- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
An Inexpensive Imaging Platform to Record and Quantitate Bacterial Swarming
Published: Vol 11, Iss 18, Sep 20, 2021 DOI: 10.21769/BioProtoc.4162 Views: 3636
Reviewed by: Alba BlesaTimo A LehtiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Development and Quantitation of Pseudomonas aeruginosa Biofilms after in vitro Cultivation in Flow-reactors
Yingdan Zhang [...] Haihua Liang
Aug 20, 2021 3681 Views

Co-culture Wood Block Decay Test with Bacteria and Wood Rotting Fungi to Analyse Synergism/Antagonism during Wood Degradation
Julia Embacher [...] Martin Kirchmair
Oct 5, 2023 1861 Views

Optimized Protocol for the Collection, Cryopreservation, and In Vitro Cultivation of Human Gut Microbiota for Toxicomicrobiomics Applications
Paulina Średnicka [...] Michał Wójcicki
Nov 5, 2025 1942 Views
Abstract
Bacterial swarming refers to a rapid spread, with coordinated motion, of flagellated bacteria on a semi-solid surface (Harshey, 2003). There has been extensive study on this particular mode of motility because of its interesting biological and physical relevance, e.g., enhanced antibiotic resistance (Kearns, 2010) and turbulent collective motion (Steager et al., 2008). Commercial equipment for the live recording of swarm expansion can easily cost tens of thousands of dollars (Morales-Soto et al., 2015); yet, often the conditions are not accurately controlled, resulting in poor robustness and a lack of reproducibility. Here, we describe a reliable design and operations protocol to perform reproducible bacterial swarming assays using time-lapse photography. This protocol consists of three main steps: 1) building a “homemade,” environment-controlled photographing incubator; 2) performing a bacterial swarming assay; and 3) calculating the swarming rate from serial photos taken over time. An efficient way of calculating the bacterial swarming rate is crucial in performing swarming phenotype-related studies, e.g., screening swarming-deficient isogenic mutant strains. The incubator is economical, easy to operate, and has a wide range of applications. In fact, this system can be applied to many slowly evolving processes, such as biofilm formation and fungal growth, which need to be monitored by camera under a controlled temperature and ambient humidity.
Keywords: Bacterial swarmingBackground
Bacteria show different motility phenotypes, such as swimming, swarming, gliding, sliding, and twitching (Kearns, 2010). Coordinated multicellular migration across a moist surface, known as bacterial swarming, is an important motility phenotype that has been studied for decades due to its relevance to pathogenicity, virulence, and antibiotic resistance (Kearns, 2010). A systematic study of colony expansion speed and morphological patterns coupled to swarming may provide insight into the correlation between bacterial motility and host health. Here, we build an environment-controlled incubator to perform a swarming assay. Inside the incubator, a digital camera is mounted on the top to take time-lapse images of the swarming activity. After the recording, images are transferred to a laptop for quantitation of the swarm expansion using the ImageJ software. The system was tested by recording over 1,000 swarming events to assure stability and reproducibility.
At first glance, one may consider a swarming assay rather simple: just inoculate bacteria on agar filled in a Petri dish, and then take an image of the plate after incubating for a certain time. However, it can be challenging to perform swarming assays with reproducible results and record clear images for further quantitative analysis. Here, we list a few technical challenges that one may encounter and provide corresponding solutions in our protocol:
A typical swarming event may take 10-20 h for the bacteria to cover a standard 9-cm Petri dish. Besides the coverage time, sometimes we need to know how long the lag phase lasts, at what time branches form at the colony edge; and at the microscopic level, when cell elongation occurs. Thus, the researcher needs to check the plate regularly, aided by microscopy. Suppose one starts the assay during the daytime; one may need to scan the plates every 10-30 min overnight. In our protocol, time-lapse photography helps to capture images so that the researcher does not have to stay up late or check every 10-30 min in order not to miss key events.
Bacterial swarming is highly sensitive to the environment. Fluctuations in ambient temperature and humidity often cause large differences in the swarming rate and colony pattern; therefore, a stable humidity- and temperature-controlled environment is critical for the assay. In our system, we utilize a thermo-insulating tent, a humidity control unit, and a temperature control unit to minimize the environmental fluctuation during the assay. One can readily set different humidities and temperatures for a swarming strain screen.
Since an agar gel typically contains over 97% water by mass, condensation readily forms on the plate lid, which obscures image taking. We designed the incubator in such a way that, when the plates are placed inverted on the platform, the temperature of the lid is slightly higher than that of the agar, which prevents condensation. Note: the agar surface architecture can vary depending on pouring methods, thickness, and percentage of agar used; these need to be individually optimized for the particular bacteria.
Taking a clear photo of the swarming plates is tricky. A swarming plate has three surfaces that affect the optical quality while imaging: the plate lid, the agar surface, and the plate bottom. When the camera flashlight or auxiliary front light is used, some light is reflected by the lid and the bottom of the plate to the camera, forming undesirable light spots on the photos. In our design, for one-plate photo shooting, we use a circular fluorescent tube as the backlight under an adjustable light shield. For multiple-plate (up to 9) assays, an LED light strip is used for sidelight illumination. Image quality is better when using the light shield because of the light field setting; however, the efficiency is higher when using a LED light since it illuminates a larger area that can fit 9 Petri dishes at a time.
Quantitation of the swarming rate takes a lot of effort; researchers generally scan the plates and measure the radius of the swarming colony using a ruler. For an irregular-shaped colony, they usually make a rough estimation of the “effective radius.” When multiple plates are involved in the assay, one should plot the swarming area vs. time instead of just calculating the average swarming rate. A lot of work is required for numerous manual measurements. In our case, we import the digital swarming images to the ImageJ software and calculate the swarming area by outlining the swarm periphery. This computerized process improves the efficiency and reduces the error.
Compared with conventional practice in this field, our protocol has distinct advantages:
(1) Affordability. In 2015, a research team headed by Dr. Joshua Shrout at the University of Notre Dame described a protocol for the preparation, imaging, and quantitation of swarming (Morales-Soto et al., 2015). In their protocol, they used the commercial equipment “Bruker in vivo imaging station.” This product is no longer available from the company, and a used one costs over $58,000. Most microbiology labs studying bacterial swarming may not need many of the complex functions that this particular equipment provides, such as x-ray imaging and in vivo fluorescence imaging. Those labs may not be able to afford or be willing to invest so much in an overqualified bacterial swarming chamber. In contrast, the total cost of our system is around $1,000, with everything included.
(2) Accuracy. For the Bruker imaging station, placing a plate of water next to each swarming plate is suggested in order to maintain humidity. In this way, however, it is hard to control the humidity within a specific range. In our design, the chamber humidity is well controlled; one can set the parameters before the assay starts, and the environment is controlled dynamically using a digital sensor and feedback controllers.
(3) Efficiency. In 2018, an independent researcher, Peñil Cobo, developed a time-lapse imaging chamber for bacterial colony morphology observation (Peñil Cobo et al., 2018), which allows for photography of one Petri dish at a time. Since the temperature is not controlled in that design, the whole system must be placed in a warm room. In our case, multiple 9-cm Petri dishes can fit inside the incubator, providing convenience and efficient use of space and energy. Having a larger box in our design, necessary to hold multiple plates, alters the optical path in a subtle way, but the geometry of our incubator and light field for photography are well calibrated to ensure the best lighting.
This protocol is divided into three steps: (1) assembly of an incubator; (2) the swarming assay; and (3) data processing. By following the procedure as detailed below, we guarantee that one can obtain a homemade bacterial plate incubator with stable swarming results and high-quality images. The overall cost depends on which camera is used in the system, but it should be within $1,000 dollars. In practice, any digital camera that has a “Manual Mode” is adequate for the purpose. A swarming assay requires researchers to be meticulous with certain details to ensure reproducibility. For instance, bacterial swarming is very sensitive to small changes in conditions such as the roughness and moisture level of the agar surface. Here, we attempt to elaborate on procedures and notes in as much detail as possible for users to follow. In the photo shooting part, we show in detail how to tune the camera settings. Finally, we explain how to process the data using ImageJ to measure the swarming area.
Materials and Reagents
To build the photographing incubator
Hydroponic Indoor Garden Grow Tent (24 in. × 24 in. × 48 in., Yaheetech, model: YT-2801)
Photography unit
Digital camera (Panasonic, model: DMC-FZ50)
LCD Timer Remote Control (JJC, model: TMD)
AAA battery (2 pcs, Duracell)
Zinc-plated slotted angle (4 pcs, 1.5 in. × 14 Gauge × 36 in., Crown Bolt)
Zinc-plated slotted angle (10 pcs, 1.5 in. × 14 Gauge × 18 in., Crown Bolt)
Aluminum flat bar (0.75 in. × 36 in. × 0.125 in., Everbilt)
Black polyester cloth (20 in. × 20 in., Dazian)
Bolts and nuts (40 pairs, M5, Crown Bolt)
Black acrylic sheet (2 pcs, 18 in. × 18 in. × 0.125 in., National Security Mirror)
LED light strip (3 meters, White, GuoTonG)
Power strip (6-outlet, Belkin)
Backlight shield
Black acrylic sheet (3 pcs, 12 in. × 12 in. × 0.118 in., National Security Mirror)
Fluorescent circline ceiling light (Sunlite, model: FC12T9/CW) with starter ballast
Zinc-threaded rod (4 pcs, 0.25 in. × 12 in., Everbilt)
Hex-plated nuts (24 pcs, 0.25 in., Everbilt)
Temperature and humidity control unit
Heated control module (Coy Lab, serial: DC1807)
Fan (AC Infinity, model: LS1225A-X)
Digital humidity controller outlet (Inkbird, model: IHC200S)
Reptile humidifier (2 L, Evergreen)
Beaker (500 ml)
Note: Other brands of humidifiers or controllers can be alternatives if their combination can well control the humidity inside the chamber.
For swarming plate preparation
Enterobacter sp. SM1, SM3, SM3_18, SM3_24 (species of bacteria for the swarming assay)
LB broth (see Recipes)
0.5% LB agar plates (see Recipes)
Equipment
Labconco Purifier Class II Biosafety Cabinet (Delta Series)
Falcon 14-ml Polystyrene Round-Bottom Tubes (17 mm × 100 mm, Corning)
Sterilized wooden sticks (2.5 in.)
Pipet-aid (Drummond, 1-100 ml) with appropriate pipette
New Brunswick Innova 4300 Incubator Shaker
Weighing paper (4 in. × 4 in., Fisher Scientific, catalog number: 09-898-12B)
Laboratory spatula (4 pcs, 6.5 in, stainless steel, Home Science Tools)
Pyrex glass bottles (250 ml, Corning, model: 1395)
Pyrex graduated cylinder (500 ml, Corning, model: 3022)
Hot plate magnetic stirrer (Barnstead International, model: SP46925)
Thermal insulating gloves
Autoclave (AMSCO Scientific, model: SV-120) with autoclavable tray
Petri dishes (100 mm × 15 mm, sterile, polystyrene, Fisher Scientific, catalog number: FB0875712)
Micropipette (0.5-10 μl, Eppendorf, catalog number: 3123000020)
Acrylic cutter (Fletcher)
Circle cutter (Bott)
Hand drill/table drill with appropriate drill bit set
Hacksaw
Software
ImageJ (ver1.59g) (https://imagej.nih.gov/ij/)
Procedure
Part I: Set up the photographing incubator
Assemble the camera stand
Set up the Yaheetech hydroponic tent according to the instruction manual. The manual comes with the product package. Assemble the skeleton first and then cover it with the polyester material.
Use M5 bolts and nuts to assemble the camera frame, as illustrated in Figure 1. Connect the Zinc-plated slotted angles first and then the aluminum bars. The slotted angles have holes by the side, but the aluminum bars do not. Drill holes by the ends of the aluminum bars using a hand or table drill. Since the aluminum bars are used to stabilize the structure, their positions do not have to be precise.
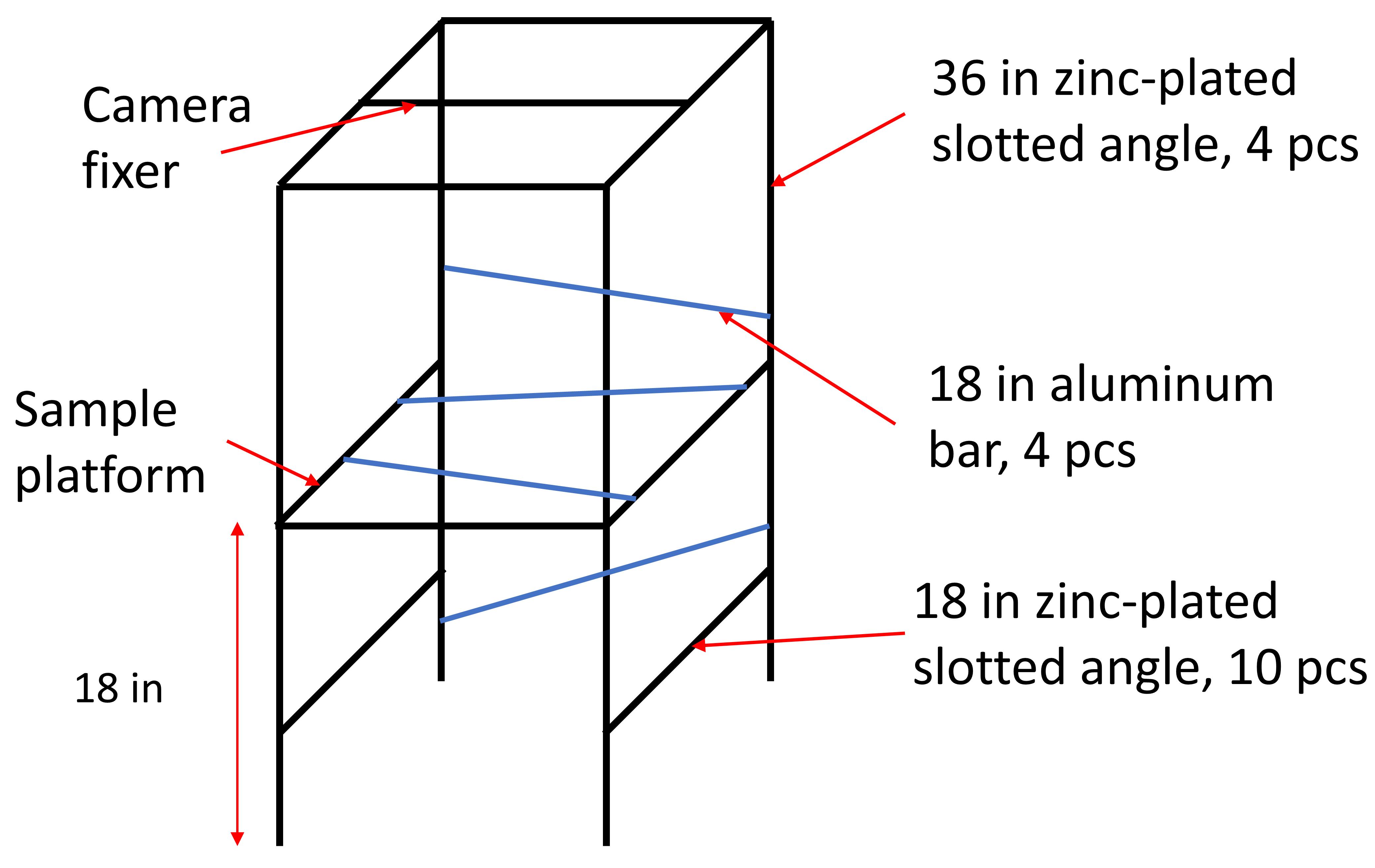
Figure 1. Schematic showing the structure of the camera frame. Four pieces of 36-in. zinc-plated slotted angle stand perpendicular to the ground and are connected by 10 pieces of 18-in. zinc-plated slotted angle using M5 bolts and nuts. The aluminum bars are to stabilize the whole structure with no specific position requirements. When using the light shield, the distance between the ground and the sample platform is 18 in. When using LED light illumination, the sample platform can be elevated by several inches for photography.Use an acrylic cutter to cut the sides of 18 in. × 18 in. acrylic sheets slightly to fit in the sample platform.
Use a circular cutter to cut a circle of 9 inches in diameter out of one of the 18 in. × 18 in. acrylic sheets “A” from the center; leave the other sheet “B” uncut.
Fix sheet “A” on the sample platform. Drill the appropriate holes on the side to allow the bolts to go through all the way down to the holes of the slotted angles.
Fix the camera on the camera fixer, with its lens facing downward. Adjust the fixer back and forth to align the camera with the circle on the acrylic sheet.
Tape the LED strip around the sample platform 1 inch above the acrylic sheet on the slotted angle. The position of the light cannot be too high because we want to avoid reflections of the light from the plates.
Cut two pieces of black cloth, around 20 in. × 20 in. Place one of them on the bottom of the tent as a black photography background. For the other piece, cut a hole in the center to allow the camera lens through, and hang the cloth onto the camera platform to shield the reflection from the top.
Place the camera stand inside the tent.
Load the batteries into the LCD timer remote control and connect it to the camera through the hole on the top of the tent. Tighten the hole using the elastic cords around it.
Make the light shield
Cut circles of 3.62 in., 5.75 in., and 9.50 in. diameter out of three pieces of acrylic sheets. Drill 0.25-in. holes in each of the corners, 1.2 in. from both edges. Assemble the light shield according to Figure 2. Place the circular fluorescent light bulb between level I and the sample platform. Stick the starter ballast under the sample platform using tape.
Pause point: There are two modes of illumination: one uses a backlight, and the other uses a side light (Figure 3). When taking close-up images of a one-plate event, it is preferable to use the light shield with its own light source below a light sheet. To take photos, place the light shield on the sample platform and align it with the camera. Turn off the LED light when using the light shield. On the other hand, when taking photos of more than one plate, the light shield is removed, and the uncut 18 in. × 18 in. acrylic sheet is placed on top of the circularly cut sample platform. As many as 9 agar plates can be placed on the uncut sheet for imaging. To take photos, turn on the LED light as the side light source.
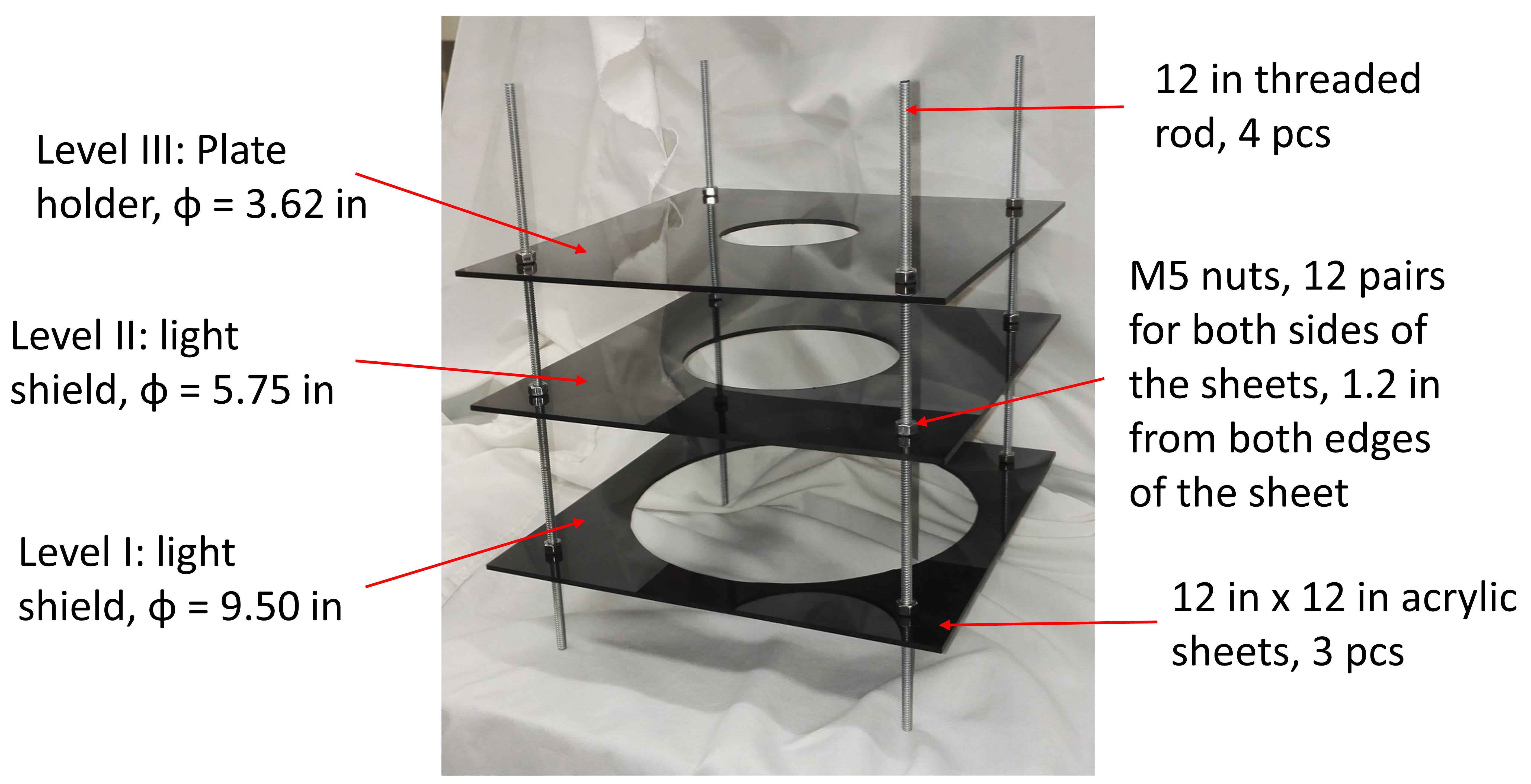
Figure 2. Image showing a light shield assembly for single-plate imaging. Three pieces of 12 in. × 12 in. a hollow acrylic sheet are connected by four pieces of 12 in. zinc-threaded rod and fixed using hex-plated nuts. The distance between the Level I sheet and the ground is about 1.5 in., leaving just enough space to place the circular fluorescent bulb to fit in. The heights of Level II and Level III can be adjusted to optimize illumination.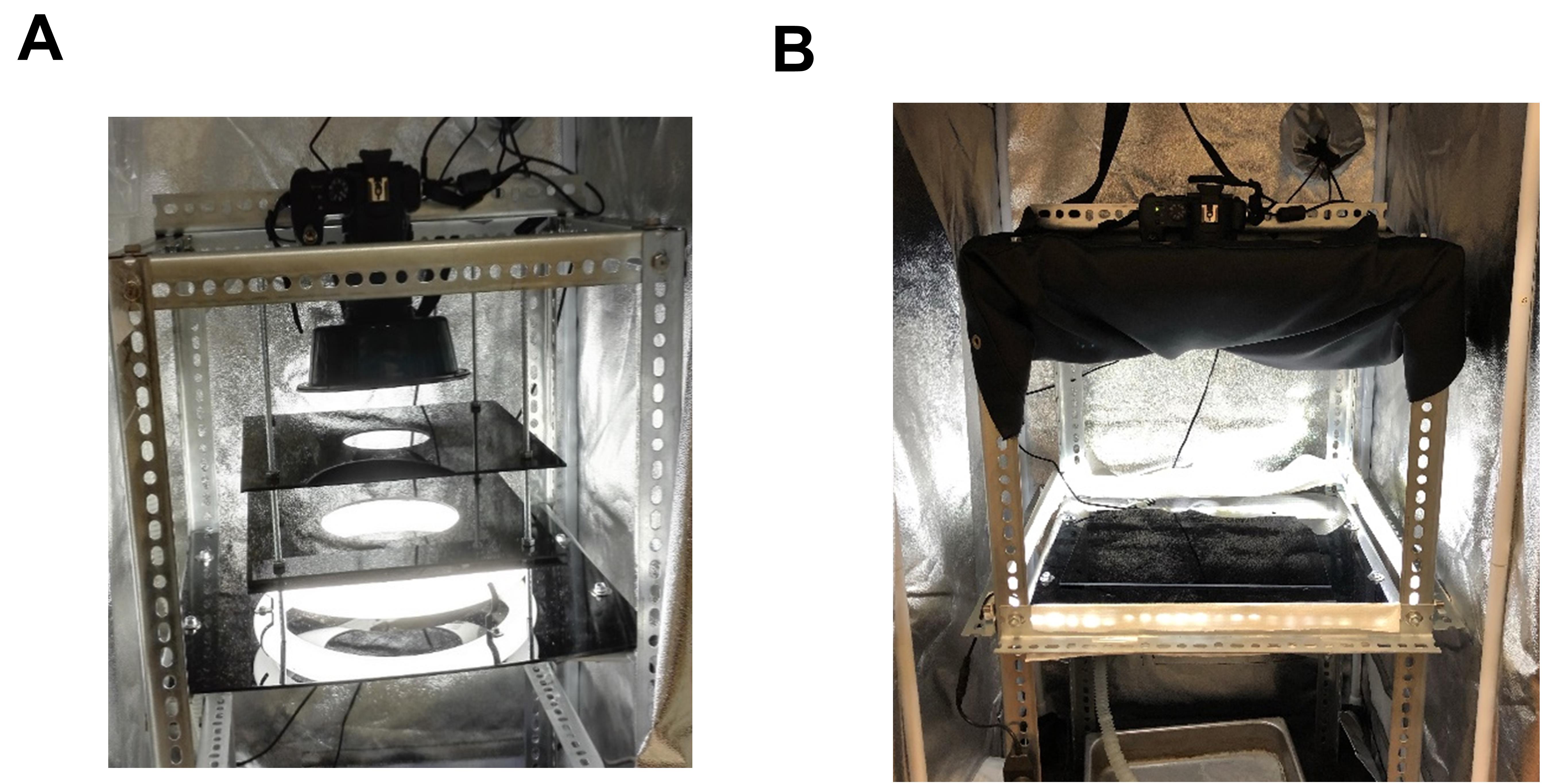
Figure 3. Taking photos using the light shield or LED light strip. (A) Taking photos using the light shield assembly. The circular fluorescent bulb is placed between Level I and the sample platform (noted in Figure 2). The distance between Level II and Level I is about 0.8 in., while the distance between Level II and Level III is 3.5 in. (B) Taking photos using the LED light strip. The LED light is fixed 1 inch above the sample platform on the horizontal zinc-plated slotted angles. If the light is too strong, one can insert a white paper belt to diffuse and reflect the light. The black cloth near the camera is necessary to block reflections from the plates.
Install the temperature and humidity control system
Place the heat control module inside the tent, on the side at the bottom, such that it will not show up in the swarming photos when using the light shield. Adjust the temperature setting to the desired temperature for the swarming assay.
Set the humidifier outside the tent and connect the power cord to the humidity controller outlet. Extend the extractable plastic mist tube through the hole on the tent wall into the tent beneath the sample platform.
Note: Check the camera preview and make sure that the tube and the mist do not show up in the image.
Fill the humidifier tank with water. Plug the humidity controller into the power strip, and adjust the humidity value with a tolerance range according to the controller manual. As an example, for Enterobacter sp. SM3 (see the corresponding main article), set the humidity to 40% ± 5% tolerance (tol).
Place a 500-ml beaker under the mist tube to collect water droplets.
Fix the AC fan on one of the slotted angle legs facing the beaker using the bolts and nuts that come with the fan. The fan is used not only to blow the mist from the humidifier to prevent fog from showing up in the photos but also to improve ventilation and uniformity of the temperature and humidity in the chamber.
Tighten all the holes and seal the zip of the tent.
Part II: Perform the swarming assay
Take the bacteria glycerol stock out of the -80°C freezer. Use a piece of sterilized wooden stick to scratch the bacteria-containing ice surface and then dip the stick into the LB broth. Place the glycerol stock back into the freezer and shake the inoculated LB broth overnight (~16 h) in a shaker. The temperature is set to 37°C and the shaking frequency to 200 revolutions per min (rpm). Start the overnight growth around 5 p.m. so that it will be ready for use around 9 a.m. the next morning.
Use a micropipette to inoculate 2 μl overnight bacterial suspension on the center of each agar plate. Transfer the plates to the incubator after the inoculation drop has been absorbed by the agar (the spread of the circular drop has become nearly invisible).
Part III: Time-lapse photo taking and swarming rate quantitation
To use the light shield, place one swarming plate inverted on the plate holder. Turn on the fluorescent light bulb and the camera. The LED light strip should stay off.
In the preview of the camera, you should see the plate sitting in the center of the screen. Otherwise, move the light shield around to align the camera with the sample.
Rotate the nuts on the threaded rod to adjust the position of the acrylic sheets. The distance between the sample platform and Level I is about 1.5 inches. The distance between Level I and Level II is about 0.8 inches, while Level II and Level III are separated by 3.5 inches. If you see a round light spot on the Petri dish, lift Level III slightly. If the Petri dish is too dark, lower Level III slightly. If you cannot get a good image by adjusting Level III, then adjust Level II slightly up or down (see Note 3).
Set the camera focal length to 35-65 mm. Adjust the zoom ring so that the sample occupies the full screen but does not exceed the border. Use “M,” the manual mode, to focus on the bacterial colonies. Set the aperture to F5.6-F7.1 and adjust the shutter speed until the resulting exposure value is 0 or -, because overexposed images will result in loss of detail in the images (Figure 4A).
For a multi-plate assay, remove the light shield, turn on the LED light strip, and place the uncut acrylic sheet on the sample platform. Place the swarming plates inverted on the acrylic sheet so that water will not form condensate on the lid. Check the camera preview to make sure that all the plates are within the range of the screen (Figure 4B).
Set the frame rate and frame number of the LCD camera timer control according to the manual. For example, in the case of Enterobacter sp. SM3, we set the frame number to 50 and the time interval between frames to 15 min. Press the “start” button to start the time-lapse photo shooting.
Zip the tent to form a good seal. The swarming assay may take about 10 h. You can leave the camera on overnight and collect the images the next morning. During the photo taking process, DO NOT shake the incubator; otherwise, the optical setup may be disturbed.
Stop the timer controller when the swarming is finished. Download the images by connecting the camera to a laptop using a USB data wire or by pulling out the memory card of the camera and placing it into a computer to download the images.
Check the images taken on your computer. If the brightness of the images varies, you can use the “Stack Deflicker” plugin in ImageJ to calibrate the brightness. If the sample position in the images varies over time, you can apply the “Image Stabilizer” plugin to fix it.
Data analysis
Representative one-plate and multi-plate swarming images are shown in Figure 4A and 4B, respectively. The time course for plates shown in Figure 4B was rendered to a video in Video 1.
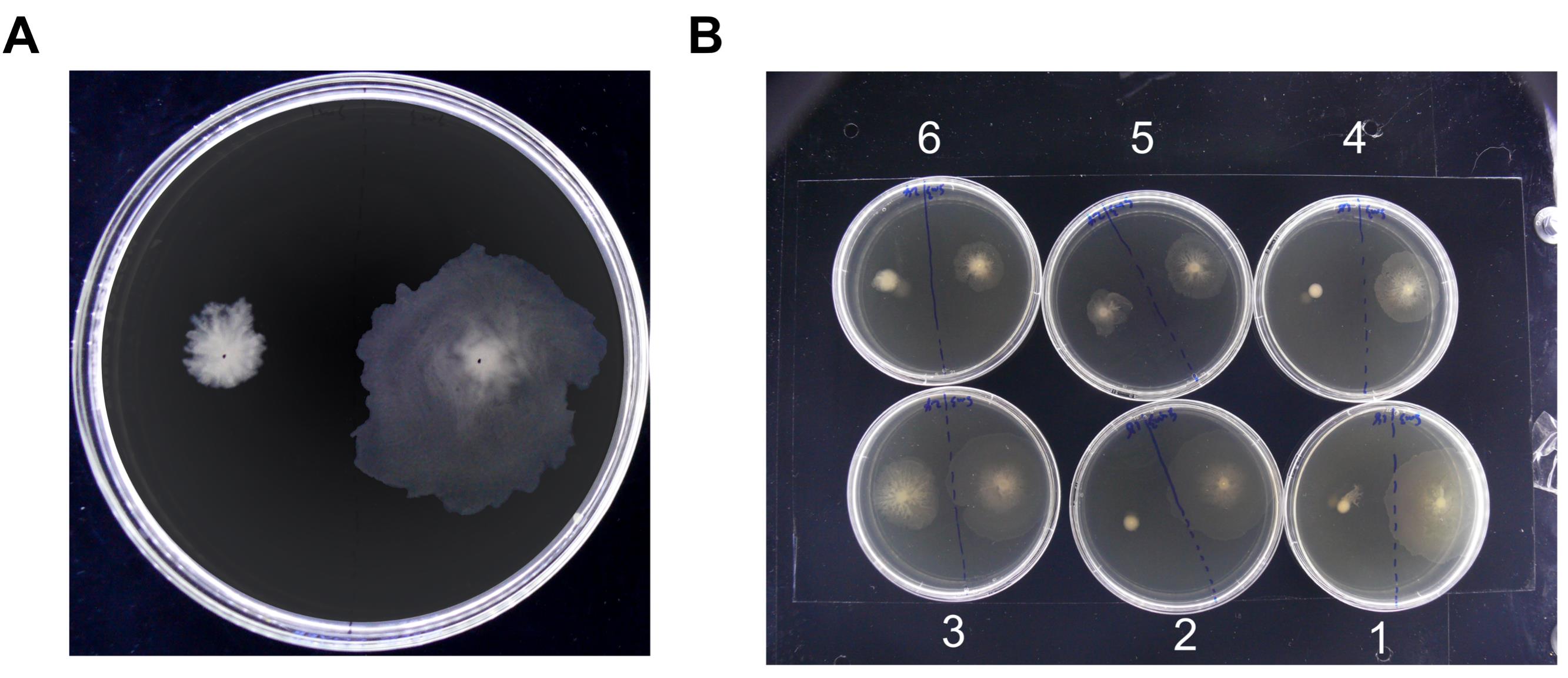
Figure 4. Swarming images taken in the incubator. (A) Plates inoculated with Enterobacter sp. SM1 (left) and SM3 (right) were incubated for 6.5 h. This image was taken using the circular fluorescent bulb and the light shield assembly. (B) Swarms of SM3 vs. a swarm-deficient mutant of SM3, SM3_18 (plates 1, 2, 4), and SM3 vs. a swarm-deficient mutant of SM3, SM3_24 (plates 3, 5, 6). Each comparison was performed in triplicate, with SM3 inoculated on the right half of each plate. The image was taken after a 6.5-h incubation. A side LED light strip was used for illumination, with all six plates imaged together.
To calculate the swarming area, open the image of interest in ImageJ. Click “Analyze” -- “Set Scale” to calibrate the ratio of pixels to the actual length. Then use the “freehand selections” tool to outline the colony edge and press “M” to calculate the area of the selected colony image by image (Figure 5). The swarming rates for Enterobacter sp. SM1 and SM3 are plotted in Figure 6 following calculation of the swarm areas.
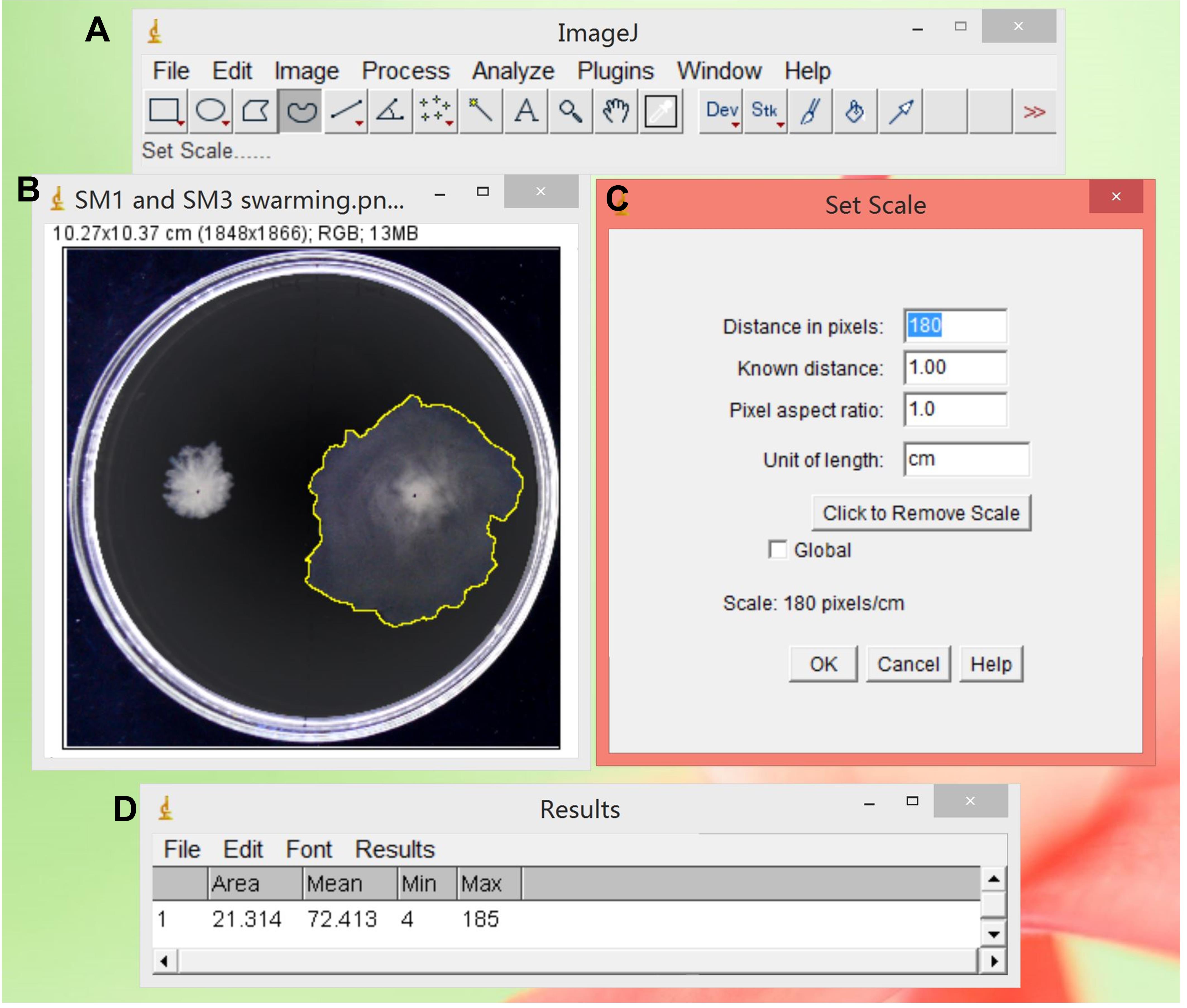
Figure 5. Calculation of the swarm area using ImageJ. (A) ImageJ user interface. The “freehand selections” tool is selected. (B) Enterobacter sp. SM3 swarming colony (on the right) is outlined by the “freehand selections” tool. (C) The scale in pixels is set to the value of the real length. (D) By pressing “M,” the area of the swarm is calculated.
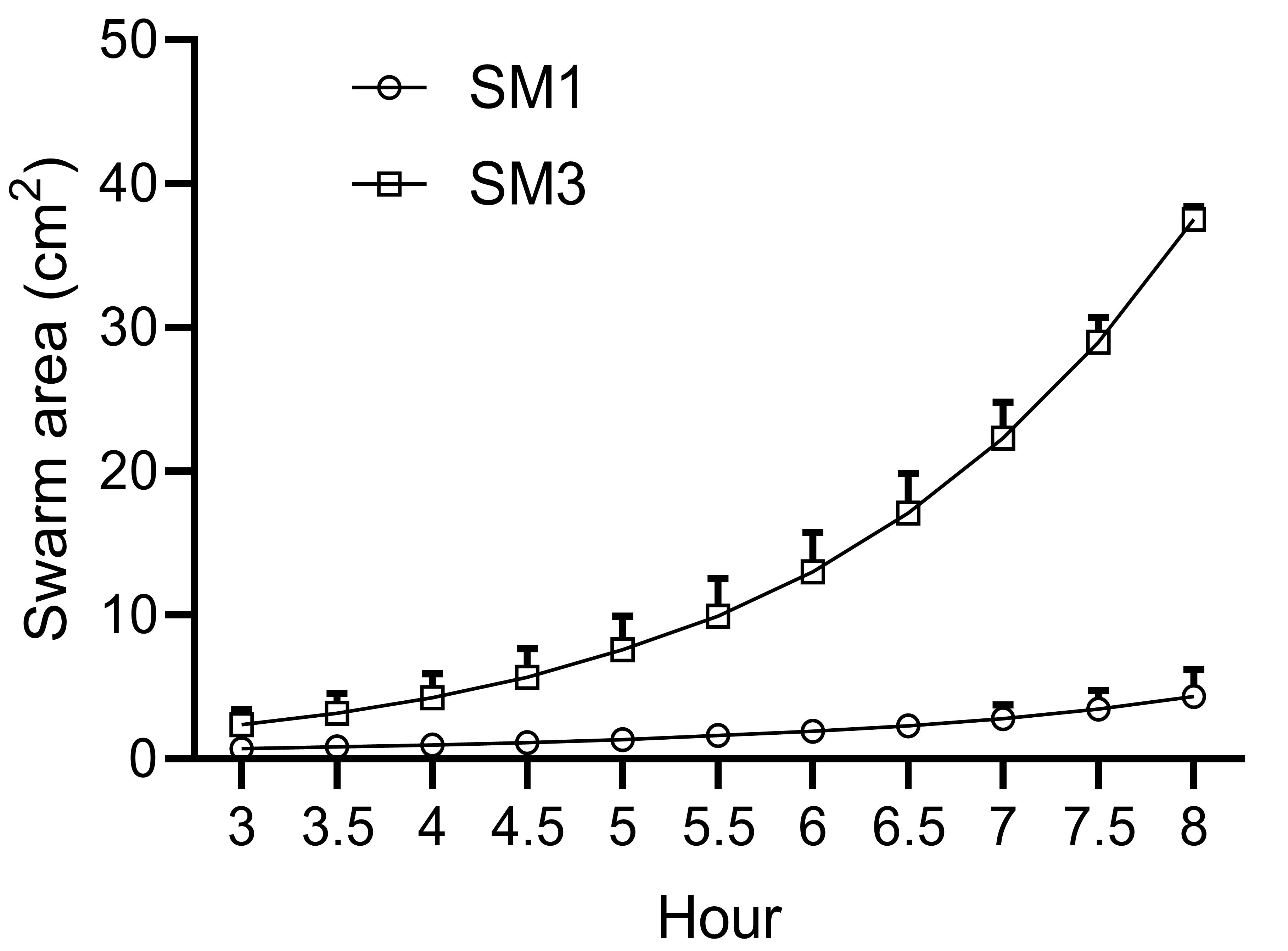
Figure 6. Quantitation of Enterobacter sp. SM1 and SM3 swarming motility. SM1 and SM3 were inoculated using 2 μl overnight bacterial culture on 0.5% LB agar plates and incubated in the swarming incubator (repeated a total of 10 times). Time-lapse images were taken every half an hour, and the swarm area was measured for each image. Data are represented as the mean, with error bars indicating the 95% confidence interval. When not visible, the errors are smaller than the size of the symbols.
Notes
The humidity and temperature inside the incubator should reach the set values within 10 min after the power is turned on and remain stable for the duration of the overnight recording.
Sometimes, certain models or specifications of products may be out of stock. If 18-in. slotted angles are not readily available, for instance, one can cut from longer angles using a hacksaw. Cutting a large circular hole out of a piece of acrylic requires some skill. To ensure safety, when using a hole saw or circle cutter, always clamp the acrylic sheet firmly on a workbench first. One alternative is to go to an engineering workshop or a carpenter’s shop to have these cuts done. They have more advanced tools such as saber saws or jigsaws to cut circular holes.
Calibration of the light shield needs practice. There is a subtle distance relationship between each acrylic sheet to achieve the best image quality, depending on the camera setting. We want the light to shine through the transparent agar and be reflected by the swarming colony. Once you find the right position, tighten all the nuts so that the position is locked for subsequent imaging.
If a swarming strain does not swarm but only forms a dense spot, the plate may be too dry; therefore, less medium may be readily available for swarm expansion. Always use fresh plates within two days from pouring them; beyond two days, the agar plate will become significantly drier and may not sustain swarming motility. The drying time in the hood should not be too long. When the lab humidity is below 30%, 10 min of drying is enough. Also, pour the plates when the agar solution is not too cold; otherwise, the agar poured on the plate may have a rough surface since solidification has already taken place in the bottle, which shows up on the plate in small clusters. Finally, double-check the tryptone or yeast. Occasionally, for certain Lot numbers, some chemicals do not dissolve thoroughly, which may change the texture of the agar surface, leading to suboptimal results.
Sometimes, non-swarming strains also appear to “swarm,” and in exceptional cases, even faster than the swarming strains. In this case, the agar concentration may be too low or the drying time too short, so the cells are swimming as opposed to swarming on the plates. Different swarming species have different agar concentration tolerance. For SM strains, 0.5% agar is the preferred concentration to distinguish swarmers from non-swarmers, whereas for B. subtilis 3610, 0.7% has proven to be the optimal agar concentration. To fix the problem of false positives, one can try to increase the drying time or raise the agar concentration slightly.
Condensation may occur on the plate lid if a different tent is used and the temperature of the lid is lower than that of the bottom. In this case, try to flip over the plates and the problem should be resolved. After flipping over the plates, adjust the camera lens accordingly to focus on the bacterial swarm.
SM strains are newly isolated bacterial strains. To request these strains under a Material Transfer Agreement, please refer to the linked main article (https://doi.org/10.1053/j.gastro.2021.03.017).
Recipes
LB broth, solution
10 g/L tryptone
5 g/L yeast extract
5 g/L NaCl
Note: The above medium recipe is designed for SM bacteria and is subject to change according to the necessities of other microbes one works with.
0.5% LB agar plates (swarm plates)
10 g/L tryptone
5 g/L yeast extract
5 g/L NaCl
0.5% agar
Weigh 0.5 g yeast extract, 1.0 g tryptone, 0.5 g NaCl, and 0.5 g agar. Place these into a 100-ml Pyrex bottle. Measure 100 ml deionized water and pour it into the bottle.
Put a magnetic stir bar of appropriate size into the bottle, loosely close the lid, and place the bottle on a hot plate magnetic stirrer. Turn on the heat and the magnetic stirrer. It takes roughly 10–15 min for the medium to boil.
After the medium starts to boil, turn off the stirrer and the heat. Wear thermal-insulated gloves to transfer the bottle to an autoclavable tray containing a thin layer of water.
Loosen the bottle lid and autoclave for 25 min under 15 psi at 121°C.
Once the autoclave has finished, place the bottle of medium back on the magnetic stirrer, with the heating function off but the stirring function on. During this step, we want the medium to cool to 40-50°C; the constant stirring is to maintain uniformity in temperature.
Once the target temperature is reached, use a pipet aid to transfer LB agar medium to 6 Petri dishes, with 15 ml in each. Perform this operation inside a laminar flow hood or near a flame to minimize the chance of contamination from the air.
Note: 15-20 ml medium is ideal for one Petri dish. If the volume is too low, the agar plate will become too dry during the incubation; if the volume is too large, the image quality will be compromised due to poor transparency.
When the agar has solidified, remove the lid of the swarming plates and dry in the hood.
Note: The plates must be dried in the hood in order to remove excess moisture from the agar surface. If there is water on the agar surface, the bacteria may be swimming rather than swarming. If the room humidity is above 50%, dry the plates for 20 min; if the room humidity is below 30%, the drying time is about 10 min; and if the humidity is 30-50%, adjust the drying time accordingly to around 15 min. Do not over-dry the plates; otherwise, the bacteria may not be able to swarm on the agar. Alternatively, use pre-stored swarming plates, which may be stored inverted in a 4°C cold room for up to 2 days; parafilm or plastic bags are not required. Using pre-stored plates, the drying time should be reduced by ~5 min.
Acknowledgments
The instrument and studies presented here were built and conducted using funds at the Albert Einstein College of Medicine, INC and supported by the Broad Medical Research Program at CCFA (Crohn’s & Colitis Foundation of America; Grant# 362520) (to S.M.); NIH R01 CA127231; CA 161879; 1R01ES030197-01; and Department of Defense Partnering PI (W81XWH-17-1-0479; PR160167) (S.M.).
Competing interests
The authors declare that they have no conflicting financial interests.
References
- Harshey, R. M. (2003). Bacterial Motility on a Surface: Many Ways to a Common Goal. Annual Rev Microbiol 57: 249-273.
- Kearns, D, B. (2010). A field guide to bacterial swarming motility. Nat Rev Microbiol 8(9): 634.
- Steager, E. B., Kim, C. B. and Kim, M. J. (2008). Dynamics of pattern formation in bacterial swarms. Phys Fluids 20: 073601
- Morales-Soto, N. Anyan, M. E., Mattingly, A. E., Madukoma, C. S., Harvey, C. W., Alber, M., Déziel, E., Kearns, D. B. and Shrout, J. D. (2015). Preparation, imaging, and quantification of bacterial surface motility assays. J Vis Exp 98: 523-538.
- Peñil Cobo, M., Libro, S., Marechal, N., D'Entremont, D., Peñil Cobo, D. and Berkmen, M. (2018). Visualizing bacterial colony morphologies using time-lapse imaging chamber MOCHA. J Bacteriol 200(2): 413-417.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chen, W., Mani, S. and Tang, J. X. (2021). An Inexpensive Imaging Platform to Record and Quantitate Bacterial Swarming. Bio-protocol 11(18): e4162. DOI: 10.21769/BioProtoc.4162.
Category
Microbiology > Microbe-host interactions > Bacterium
Microbiology > Microbial biofilm > Biofilm culture
Biological Sciences > Microbiology > Microbial communities > Microbiome
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










