9P)
Human induced pluripotent stem cell (hiPSC) strains 253G1 and 201B7 (iPS Academia Japan)
Maintenance culture media mTeSR1 (Stemcell Technologies, catalog number: 5850)
Biolaminin 521 LN (LN521) (BioLamina, catalog number: LN521-03)
DPBS (FUJIFILM Wako Pure Chemical, Wako, catalog number: 045-29795)
HBSS, no calcium (Thermo Fisher Scientific, Gibco, catalog number: 14170112)
DMEM (4.5g/L Glucose) with L-Gln, without Sodium Pyruvate, liquid (Nacalai, catalog number: 08459-35)
0.5 mol/L-EDTA solution (pH 8.0) (Nacalai, catalog number: 06894-14)
Fetal bovine serum, FBS (MP Biomedicals, CELLECT, catalog number: 2916154)
Accutase (Merck, Sigma-Aldrich, catalog number: A6964-100ML)
AccuMax (Innovative Cell Technologies, catalog number: AM105)
TrypLE select Enzyme (1×), no phenol red (Thermo Fisher Scientific, Gibco, catalog number:12563011)
StemProTM-34 SFM (1×) (Thermo Fisher Scientific, Gibco, catalog number: 10639011)
Penicillin-Streptomycin (10,000 U/ml) (Thermo Fisher Scientific, Gibco, catalog number: 15140122)
L-Glutamine (200 mM) (Thermo Fisher Scientific, Gibco, catalog number: 25030081)
holo-Transferrin human (Merck, Sigma-Aldrich, catalog number: T0665-100MG)
L-Ascorbic acid (Merck, Sigma-Aldrich, catalog number: A92902-100MG)
1-Tioglycerol (Merck, Sigma-Aldrich, catalog number: M6145-25ML)
ROCK inhibitor Y-27632 (Wako Pure Chemical Industries, Wako, catalog number: 253-00513)
Recombinant Human BMP-4 Protein (Bio-Techne, R&D Systems, catalog number: 314-BP-010)
Fibroblast Growth Factor (basic), Human, recombinant (154 aa) (FUJIFILM Wako Pure Chemical, Wako, catalog number: 064-04541)
Recombinant Human VEGF 165 Protein (Bio-Techne, R&D Systems, catalog number: 293-VE-010)
Recombinant Human/Mouse/Rat Activin A Protein (Bio-Techne, R&D Systems, catalog number: 338-AC-010)
Stemolecule Wnt Inhibitor IWP-4 (REPROCELL USA, Stemgent, catalog number: 04-0036)
DNase I, Bovine Pancreas (Merck, Millipore, catalog number: 260913)
30 w/v% Albumin D-PBS (-) Solution, from Bovine Serum (BSA), Fatty Acid Free (FUJIFILM Wako Pure Chemical, Wako, catalog number: 015-23871)
4% Paraformaldehyde Phosphate Buffer Solution (Wako Pure Chemical Industries, Wako, catalog number: 163-20145)
Antibodies used in flow cytometry analysis (Table 1)
Table 1. Antibodies used in flow cytometry analysis
Antibodies Distributor Cat. No. Dilution ratio APC-KDR BioLegend 359915 2.5:100 APC-Mouse IgG1, kappa Isotype control BioLegend 400121 2.5:100 PE-PDGFRα BioLegend 323505 5:100 PE-Mouse IgG1, kappa Isotype control BioLegend 400113 5:100 PE anti-cardiac Troponin T BD Pharmingen 564767 2.5:100 PE-Isotype control BD Pharmingen 554680 2.5:100 Cell culture Reagents (see Recipes)
Cytokines and small molecular compounds (see Recipes)
Modified StemPro-34 (see Recipes)
Spheroid formation medium (see Recipes)
Stage-1 medium (2×) (see Recipes)
Stage-2 medium (see Recipes)
Stage-3 medium (see Recipes)
Equipment
Laminar flow hood (Hitachi, model: SCV-Class II type A/B)
Water bath (AS ONE Corporation, model: TR-1AR)
TC10 Automated Cell Counter (Bio-Rad, catalog number: 145-0001)
EVOS FL Auto Cell Imaging System (Life Technologies, EVOS, catalog number: AMAFD1000)
EVOS Onstage Incubator (Life Technologies, EVOS, catalog number: AMC1000)
BD FACSVerse flow Cytometer (BD Bioscience, catalog number: 651154)
Pipet-Aid XPress 110V (Drummond Scientific Company, catalog number: 4-040-135)
Centrifuge (Himac, CF16RX, model: S101812)
Vortex mixer Vortex Gwnie 2 (Scientific Industries, Inc., catalog number: SI-0236)
HERAcell CO2 incubator (Kendro Laboratory Products)
Vacuum pump (Model JN726 FT.18)
Software
ImageJ (NIH, https://imagej.nih.gov/ij/)
AGDRec AG-Desktop recorder (AmuseGraphics, http://t-ishii.la.coocan.jp/hp/ag/index.html)
FlowJo software (FlowJo LLC, https://www.flowjo.com/)
Procedure
Note: Perform the culture operation in a biosafety level 2 laboratory in a dedicated laminar flow hood. All waste (tubes, tips, plates) should be inactivated by autoclaving and then properly disposed of according to the rules of your facility.
High-throughput spheroid formation from hiPSCs
Culture the hiPSCs in the medium mTeSR1 in Flasks coated with Biolaminin 521 LN.
Note: Subculture the hiPSCs as single cells, not cell clumps, without adding the ROCK inhibitor.
Prepare Spheroid formation medium (see Recipes: Table 5) and pre-warm at 37°C in a water bath.
Use 90% confluent hiPSCs.
Aspirate medium from the cells and wash with 2.5 ml of room temperature DPBS. For aspiration, use the vacuum pump with the pipette and an appropriate tube.
Add 2.5 ml of TrypLE select and incubate at 37°C for 4 min.
Aspirate TrypLE select and add 2.5 ml of Spheroid formation medium.
Detach the cells by flushing the medium to the cells using the Pipet-Aid with a 5 ml pipette. If the cells do not detach, scrape them with a Cell Scraper.
Flush the cells 2-8 times by pipetting, to dissociate the cell clumps into single cells, and collect them into a 15 ml centrifuge tube.
Centrifuge at 100 × g for 4 min at 20°C.
Aspirate supernatant and resuspend pellet with 1-5 ml of Spheroid formation medium.
Measure cell density using the TC10 Automated Cell Counter.
Dilute cells to the density of 3 × 105 cells/ml with Spheroid formation medium, and inoculate 10 ml of the suspended cells to the EZSPHERE Dish 100 mm Type 900 (see Figure 1).
Place the EZSPHERE Dish in a 37°C, 5% CO2 incubator and culture for 24 h.
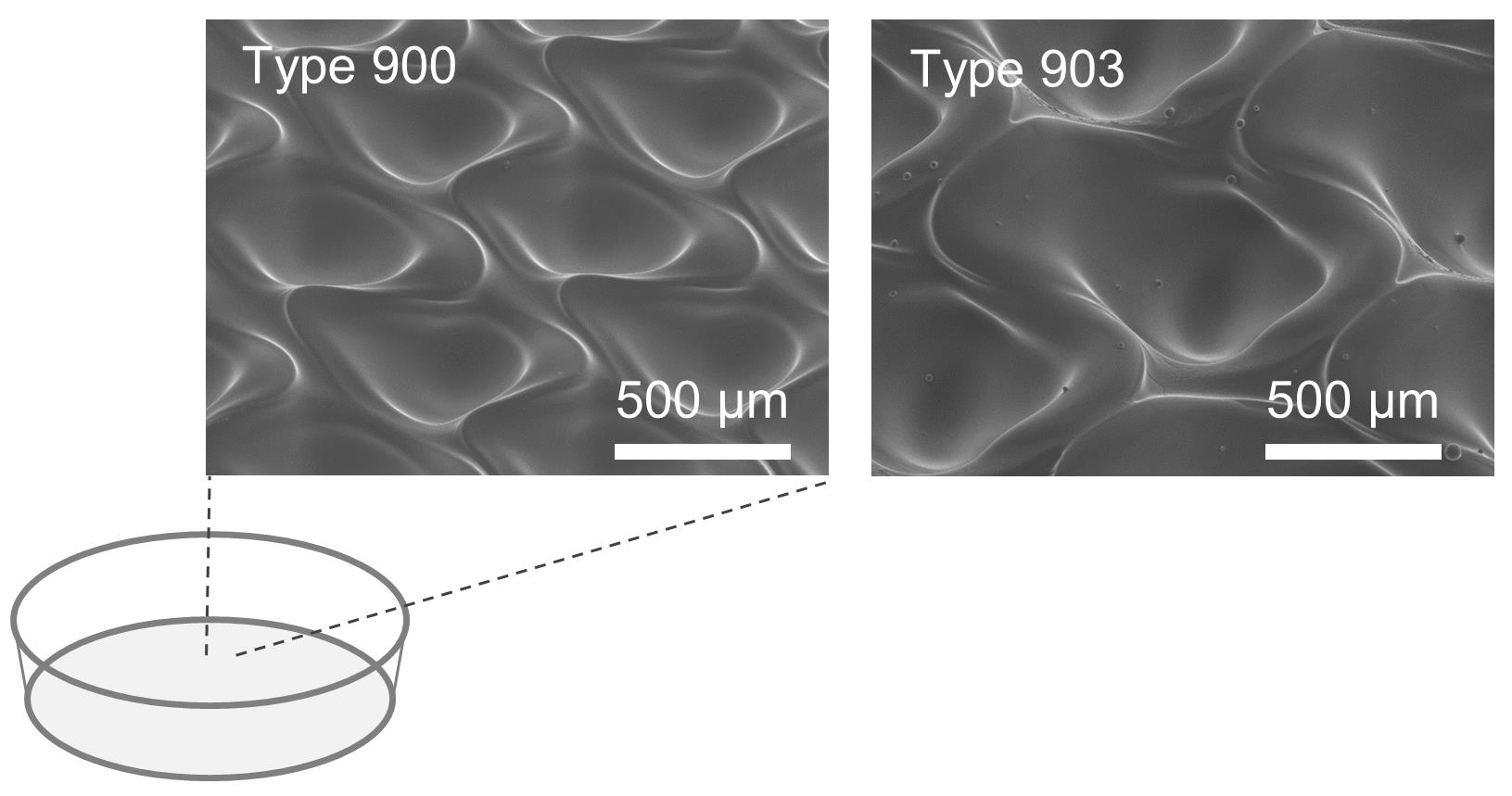
Figure 1. Images of the microfabric vessels “EZSPHERE” with a dish type
Stage-1 of cardiac differentiation
Move on to Stage-1 of the cardiac differentiation process, 24 ± 2 h after spheroid formation.
Prepare Stage-1 medium (2×) (see Recipes: Table 6) and pre-warm at 37°C in a water bath.
Take out the EZSPHERE dish from the CO2 incubator and check if spheroids have formed in each micro-well of the EZSPHERE, under a phase contrast microscope.
Carefully add an equal volume (10 ml) of Stage-1 medium (2×) to the spheroids.
Note: We recommend taking 10 ml of Stage-1 medium (2×) with a 10 ml pipette, placing the tip of the pipette on the liquid surface around the center of the EZSPHERE dish, standing the pipette vertically, and pouring the medium carefully.
Return the EZSPHERE dish to the 37°C, 5% CO2 incubator, and culture for 89 h.
Stage-2 of cardiac differentiation
Move on to Stage-2 of the cardiac differentiation process, 89 ± 2 h after the Stage-1 mentioned above.
Prepare Stage-2 medium (see Recipes: Table 7) and IMEM medium, then pre-warm at 37°C in a water bath.
Harvest spheroids from EZSPHERE dish into a 15 ml or 50 ml tube.
(Optional)
Transfer spheroids to a 100-mm flat bottom dish and place on the stage of the microscope.
Shake the dish finely in a circular motion to place the spheroids in the center of the dish.
Take a picture of spheroids with the microscope (see Figure 2).
Measure the size of spheroids using the ImageJ software (see Data analysis A).
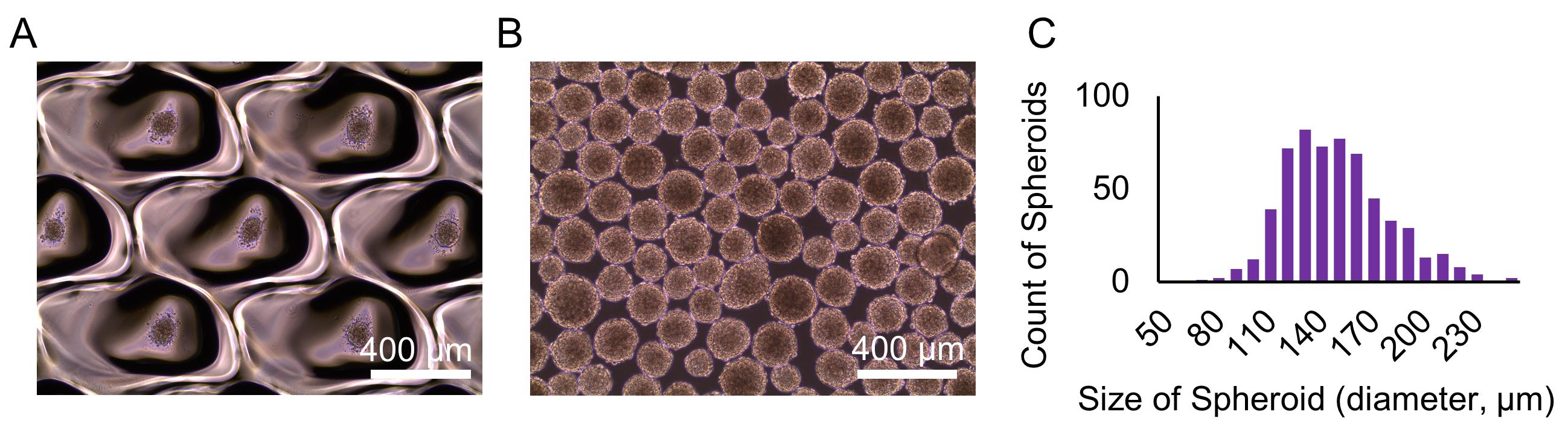
Figure 2. Images and size of the hiPSC spheroids generated at Procedure C. Spheroids on the EZSPHERE dish (A) and transferred onto a flat bottom dish (B). Histogram of the size (diameter) of the spheroids (μm) (C).Loosen the cap of the tube for ventilation, and place the tube in the 37°C CO2 incubator for 10 min to precipitate the spheroids.
Carefully remove the supernatant from spheroids as much as possible using a pipette, and add 5 ml of pre-warmed IMEM medium to wash the spheroids.
Centrifuge spheroids at 50 × g for 3 min at 20°C, removing the supernatant as much as possible.
Add 20 ml of pre-warmed Stage-2 medium to spheroids, and transfer spheroids to a 100-mm low cell adhesion dish (EZ-BindShut II).
Place the dish into the 37°C CO2 incubator.
Note: We recommend shaking the dish three times back and forth and left and right to distribute the spheroids evenly in the dish.
Shake the dish every morning and evening as described above to equally disperse the spheroids.
Stage-3 of cardiac differentiation and re-aggregation of spheroids
Note: The re-aggregation of spheroids increases the purity in the cardiac differentiation process. As a negative control, do not re-aggregate the spheroids at this step. To confirm the effect of re-aggregation, analyze the purity of CTNT positive iPS-CMs by flow cytometry after Procedure G.
On day 6 of cardiac differentiation, move on to Stage-3 for re-aggregation of the spheroids.
Note: While it is recommended that this re-aggregation step be performed on day 6, day 7 is also acceptable.
Prepare Stage-3 medium (see Recipes: Table 8) and pre-warm at 37°C in a water bath.
Collect spheroids from the 100-mm EZ-BindShut II dish into a 50 ml tube and place it in the 37°C CO2 incubator for 10 min to precipitate the spheroids.
Remove supernatant from the collected spheroids and add 5 ml of room temperature DPBS to wash them.
Centrifuge at 50 × g for 3 min at 20°C and remove the supernatant.
Note: For a negative control, skip Steps D6-D9 and proceed to Step D10.
Add 1 ml of Accutase to the spheroids and incubate at 37°C in the water bath for 8 min to dissociate the spheroids into single cells. During this incubation, mix the cells vigorously by vortexing the tube for 10 s every 4 min.
If cell aggregates remain after 8 min of incubation, as mentioned above, enforce additional incubation: collect only the remaining cell aggregates from single dissociated cells using a pipette and transfer to a new tube. Then, add double the amount (2 ml) of Stage-3 medium to the single dissociated cells and add 1 ml of Accutase to the aggregates. Incubate the aggregates at 37°C in the water bath for 8 min again. During the incubation, mix the cells vigorously, as mentioned above.
Add double the amount (2 ml) of Stage-3 medium to the single dissociated cells in Accutase, centrifuge at 200 × g for 4 min at 20°C, and then remove supernatant.
Resuspend the collected cell pellet with 5 ml of Stage-3 medium, and count the number of cells with diameter ranging 8-27 μm. If cell aggregates remain, remove them using a 70 μm Cell Strainer.
Dilute the cells with Stage-3 medium to the density of 1 × 105 cells/ml and inoculate 3 × 105 cells into a 35-mm EZSPHERE Type 903 dish.
Note: For a negative control, add 20 ml of Stage-3 medium to the spheroids obtained from the one 100-mm EZ-BindShut II dish at step 5 and dispense 3 ml each into 35-mm EZ-BindShut II dish.
Change half volume of medium every 2 or 3 days.
Harvest cardiomyocyte differentiated cells
Around day 10 to 14 of the cardiac differentiation process, harvest the obtained hiPSC-CMs or negative control.
Prepare 4 ml of dissociation buffer (0.05% Trypsin-EDTA:AccuMax = 3:1) and 4 ml of quench buffer (DMEM:FBS 1:1 + 200 units ml-1 of DNase I).
Observe the beating state of the cardiac spheroids under the microscope (see Videos 1 and 2).
Note: Beating of the cardiac spheroids might decay as the EZSPHERE dish grows colder at room temperature. Therefore, we recommend using an on-stage incubator to keep the dish warm at 37°C during microscopic observation. The EVOS FL Auto Cell Imaging System and EVOS on-stage incubator were used for observation, and an AGDRec AG-Desktop recorder was used for capturing videos.
Video 1. Representative video of the cardiac spheroids re-aggregated in the micro-wells of EZSPHERE. The spheroids were re-aggregated on day 6 and began beating from day 8. This video was recorded on day 14.Video 2. Video of a population of beating cardiac spheroids recovered after the re-aggregation process. The cardiac spheroids were transferred from the EZSPHERE dish to a flat dish and then videotaped as they were beating.Collect the spheroids into a 15 ml tube and let stand for 2-3 min until they settle.
Remove supernatant and add 5 ml of DPBS to wash the spheroids.
Centrifuge the tube at 50 × g for 3 min and remove the supernatant, taking care not to lose the spheroids.
Add 1 ml of dissociation buffer to the collected spheroids and incubate at 37°C in the water bath for 8 min, to dissociate the spheroids into single cells. During the incubation, mix the cells vigorously, as mentioned above.
If cell aggregates still remain after 8 min of incubation, enforce an additional incubation step. Collect only the remaining cell aggregates from single dissociated cells using a pipette and transfer into a new tube. Then, add equal volume (1 ml) of quench buffer to the dissociated cells and add 1 ml of dissociation buffer to the aggregates. Incubate the aggregates again at 37°C in the water bath for 8 min. During the incubation, mix the cells vigorously, as mentioned above.
Add equal volume (1 ml) of quench buffer to the dissociated cells.
Centrifuge the tube at 200 × g for 5 min and remove the supernatant by careful pipetting. Do not use an aspirator to prevent the inhalation of cell aggregates entwined with some broken cell-derived DNA.
For cytometry analysis, resuspend the harvested cells with DPBS. For cultivation in the next step, resuspend the cells with DMEM containing 10% FBS.
Count the number of cells within 8-30 μm in diameter.
Flow cytometry analysis of Cardiac Mesoderm.
Use the cells harvested on day 6 (Step D8).
Prepare two tubes (#1 and #2) and transfer 0.5 × 106 cells into each tube respectively for cell staining and its isotype control.
Prepare 20 ml of flow cytometry buffer [HBSS (-), 3% FBS, 0.03 mM EDTA].
Add 2 ml of flow cytometry buffer to the cells.
Centrifuge the cells at 300 × g for 5 min at 20-25°C and aspirate supernatant.
Repeat Steps F4-F5.
Resuspend 0.5 × 106 cells in 100 μl of flow cytometry buffer.
Add APC-KDR and PE-PDGFRα antibodies into tube #1 for cell staining, and add APC- and PE-isotype control into tube #2 for isotype control (Table 1).
Incubate each tube at room temperature for 45 min in the dark.
Add 2 ml of flow cytometry buffer into each tube.
Centrifuge the cells at 300 × g for 5 min at 20-25°C and aspirate the DPBS.
Repeat Steps F10-F11.
Resuspend the cells in 300 μl of flow cytometry buffer.
Measure the cells using the flow cytometer (BD FACS Verse) and analyze the data using the FlowJo software (Figure 3).
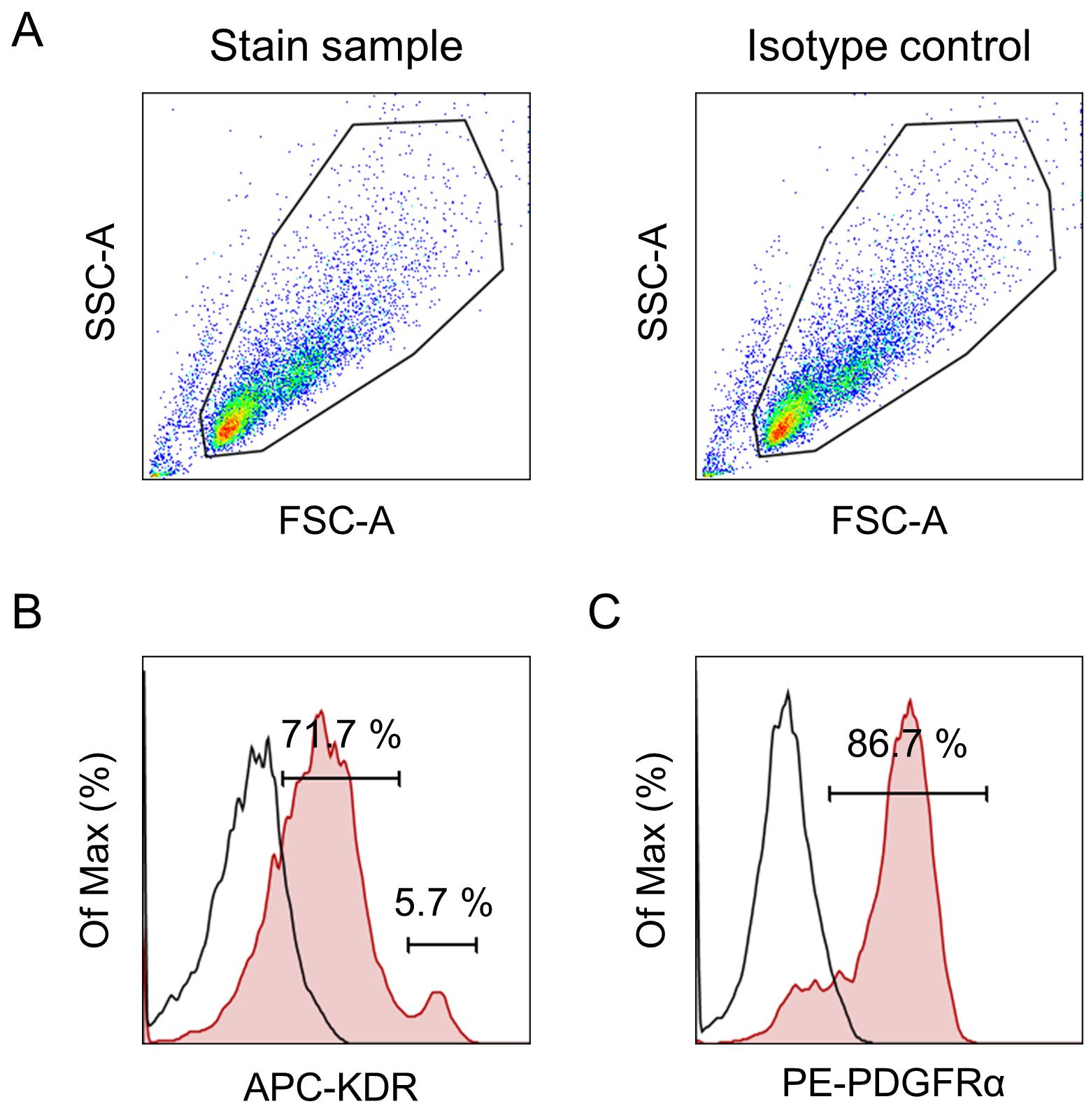
Figure 3. Representative flow cytometry profile of iPSC-derived cardiac mesoderm on day 6. Events were acquired using the flow cytometer (BD FACS Verse) and analyzed with the FlowJo software. First, the major population was selected by gating in the SSC vs. FSC dot plots (A). Histograms are shown for each antibody: APC-KDR (B) and PE-PDGFRα (C). The plots show isotype control (open histogram) versus stained sample (red histogram). The main population present on day 6 is KDR and PDGFRα positive (5.7% of the cells were KDRhigh, 71.7% of the cells were KDRlow, and 86.7% PDGFRα+). This result suggests that the main population present on day 6 is considered as cardiac mesoderm-like cells.
Flow cytometry analysis of the hiPSC-derived cardiomyocytes (hiPSC-CMs)
Use 1 × 106 of the differentiated or negative control cells obtained in Step E10.
Note: If the cells were harvested in the medium, wash them with DPBS.
Centrifuge the cells at 300 × g for 5 min at 20-25°C and aspirate the supernatant.
Add 200 μl of 4% paraformaldehyde phosphate buffer solution and incubate at room temperature for 20 min to fix the cells.
Add 2 ml of DPBS to the cells and centrifuge at 300 × g for 5 min at 20-25°C.
Remove the supernatant containing paraformaldehyde solution and discard it into an appropriate waste bottle.
Resuspend the cell pellets in 2 ml of DPBS. The cells could be stored at this step for up to 7 days.
Centrifuge the cells at 300 × g for 5 min at 20-25 °C and aspirate the supernatant.
Resuspend the cell pellets in 200 μl (0.5 × 106 cells per 100 μl) of Flow Cytometry Permeabilization/Wash Buffer I and incubate at room temperature for 30 min.
Transfer 100 μl (0.5 × 106 cells) of resuspended cells into one well of a 96-well round-bottom plate as a staining sample. Transfer remaining 100 μl (0.5 × 106 cells) of resuspended cells into one well of another 96-well round-bottom plate as an isotype control.
Add 2.5 μl of PE anti-cardiac Troponin T antibodies to the sample and 2.5 μl of PE-isotype control antibodies to the isotype control (Table 1) and incubate at room temperature for 30 min in the dark.
Add 100 μl of Flow Cytometry Permeabilization/Wash Buffer I to each well and shake the plate gently for 5 min at room temperature in the dark.
Centrifuge the plates at 300 × g for 5 min at 20-25°C and remove the supernatant by decanting or pipetting.
Carefully add 200 μl of Flow Cytometry Permeabilization/Wash Buffer I to each plate without breaking the cell pellets.
Centrifuge the plates at 300 × g for 5 min at 20-25°C and remove the supernatant of each plate by decanting or pipetting.
Repeat Steps G13-G14.
Resuspend the cell pellets with 200 μl of DPBS in each plate and mix well.
Measure the cells in each plate with the flow cytometer (BD FACSVerse) and analyze the data using FlowJo software (Figure 4).
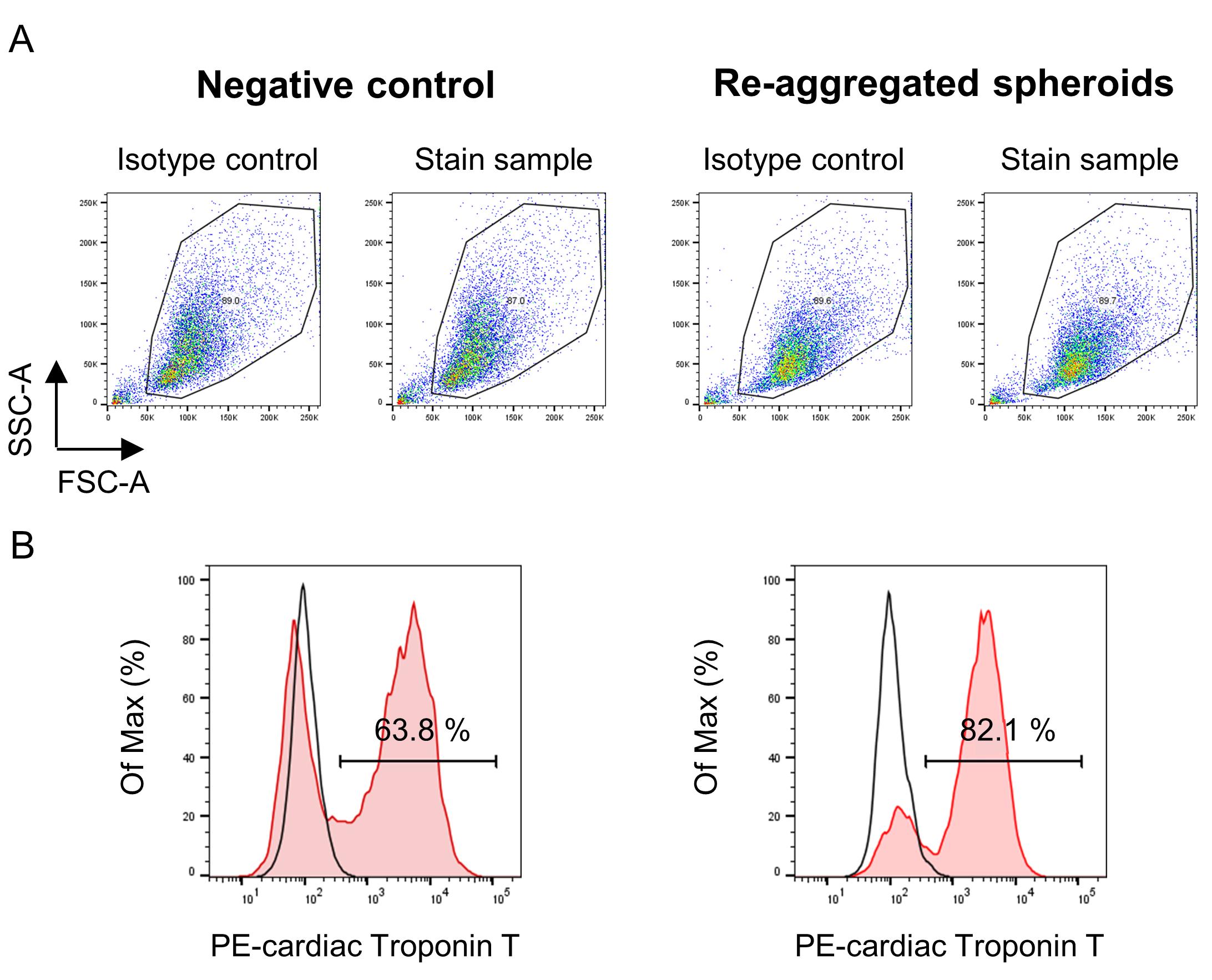
Figure 4. Representative flow cytometry profile of hiPSC-CMs. Events were acquired using the flow cytometer (BD FACS Verse) and analyzed with the FlowJo software. First, the major population was selected by gating in the SSC vs. FSC dot plots (A). The histograms show the Cardiac Troponin T positive hiPSC-CM populations for no re-aggregated negative control (left) and re-aggregated spheroids (right), respectively (B). The plots show isotype control (open histogram) versus stained sample (red histogram). The histograms show that the purity of cardiac Troponin-T hiPSC-MS in the re-aggregated spheroids increased when compared with the negative control.
Data analysis
Measurement of spheroid size using ImageJ software
Figure 5 shows the flow of spheroid size analysis.
Open an image of spheroids with the ImageJ software.
If the distance is shown in pixel, set the known distance using the “Set Scale” command in ImageJ. For example, 450 pixels = 1,000 μm.
Convert the image type to 8-bit.
(Option) Filter the image using “Gaussian Blur (Sigma 2.0)” command if the next step of “bipolarization” does not work.
Bipolarize the image using the “Threshold” command.
(Option) Divide overlapping spheroids using the “Watershed” command.
To measure the surface area for each spheroid, use “Analyze Particles” with parameters “Size: 706.5 (Φ30 μm)” – infinity, “Circularity:” 0.50-1.00, “Show:” Outlines; check “Display results,” “Exclude on edges,” and “Include holes.”
(Option) Overlay the Outlines in the original image to ensure the spheroids are recognized correctly.
Change the color of the Outlines using “Lookup Tables > Green” command.
Invert the color of the Outlines using “Lookup Table > Invert LUT” command.
Overlay the Outlines on the original image using “Image Calculator (Image 1: original image, Image 2: outlines)” command.
Analyze the data set consisting of the surface area of each spheroid with Excel.
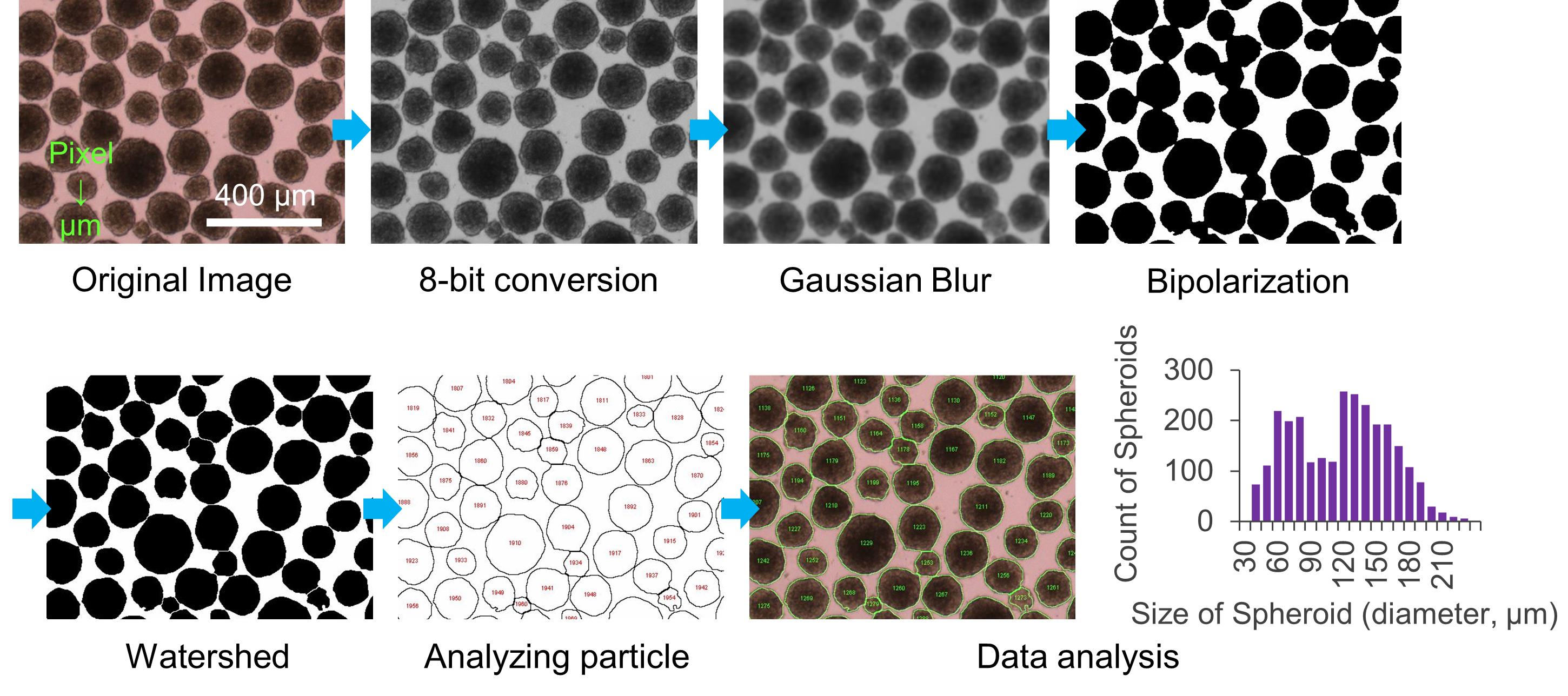
Figure 5. Flow of spheroid size analysis with ImageJ at Data analysis A
Recipes
Cell culture reagents and media preparation
Reagents (Table 2)
Table 2. Reagent preparation
Reagents Preparation method Stock concentrations L-Ascorbic Acid Dissolve L-Ascorbic Acid at 4°C with pre-cooled ultrapure water at 5 mg/ml and mix using a vortex mixer until completely dissolved. Sterilize using a 22 μm syringe filter. Aliquot the solution into 1 ml volumes and freeze at -20°C. Freshly thaw the solution at 4°C or on ice in the dark when preparing the medium. 5 mg/ml Monothioglycerol (MTG) Aliquot MTG to 1 ml volumes and freeze at -20°C. Use thawed MTG within 3 months. To prevent oxidation of MTG, minimize the opening and closing of the cap of the stock tube. On the day of media preparation, dilute 13 μl MTG in 1 ml StemPro-34. 100%
(1.3%(v/v) in use)Transferrin Dissolve Transferrin at the concentration of 30 mg/ml with sterilized ultrapure water. Aliquot the transferrin solution to 1 ml volumes and freeze at -20°C. 30 mg/ml 4 mM Hydrochloric acid (HCl), 0.1% BSA Buffer In a draft chamber, transfer 30 μl of 6.0 N HCl into 50 ml of sterilized ultrapure water. Sterilize using a 22 μm syringe filter. Add 1 ml of 30% BSA into the 30 ml of HCl solution. 4 mM HCl
0.1% BSADPBS, 0.1% BSA Buffer Add 10 μl of 30% BSA into 3 ml of DPBS. 0.1% BSA Cytokines and small molecular compounds (Table 3)
Table 3. Stock preparation of cytokines and small molecular compounds
Growth factors/small molecular compounds Buffers Stock concentrations BMP4 4 mM Hydrochloric acid, 0.1% BSA 10 μg/ml bFGF PBS, 0.1% BSA 10 μg/ml VEGF PBS, 0.1% BSA 5 μg/ml Activin A PBS, 0.1% BSA 10 μg/ml IWP-4 Dimethyl sulfoxide 1.2 mM Dissolve cytokines or small molecular compounds using their appropriate buffers at the stock concentration shown in Table 2. Aliquot each solution if needed and freeze at -20°C.
Modified StemPro-34 (Table 4)
Table 4. Preparation of Modified StemPro-34
Compositions Final concentration Volume StemPro-34 - 48.5 ml Pen/Strep (100×) 1× 500 μl L -glutamine (100×) 1× 500 μl Transferrin (30 mg/ml) 150 μg/ml 250 μl Ascorbic Acid (5 mg/ml) 50 μg/ml 500 μl 1.3% (v/v) MTG 0.0039% (v/v) 150 μl Spheroid formation medium (Table 5)
Table 5. Preparation of Spheroid formation medium
Compositions Final concentration Volume Modified StemPro-34 - 50 ml BMP4 (10 μg/ml) 1 ng/ml 5 μl Y-27632 (10 mM) 10 μM 50 μl Stage-1 medium (2×) (Table 6)
Table 6. Preparation of Stage-1 medium (2×)
Compositions Final concentration Volume Modified StemPro-34 - 50 ml BMP4 (10 μg/ml) 20 ng/ml 100 μl bFGF (10 μg/ml) 10 ng/ml 50 μl Activin A (10 μg/ml) 12 ng/ml 60 μl Stage-2 medium (Table 7)
Table 7. Preparation of Stage-2 medium
Compositions Final concentration Volume Modified StemPro-34 - 50 ml VEGF (5 μg/ml) 10 ng/ml 100 μl IWP-4 (1.2 mM) 2.5 μM 104 μl Stage-3 medium (Table 8)
Table 8. Preparation of Stage-3 medium
Compositions Final concentration Volume Modified StemPro-34 - 50 ml VEGF (5 μg/ml) 10 ng/ml 100 μl bFGF (10 μg/ml) 5 ng/ml 25 μl
Acknowledgments
A part of this work was supported by the “Network Program for Realization of Regenerative Medicine” from the Japan Agency for Medical Research and Development (AMED). This protocol was adapted from a previously published work (Miwa et al., 2020).
Competing interests
There are no conflicts of interest or competing interests.
References
- Bauwens, C. L. and Ungrin, M. D. (2014). Scalable cardiac differentiation of human pluripotent stem cells as microwell-generated, size controlled three-dimensional aggregates. Methods Mol Biol 1181: 15-25.
- Birket, M. J., Ribeiro, M. C., Verkerk, A. O., Ward, D., Leitoguinho, A. R., den Hartogh, S. C., Orlova, V. V., Devalla, H. D., Schwach, V., Bellin, M., et al. (2015). Expansion and patterning of cardiovascular progenitors derived from human pluripotent stem cells. Nat Biotechnol 33(9): 970-979.
- Blinova, K., Stohlman, J., Vicente, J., Chan, D., Johannesen, L., Hortigon-Vinagre, M. P., Zamora, V., Smith, G., Crumb, W. J., Pang, L., et al. (2017). Comprehensive Translational Assessment of Human-Induced Pluripotent Stem Cell Derived Cardiomyocytes for Evaluating Drug-Induced Arrhythmias. Toxicol Sci 155(1): 234-247.
- Chen, M., Qian, C., Bi, L. L., Zhao, F., Zhang, G. Y., Wang, Z. Q., Gan, X. D. and Wang, Y. G. (2015). Enrichment of cardiac differentiation by a large starting number of embryonic stem cells in embryoid bodies is mediated by the Wnt11-JNK pathway. Biotechnol Lett 37(2): 475-481.
- Cohen, E. D., Miller, M. F., Wang, Z., Moon, R. T. and Morrisey, E. E. (2012). Wnt5a and Wnt11 are essential for second heart field progenitor development. Development 139(11): 1931-1940.
- Hwang, Y. S., Chung, B. G., Ortmann, D., Hattori, N., Moeller, H. C. and Khademhosseini, A. (2009). Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11. Proc Natl Acad Sci U S A 106(40): 16978-16983.
- Kattman, S. J., Witty, A. D., Gagliardi, M., Dubois, N. C., Niapour, M., Hotta, A., Ellis, J. and Keller, G. (2011). Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8(2): 228-240.
- Kempf, H., Olmer, R., Kropp, C., Ruckert, M., Jara-Avaca, M., Robles-Diaz, D., Franke, A., Elliott, D. A., Wojciechowski, D., Fischer, M., et al. (2014). Controlling expansion and cardiomyogenic differentiation of human pluripotent stem cells in scalable suspension culture. Stem Cell Reports 3(6): 1132-1146.
- Loh, K. M., Chen, A., Koh, P. W., Deng, T. Z., Sinha, R., Tsai, J. M., Barkal, A. A., Shen, K. Y., Jain, R., Morganti, R. M., et al. (2016). Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell 166(2): 451-467.
- Pal, R., Mamidi, M. K., Das, A. K. and Bhonde, R. (2013). Comparative analysis of cardiomyocyte differentiation from human embryonic stem cells under 3-D and 2-D culture conditions. J Biosci Bioeng 115(2): 200-206.
- Panakova, D., Werdich, A. A. and Macrae, C. A. (2010). Wnt11 patterns a myocardial electrical gradient through regulation of the L-type Ca2+ channel. Nature 466(7308): 874-878.
- da Rocha, A. M., Campbell, K., Mironov, S., Jiang, J., Mundada, L., Guerrero-Serna, G., Jalife, J. and Herron, T. J. (2017). hiPSC-CM Monolayer Maturation State Determines Drug Responsiveness in High Throughput Pro-Arrhythmia Screen. Sci Rep 7(1): 13834.
- Sato, H., Idiris, A., Miwa, T. and Kumagai, H. (2016). Microfabric Vessels for Embryoid Body Formation and Rapid Differentiation of Pluripotent Stem Cells. Sci Rep 6: 31063.
- Shiba, Y., Gomibuchi, T., Seto, T., Wada, Y., Ichimura, H., Tanaka, Y., Ogasawara, T., Okada, K., Shiba, N., Sakamoto, K., et al. (2016). Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 538(7625): 388-391.
- WHO. (2018). World health statistics 2018: monitoring health for the SDGs, sustainable development goals.
- Yamazaki, D., Kitaguchi, T., Ishimura, M., Taniguchi, T., Yamanishi, A., Saji, D., Takahashi, E., Oguchi, M., Moriyama, Y., Maeda, S., et al. (2018). Proarrhythmia risk prediction using human induced pluripotent stem cell-derived cardiomyocytes. J Pharmacol Sci 136(4): 249-256.
- Yang, L., Soonpaa, M. H., Adler, E. D., Roepke, T. K., Kattman, S. J., Kennedy, M., Henckaerts, E., Bonham, K., Abbott, G. W., Linden, R. M., et al. (2008). Human cardiovascular progenitor cells develop from a KDR+ embryonicstem-cell-derived population. Nature 453(7194): 524-8.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Miwa, T., Idiris, A. and Kumagai, H. (2021). High-throughput 3D Spheroid Formation and Effective Cardiomyocyte Differentiation from Human iPS Cells Using the Microfabric Vessels EZSPHERETM. Bio-protocol 11(21): e4203. DOI: 10.21769/BioProtoc.4203.
Category
Stem Cell > Pluripotent stem cell > Cell differentiation
Cell Biology > Cell isolation and culture > Cell differentiation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









