- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of the Effects of Hexokinase 2 Detachment From Mitochondria-Associated Membranes with the Highly Selective Peptide HK2pep
(*contributed equally to this work) Published: Vol 11, Iss 14, Jul 20, 2021 DOI: 10.21769/BioProtoc.4087 Views: 4427
Reviewed by: Julie WeidnerAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of Autophagosomes in Human Fibroblasts Using Cyto-ID® Staining and Cytation Imaging
Barbara Hochecker [...] Jörg Bergemann
Jul 5, 2024 1658 Views

Microfluidic Cultures of Basal Forebrain Cholinergic Neurons for Assessing Retrograde Cell Death by Live Imaging
Srestha Dasgupta [...] Wilma J. Friedman
Jan 5, 2025 1846 Views

Real-time IncuCyte® Assay for the Dynamic Assessment of Live and Dead Cells in 2D Cultures
Arlene K. Gidda [...] Sharon M. Gorski
Feb 5, 2025 2817 Views
Abstract
The crucial role of hexokinase 2 (HK2) in the metabolic rewiring of tumors is now well established, which makes it a suitable target for the design of novel therapies. However, hexokinase activity is central to glucose utilization in all tissues; thus, enzymatic inhibition of HK2 can induce severe adverse effects. In an effort to find a selective anti-neoplastic strategy, we exploited an alternative approach based on HK2 detachment from its location on the outer mitochondrial membrane. We designed a HK2-targeting peptide named HK2pep, corresponding to the N-terminal hydrophobic domain of HK2 and armed with a metalloprotease cleavage sequence and a polycation stretch shielded by a polyanion sequence. In the tumor microenvironment, metalloproteases unleash polycations to allow selective plasma membrane permeation in neoplastic cells. HK2pep delivery induces the detachment of HK2 from mitochondria-associated membranes (MAMs) and mitochondrial Ca2+ overload caused by the opening of inositol-3-phosphate receptors on the endoplasmic reticulum (ER) and Ca2+ entry through the plasma membrane leading to Ca2+-mediated calpain activation and mitochondrial depolarization. As a result, HK2pep rapidly elicits death of diverse tumor cell types and dramatically reduces in vivo tumor mass. HK2pep does not affect hexokinase enzymatic activity, avoiding any noxious effect on non-transformed cells. Here, we make available a detailed protocol for the use of HK2pep and to investigate its biological effects, providing a comprehensive panel of assays to quantitate both HK2 enzymatic activity and changes in mitochondrial functions, Ca2+ flux, and cell viability elicited by HK2pep treatment of tumor cells.
Graphical abstract:
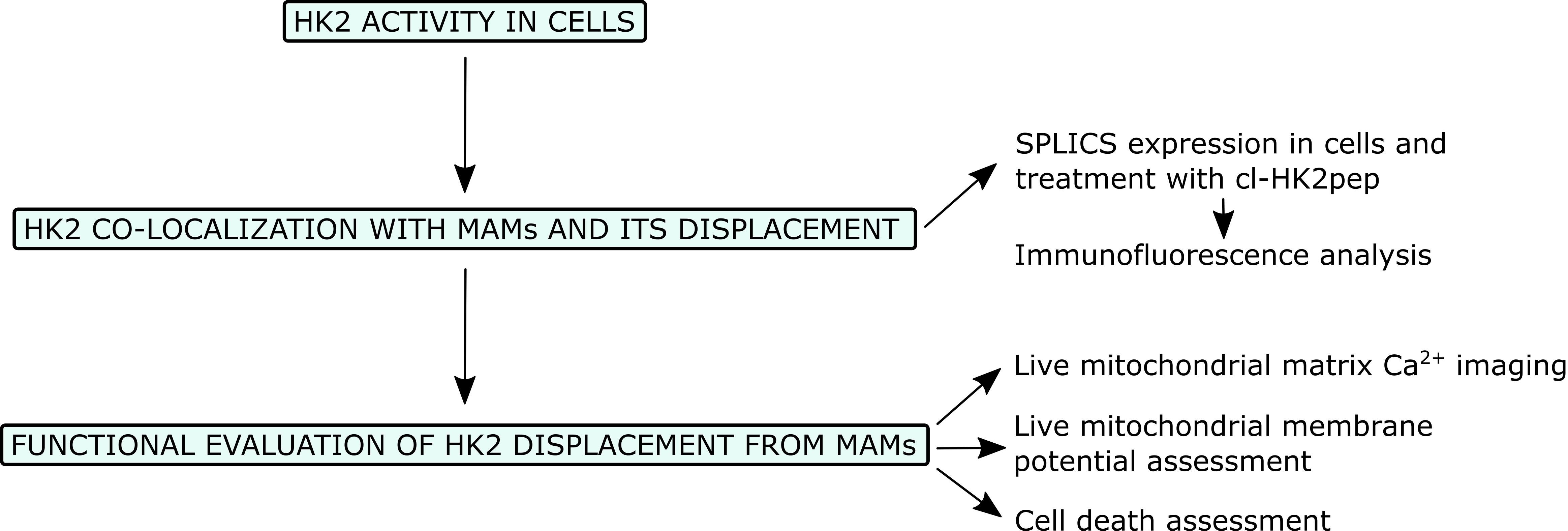
Flowchart for the analysis of the effects of HK2 detachment from MAMs.
Background
Metabolic rewiring in tumor cells (Boroughs and DeBerardinis, 2015; Vander Heiden and DeBerardinis, 2017) encompasses increased uptake and usage of glucose and decreased oxidative phosphorylation (OXPHOS) (Cannino et al., 2018; Faubert et al., 2020), conferring a selective advantage to neoplastic cells to survive and thrive under shortage of both nutrients and oxygen (Nakazawa et al., 2016). Hence, targeting metabolic components offers promising therapeutic perspectives, and the selective inhibition of glucose utilization has been considered for clinical cancer therapy (Hay, 2016). Hexokinases convert glucose to glucose-6-phosphate and make it available for utilization in glycolysis, the pentose phosphate pathway, glycogenesis, and hexosamine biosynthesis (Wilson, 2003). The most active isozyme, hexokinase 2 (HK2), is overexpressed in numerous types of cancer and constitutes a promising target for the development of anti-neoplastic strategies (Roberts and Miyamoto, 2015). Indeed, HK2 plays a major role in the metabolic rewiring of tumors and is induced by oncogenic K-Ras activation (Patra et al., 2013) or in response to hypoxia (Semenza, 2013). HK2 overexpression is related to stage progression, acquisition of invasive and metastatic capabilities, and poor prognosis (Mathupala et al., 2010). In tumor cells, HK2 binds to the outer mitochondrial membrane following Akt-dependent phosphorylation (Miyamoto et al., 2008), and mitochondrial binding of HK2 has been associated with the protection of cancer cells from noxious stimuli (Roberts and Miyamoto, 2015). The natural consequence of these observations was the development of therapeutic strategies aimed at inhibiting the activity of HKs; however, the clinical use of hexokinase inhibitors was hampered by the lack of specificity or the side effects (Akins et al., 2018) elicited by the conserved nature of the active sites among the ubiquitously expressed hexokinase isozymes (Roberts and Miyamoto, 2015).
We have previously exploited an alternative approach to target HK2, based on detaching it from mitochondria with a peptide corresponding to the N-terminal hydrophobic domain of the enzyme. This treatment causes the opening of a mitochondrial channel, the permeability transition pore (PTP) (Rasola and Bernardi, 2011), and consequent cell death (Chiara et al., 2008; Masgras et al., 2012; Pantic et al., 2013). Recently, we established that HK2 localizes to domains of interaction between the ER and mitochondria called mitochondria-associated membranes (MAMs), and its displacement elicits a massive Ca2+ flux from the ER and across the plasma membrane into mitochondria, inducing mitochondrial depolarization and death in a variety of cancer models, both in vitro and in vivo, in a calpain-dependent manner (Ciscato et al., 2020). We have improved HK2 peptide efficacy and specificity by adding a polycation stretch required for plasma membrane permeation linked to a shielding polyanion sequence through a matrix metalloprotease 2 and 9 (MMP2/9) target sequence. This novel tool, called HK2pep, is specifically taken up by neoplastic cells when its polycation sequence is unmasked by removal of the polyanion stretch through MMP2/9 cleavage in the tumor microenvironment, where these proteases are highly induced (Figure 1). HK2pep has proven to be an excellent tool to dissect the precise localization of HK2 in MAMs and the cascade of events leading to tumor cell death, in addition to being effective in tumors allografted in mice, while leaving hexokinase enzymatic activity unaffected, thus protecting healthy tissues from any off-target effects of the treatment (Ciscato et al., 2020).
Taken together, our data disclose novel signaling pathways primed by HK2 displacement from MAMs and open possibilities for the development of effective anti-neoplastic strategies, alone or in combination with other chemotherapies.
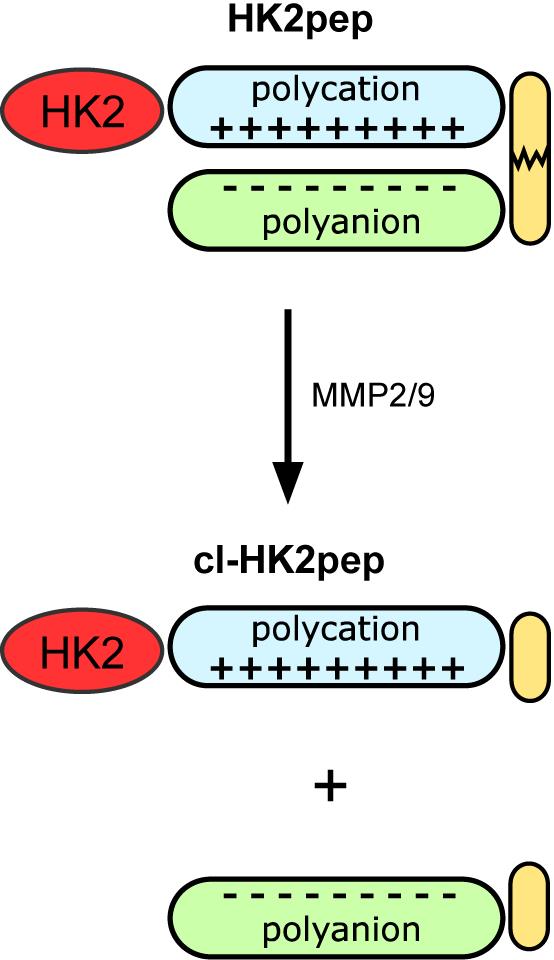
Figure 1. Modified from Ciscato et al., 2020. Functional unit composition of HK2pep. The HK2-targeting sequence is in red; the polycation and polyanion stretches are in light blue and light green, respectively; the MMP2/9 target sequence is in yellow. Cl-HK2pep is the active peptide after MMP2/9 cleavage.
Materials and Reagents
Flasks, 25 cm2 (Falcon, catalog number: 353108)
6-well plates (Falcon, catalog number: 353046)
12-well plates (Falcon, catalog number: 353103)
24-well plates (Falcon, catalog number: 353047)
96-well plates (Falcon, catalog number: 353916)
13-mm diameter coverslips (VWR, catalog number: 631-0149P)
18-mm diameter coverslips (VWR, catalog number: 631-0153P)
24-mm diameter coverslips (VWR, catalog number: 631-0161)
PBS (Sigma-Aldrich, catalog number: D8537, storage RT)
Trypsin-EDTA (GIBCO, catalog number: 25200-056, storage 4°C)
NaCl (Sigma-Aldrich, catalog number: S9888, storage RT)
Trizma® base (Sigma-Aldrich, catalog number: T6066, storage RT)
EDTA (Sigma-Aldrich, catalog number: E5134, storage RT)
Triton X-100 (Sigma, catalog number: 9002-93-1, storage RT)
Glycerol (Sigma-Aldrich, catalog number: G5516, storage RT)
PierceTM BCA protein assay (Thermo ScientificTM, catalog number: 23225, storage RT)
MgCl2 (Sigma-Aldrich, catalog number: 63068, storage RT)
ATP (Sigma-Aldrich, catalog number: A2383, storage -20°C)
Glucose (Sigma-Aldrich, catalog number: G8270, storage RT)
Glucose 6-phosphate-dehydrogenase (Sigma-Aldrich, catalog number: SRP6505, storage -20°C)
NADP (Sigma-Aldrich, catalog number: 93205, storage -20°C)
SPLICSs (split-GFP-based contact site sensor short) and SPLICSL (split-GFP-based contact site sensor long) plasmids (Cieri et al., 2018, available from the authors) (storage -20°C)
OptiMEM (GIBCO, catalog number: 51985026, storage 4°C)
TransIT-LT1 (Mirus, catalog number: MIR 2304, storage 4°C)
KCl (Sigma-Aldrich, catalog number: P3911, storage RT)
KH2PO4 (Merck, catalog number: 1551139, storage RT)
CaCl2 (Sigma-Aldrich, catalog number: 21097, storage RT)
HEPES (Sigma-Aldrich, catalog number: H3375, storage RT)
HK2pep: (Chemical synthesis, seq. MIASHLLAYFFTELN-bA-RRRRRRRRR-PLGLAG-Ahx-EEEEEEEE, storage -20°C)
SCRpep: (Chemical synthesis, seq. VGAHAGEYGAEALER-bA-RRRRRRRRR-PLGLAG-Ahx-EEEEEEEE, storage -20°C)
cl-HK2pep (Chemical synthesis, seq. MIASHLLAYFFTELN-βA-RRRRRRRRR-PLG, storage -20°C)
cl-SCRpep (Chemical synthesis, seq. VGAHAGEYGAEALER-βA-RRRRRRRRR-PLG storage -20°C). All peptides were synthesized by automatic solid-phase procedures (for details, see Ciscato et al., 2020). Suggested purity ≥ 95% measured by analytical reversed-phase HPLC. Peptides are not commercially available but can be synthesized on demand by specialized companies.
PFA (Sigma-Aldrich, catalog number: P6148, storage 4°C)
NH4Cl (Sigma-Aldrich, catalog number: A9434, storage RT)
BSA (Sigma-Aldrich, catalog number: AG4503, storage 4°C)
Goat serum (Sigma-Aldrich, catalog number: G6767, storage -20°C)
Gelatin (Sigma-Aldrich, catalog number: G2500, storage -20°C)
Rabbit monoclonal anti-HK2 antibody (Thermo Scientific, catalog number: H.738.7, storage -20°C)
AlexaFluor555 donkey anti-rabbit IgG (Thermo Fisher Scientific, catalog number: A-31572, storage -20°C)
Mowiol 4-88 (Sigma-Aldrich, catalog number: 81381, storage RT)
Plasmids encoding mitochondria-targeted GCAMP6f (Filadi et al., 2018, available from the authors, storage -20°C)
Cyclosporin H (Vinci-Biochem, catalog number: AG-CN2-0447-M005, storage -20°C)
TetraMethyl-Rhodamine Methyl ester (TMRM) (Invitrogen, MitoProbeTM, catalog number: M20036, storage -20°C)
Annexin V-FITC - Annexin-V-FLUOS labeling reagent (Roche - now sold by MERCK, catalog number: 11828681001, storage -20°C)
7-aminoactinomycin D (7-AAD; Sigma-Aldrich, catalog number: A9400, storage -20°C)
Equipment
Cell culture incubators (Thermo Scientific FormaTM Steri-CycleTM CO2 Incubator)
Equipped cell culture hoods (Angelantoni Industries, catalog number: VBH 48 C2)
Micropipettes (Gilson Pipetman Classic, P1,000, P200, P20, P10, P2)
Burker chamber (VWR, catalog number: HECH40443703)
Centrifuges from 200 to 2,000 × g, fixed bucket, no specific rotors needed (e.g., MPW Med. Instruments, catalog number: MPW 251)
Thermo-shaker (Biosan, catalog number: TS-100)
Centrifuges from 200 to 16,000 × g, fixed bucket, no specific rotors needed (e.g., Eppendorf, catalog number: 5417 R)
Plate-Reader Spectrophotometer (TECAN, model: Infinite M200 spectrophotometer)
Confocal microscope (Leica SP5-II equipped with a 100×/1.4 N.A. plan apochromat objective, a WLL laser to excite each specific dye, and a HyD detector for signal collection)
Inverted fluorescence microscope (Zeiss Axiovert 100, Fluar 40× oil objective, NA 1.30)
Inverted fluorescence microscope (Leica, model: DMI6000 B)
Cytofluorimeter (BD FACSCantoTM II)
Software
i-controlTM (for TECAN, Infinite M200 spectrophotometer, https://lifesciences.tecan.com/plate_readers/infinite_200_pro?p=tab--3)
Excel data analysis software (Microsoft Office)
Leica Application Suite LAS-AF (for Leica SP5-II https://www.leica-microsystems.com/products/confocal-microscopes/p/leica-tcs-sp5-ii)
ImageJ (NIH, https://imagej.nih.gov/ij/download.html)
For the Zeiss microscope, synchronization of the cool camera with the excitation source is performed by a custom-made software package (Roboscope - developed by Catalin Dacian Ciubotaru at VIMM, Padova, Italy); alternatively, the specific manufacturer’s microscope software is suitable for kinetic image acquisition
Leica Application Suite LAS-AF for the Leica fluorescence microscope (https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/)
BD FACSDivaTM for the BD FACSCantoTM II cytoflurimeter (https://www.bdbiosciences.com/en-us/instruments/research-instruments/research-software/flow-cytometry-acquisition/facsdiva-software)
Procedure
HK2 activity in cells
Note: To carry out these measurements, we used mouse 4T1 breast carcinoma cells, mouse CT26 colorectal carcinoma cells, and human HeLa cervix carcinoma cells (Ciscato et al., 2020). It is possible to perform this protocol on both adherent and suspended cell lines and on primary cells.
Cell culture and sample collection
Plate 1-4 × 106 cells (depending on the cell type; e.g., 2 × 106 HeLa cells, 2 × 106 4T1 and CT26 cells) in 25 cm2 flasks at least 24 h before harvesting/treatment.
Harvest the cells by washing the flask with PBS, detaching them with trypsin-EDTA, and blocking the reaction with fresh media (DMEM for HeLa and CT26 cells, RPMI for 4T1 cells) containing 10% FBS.
Pellet the cells (adjust the centrifugal force and time according to your cell lines to safely pellet them; e.g., 5 min, 400 × g for HeLa, 4T1, and CT26 cells).
Remove the supernatant.
Lyse cells in 100-200 µl Lysis Buffer.
Quantitate the protein content (PierceTM BCA protein assay kit).
Calculate the volume corresponding to 20 μg protein (this amount of protein can be modified according to the cell line under analysis), which is sufficient to detect hexokinase activity.
Sample preparation
Prepare 2 vials containing 20 μg protein for each experimental condition.
Incubate one vial at 4°C and the other at 46°C for 30 min. Heating 30 min at 46°C is sufficient to remove the catalytic activity of HK2, the only heat-sensitive hexokinase, preserving the activity of the other isozymes; HK2 is not degraded by this treatment.
After incubation, store the samples on ice.
Sample treatment with cl-HK2pep (and/or your compound of interest) and measurement of HKs/HK2 activity
Prepare fresh Reaction Buffer at room temperature.
The final reaction volume is 100 μl. Prepare the proper amount of buffer spotted in a 96-well plate and add 10 μM cl-HK2pep (and/or your compound of interest - use cl-SCRpep as a control peptide).
Mix for 5 s using a plate mixer (generally, we shake using the plate reader, but any plate mixer is suitable).
Add 20-100 μg cell extract (heated or not) and mix for 10 s.
Read the 96-well plate in kinetic mode (with a minimum 30 s interval) at 340 nm using a spectrophotometric plate reader (Infinite M200 spectrophotometer, TECAN) for 15 min at 37°C.
Data analysis
HK activities were calculated in nmol/min/mg: nmol per minute was obtained by calculating the slope (m) of the plotted absorbance at 340 nm during the first minute of recording and dividing it by the extinction coefficient of NADH (ϵ = 6.22) and the cuvette path length (l = 0.4 cm for the 96-well plate). Then, values were normalized to total mg of protein added to the reaction (0.08 mg 4T1 total lysate).
To calculate HK2 activity, subtract the activity obtained in the samples pre-incubated at 46°C from that in the samples incubated at 4°C (Figure 2).
Apply the proper statistical analysis (e.g., Student’s t-test to compare two groups or one-way ANOVA to compare more than two groups).
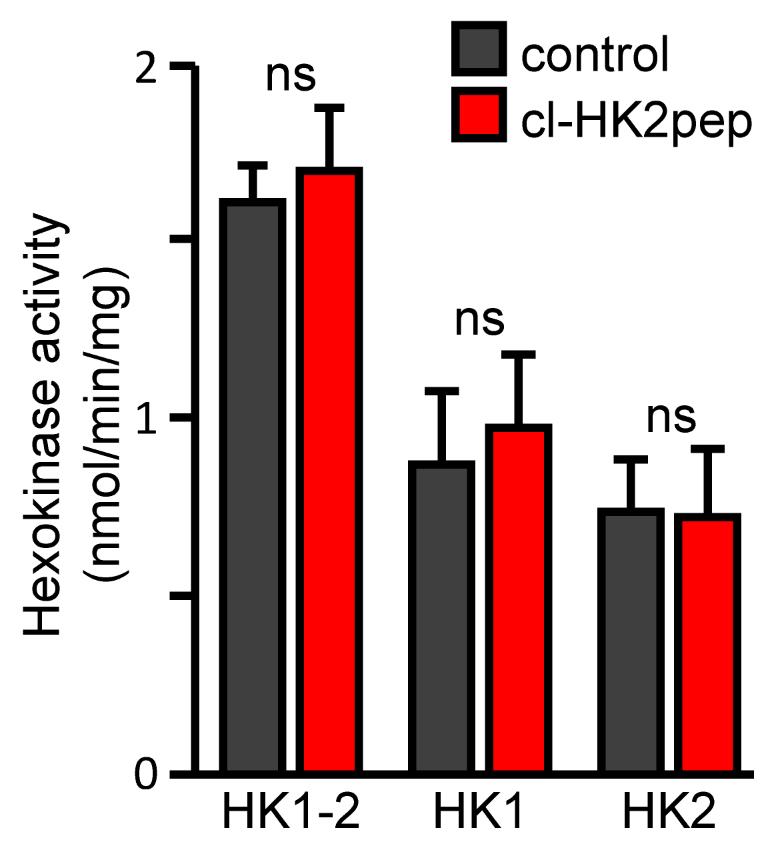
Figure 2. Modified from Ciscato et al., 2020. Measurement of hexokinase enzymatic activities. HK1+HK2, HK1, and HK2 enzymatic activity calculated after treatment with cl-HK2pep in 4T1 cell extract as described above. cl-SCRpep is used as a negative control.
HK2 co-localization with MAMS and its displacement by cl-HK2pep
Cell seeding and culture
Plate 0.5-1 × 105 cells in 24-well plates containing a 13-mm (diameter) coverslip and culture (37°C, 5% CO2, humidified atmosphere) for at least 24 h. Cells can be grown for 48 h but must be at 60-70% confluence at the time of transfection (next step).
Cell transfection with SPLICS-encoding plasmids
Transfect cells with plasmids encoding the two subunits of SPLICSS (to evaluate ER-mitochondria contacts < 10 nm) or the SPLICSL probe (for contacts < 40-50 nm) (Cieri et al., 2018). Mix 0.5 µg each plasmid/well in 100 µl OptiMEM together with 2 µl TransIT-LT1 (MIR 2304 - Mirus) and incubate for 15 min.
Add the transfection mix dropwise to the cells (leaving 0.5 ml culture medium) and incubate for 5 h.
Eliminate the medium containing the transfection mix and replace with 0.5 ml fresh medium.
Leave cells in the incubator overnight.
Cell treatment(s) with cl-HK2pep (and/or your compound of interest)
Prepare a solution containing mKRB Buffer and 2 μM cl-HK2pep (and/or your compound of interest; use cl-SCRpep as a control peptide). This amount of cl-HK2pep is able to detach part of HK2 from MAMs in less than 2 min, so please consider pre-incubating other compounds before using cl-HK2pep if they take longer to be effective in intact cells.
Substitute cell media with 500 μl solution containing 1-2 μM cl-HK2pep (and/or your compound of interest; use cl-SCRpep as a control peptide). Consider using different media according to experimental conditions; cl-HK2pep is effective in a broad range of physiological solutions.
Incubate at room temperature for 2 min. The 2 μM cl-HK2pep treatment can lead to mitochondrial depolarization in a few minutes and apoptotic cell death in 15 min (Ciscato et al., 2020). Consider not exceeding a 5-min treatment; otherwise, decrease the cl-HK2pep concentration. Depending on the cell type, different media can accelerate or delay the activity of cl-HK2pep.
Remove media and add 500 μl PBS using a pipette to wash the cells.
Remove PBS and add 500 μl 4% PFA (prepared in PBS) for 10 min to fix the cells for the immunofluorescence protocol.
Remove the PFA solution and wash the cells 3 × 5 min with 0.5 ml PBS during gentle agitation (100-120 rpm on an orbital shaker).
Add 0.5 ml quenching solution for 20 min.
Remove the quenching solution and wash the cells 3 × 5 min with 0.5 ml PBS during gentle agitation (100-120 rpm on an orbital shaker). Then proceed with immunofluorescence (step B4 below). Cells can be stored at 4°C in PBS for 1 week if it is not possible to proceed immediately with step B4.
HK2 immunofluorescence protocol and image acquisition
Remove the PBS and permeabilize the cells with 0.5 ml Triton X-100 for 3 min.
Remove the Triton X-100 and wash the cells with 0.5 ml PBS. The solutions must be added and removed using a pipette.
Incubate cells with 0.5 ml blocking solution for at least 30 min. Add the solution using a pipette. Protect from light.
Dilute the primary antibody (rabbit monoclonal anti-HK2 antibody, H.738.7, Thermo Scientific) 1:150 in blocking solution and incubate cells for 90 min at room temperature, protected from light.
Remove the antibody solution and wash the cells 3 × 5 min with blocking solution during gentle agitation (100-120 rpm on an orbital shaker).
Dilute the secondary antibody (AlexaFluor555 donkey anti-rabbit IgG, A-31572 Thermo Fisher Scientific) 1:300 in blocking solution and centrifuge at 13,000 × g for 10 min.
Incubate cells with this solution for 45 min at room temperature. Protect from light.
Remove the antibody solution and wash the cells 3 × 5 min with blocking solution during gentle agitation (100-120 rpm on an orbital shaker). Perform an additional wash with PBS during gentle agitation (100-120 rpm on an orbital shaker).
Wash coverslips with H2O and immediately mount with Mowiol (or a different mounting solution) on an object glass. Allow to dry overnight protected from light at room temperature, then store at 4°C until image acquisition.
Collect images using a confocal microscope (Leica SP5-II equipped with a 100×/1.4 N.A. plan apochromat objective, a WLL laser to excite each specific dye, and a HyD detector for signal collection). The power of the lasers should be set on the sample with the highest signal intensity to avoid signal saturation and kept the same for all samples/experimental conditions. Export images in TIFF format.
Data analysis
Open TIFF images of the green (SPLICS probes) and red (HK2 staining) channels in ImageJ (version 1.47b or later, NIH).
Subtract the background. Trace a region of interest (ROI) in a background region; in Plugins, select ROI, BG subtraction from ROI, and press OK. Repeat for both channels.
Calculate the Pearson’s co-localization coefficient in Plugins, Co-localization analysis, Manders coefficients, choose the appropriate combination of the red and green channels, select exclude zero-zero pixels, and press OK. The Mander’s and Pearson’s co-localization coefficients will be calculated by the software (Figure 3).
Repeat for all images and apply the appropriate statistical tests. We recommend analyzing at least 10 different cells for each coverslip and repeating the experiment at least three times on three different days.
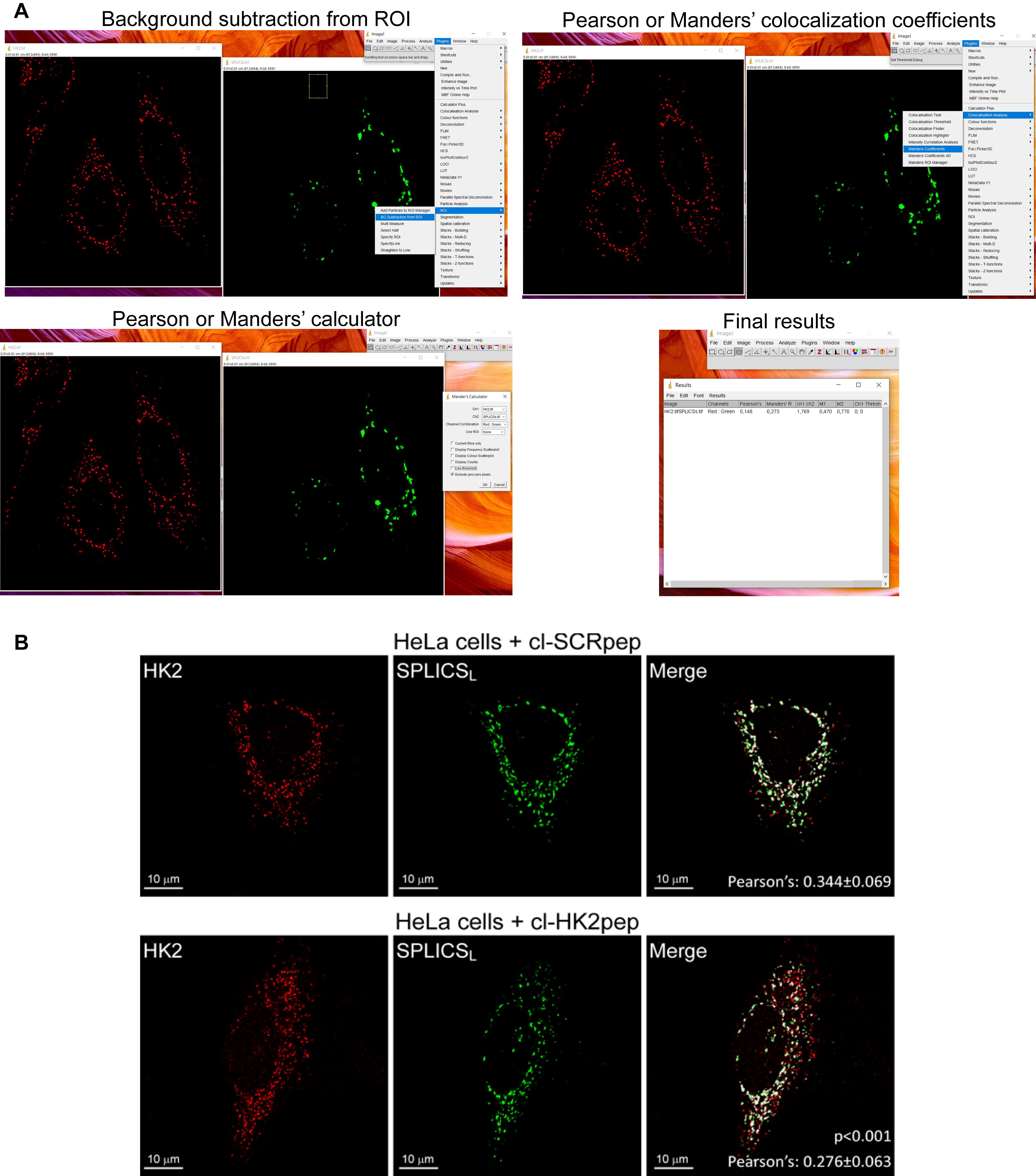
Figure 3. Pearson’s co-localization coefficient calculation, modified from Ciscato et al. (2020). (A) Step-by-step representative confocal image analysis in HeLa cells performed using ImageJ (version 1.47b or later, NIH). (B) Representative results from confocal image analysis: HK2 displacement from HeLa MAMs after treatment with cl-HK2pep is shown by the loss of the merged signal between HK2 (red) and the split-GFP-based probe for ER-mitochondria contacts (SPLICSL, green); the merged signal is white. cl-SCRpep is used as a negative control, and Pearson’s co-localization coefficient is indicated (cl-SCRpep n = 26 cells; cl-HK2pep n = 24 cells; P < 0.001 with a Student’s t-test). Scale bar: 10 μm.
Functional evaluation of HK2 displacement from MAMS by cl-HK2pep
Cell seeding and culture
Note: Consider adjusting the number of cells to be plated depending on the specific cell type under analysis. The following analyses must be performed on adherent cells.
For live mitochondrial matrix Ca2+ imaging, plate 1 × 105 cells in 12-well plates containing 18-mm-diameter coverslips at least 24 h before transfection.
For live TMRM (tetramethylrhodamine methyl ester) imaging, plate 2.5 × 105 cells in 6-well plates containing 24-mm-diameter coverslips at least 24 h before the experiment.
For cell death assessment, plate 1.5 × 106 cells in 25 mm2 flasks for each condition the day before the experiment.
Live mitochondrial matrix Ca2+ imaging and data analysis
Transfect cells with the plasmids encoding mitochondria-targeted GCAMP6f (see point B.2 for the transfection protocol; use 0.75 µg DNA for each coverslip and 1.5 µl TransIT-LT1 reagent).
24 h after transfection, replace the medium with mKRB buffer supplemented with 1 µM cyclosporin H, and place the coverslip in a proper holder for live microscope imaging. Incubate for 10-20 min with 500 μl mKRB at room temperature. Specific drugs can be added and incubated for 30-60 min before the experiment.
Sequentially excite GCAMP6f at 475 nm (the Ca2+-sensitive wavelength) and 410 nm (the isosbestic point) on an inverted fluorescence microscope (Zeiss Axiovert 100, Fluar 40× oil objective, NA 1.30), producing the excitation light from a monochromator (polychrome V; TILL Photonics) and filtering it with a 505 nm DRLP filter (Chroma Technologies). Collect the emitted fluorescence in the 500-530 nm range (using a band-pass filter, Chroma Technologies) and repeat this cycle every 5 s (Figure 4A). Imaging can be performed for up to 90 min.
Add 2 μM cl-HK2pep (and/or your compound of interest) 2-4 min after starting the recording. For a homogeneous distribution of added compounds, collect 100 μl solution on the holder, dissolve cl-HK2pep (and/or your compound of interest), and carefully add the already-made solution on top of the coverslip by mixing using a pipette. On a single experimental day, we recommend analyzing at least 3 different coverslips for each condition, with 4-10 cells in the acquired field. Repeat the experiment at least three times on three different days.
Export images in TIFF format and open in the ImageJ software.
Subtract the background (see above point B5.b), trace the appropriate ROIs including the mitochondrial network of each single cell, and measure the emitted fluorescence intensity for each frame upon excitation at 475 nm and 410 nm.
Calculate the ratio (R) between the generated emissions by exciting the cells at 475 and 410 nm. R is proportional to [Ca2+]. The variation in R between conditions in which [Ca2+] in the mitochondrial matrix is relatively low (< 100 nM, resting conditions) and those in which the probe is saturated (high [Ca2+], > 10 µM) should be around 15-20 fold (Figure 4B).
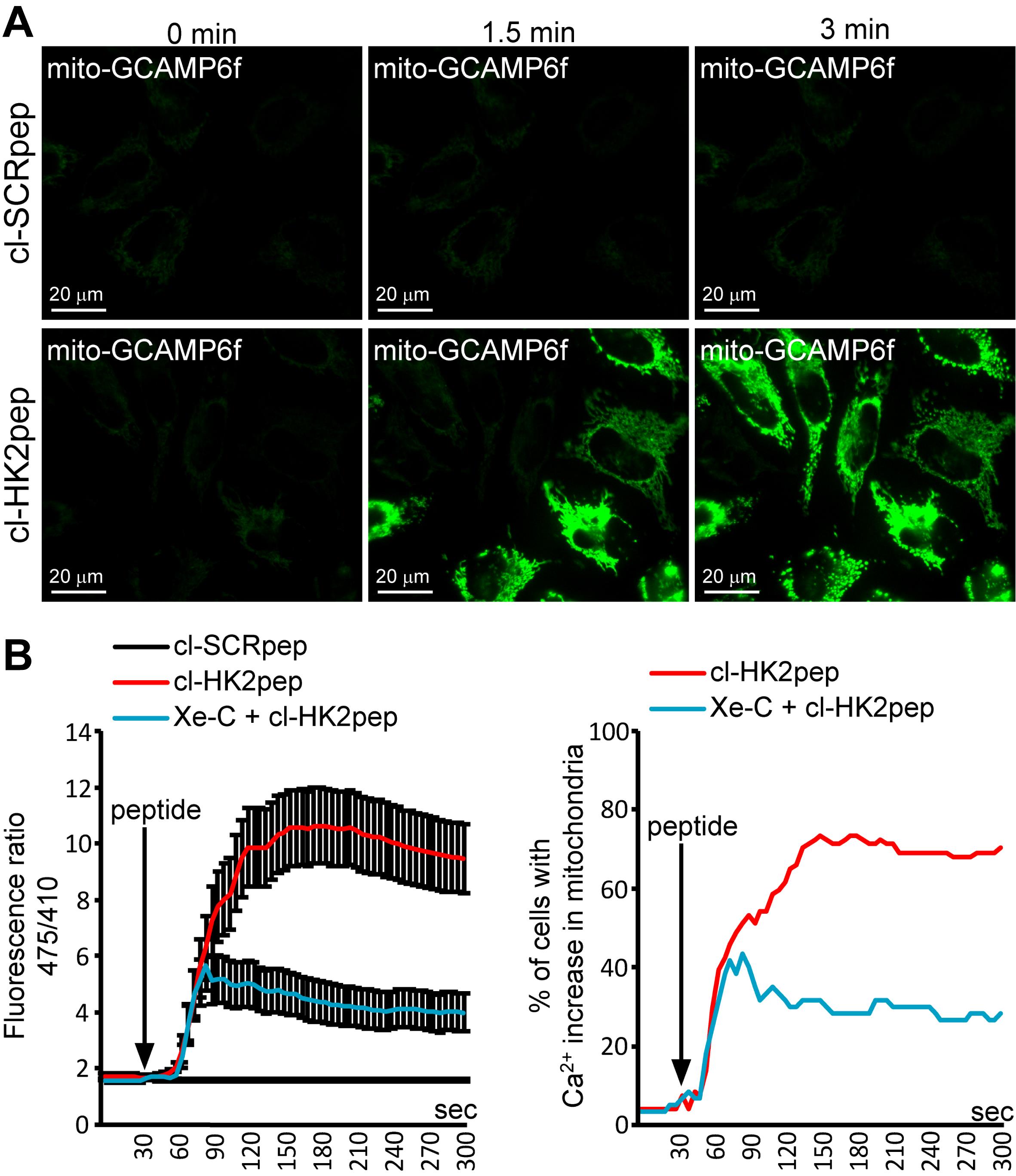
Figure 4. Modified from Ciscato et al., 2020. Measurement of changes in mitochondrial Ca2+ levels. (A) Representative images of kinetic analysis of mitochondrial matrix Ca2+ variations (assessed with a mitochondria-targeted GCAMP6f probe) after cl-HK2pep treatment in HeLa cells. (B) Quantitation of mitochondria-targeted GCAMP6f signals as the mean ± SEM of the 475/410 nm ratio (left panel) or the percentage of cells with increased Ca2+ in the mitochondria (right panel – threshold 475/410 ratio for positivity > 3; baseline mean ratio = 1.84 ± 0.54). cl-SCRpep is used as a negative control.Live mitochondrial membrane potential assessment and data analysis
Prepare a solution containing mKRB (or the solution adapted to the cells/experimental conditions) plus 20 nM TMRM and 1 μM cyclosporin H (to inhibit probe extrusion through multidrug resistance pumps).
Wash the coverslip with PBS in another 6-well plate.
Place the coverslip in a proper holder for live microscope imaging (according to your microscope set up) and incubate for 20-40 min with 500 μl TMRM-containing solution at room temperature. The incubation time is dependent on cell type. TMRM accumulates in the mitochondrial matrix and takes time to equilibrate; at the beginning of the experiment, TMRM accumulation must be in the steady state (Figures 5 and 6).
After incubation, use an inverted fluorescence microscope (Leica DMI6000 B) to record TMRM fluorescence in kinetic mode every 30 s - 3 min (depending on cell type). In some cell types, TMRM fluorescence can be phototoxic with consequent mitochondrial membrane depolarization. Therefore, before starting the experiments, monitor TMRM fluorescence in kinetic mode for 1 h: if the TMRM emission signal decreases, increase the lag time between acquisitions and/or decrease excitation intensity; experiments must be performed only under conditions in which cells display a stable signal during the entire hour of recording.
Add 2 μM cl-HK2pep (and/or your compound of interest; use cl-SCRpep as a control peptide) 2-4 min after starting the recording. For homogeneous distribution of added compounds, collect 100 μl solution on the holder, dissolve cl-HK2pep (and/or your compound of interest), and carefully add the mix on top of the coverslip. On a single experimental day, we recommend analyzing at least 3 different coverslips for each condition, with 5 -30 cells in the acquired field. Repeat the experiment at least three times on three different days.
For image analysis: Open TIFF images in ImageJ (1.47b, NIH).
Subtract the background (see above point B5.b), trace the appropriate ROIs including the mitochondrial network of each single cell, and measure the emitted fluorescence intensity in kinetic mode (Figure 5).
Repeat for all kinetics and apply the appropriate statistical tests.
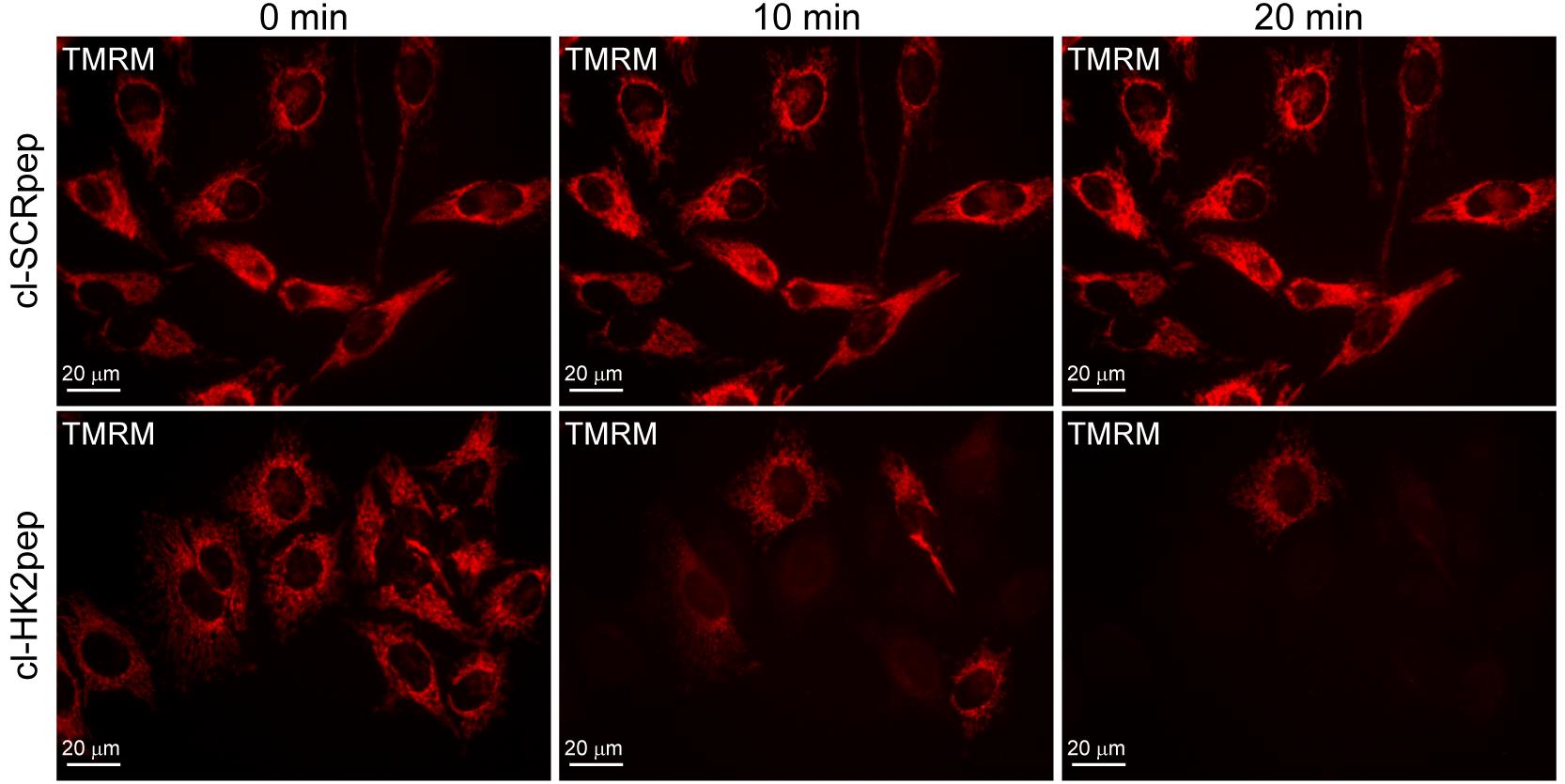
Figure 5. Modified from Ciscato et al., 2020. Assessment of changes in mitochondrial membrane potential. Effect of cl-HK2pep treatment on mitochondrial membrane potential assessed with the TMRM probe in HeLa cells.
Figure 6. Representative set up with a coverslip chamber for live cell imagingCell death assessment and data analysis
Harvest cells by washing flasks one at a time with PBS, detaching cells with trypsin-EDTA, and blocking the reaction with 5 ml fresh medium containing 10% FBS. Collect the detached cells.
Count cells using a Burker chamber.
Collect 1.5 × 106 cells for each experimental condition and pellet them.
Prepare mKRB buffer containing 1:100 Annexin-V-FLUOS labeling reagent pre-made by Roche (alternatively, it is possible to use other Annexin V-FITC probes following the manufacturer’s instructions) and 1 μg/ml 7-aminoactinomycin D (7-AAD).
Note: It is possible to substitute mRKB buffer with other culture media if needed for the survival of some specific cell types (e.g., we used DMEM without phenol red and added 10 mM HEPES and 0.1% FBS (pH 7.4) for freshly isolated B cells or freshly isolated B-CLL cancer cells).
Resuspend 1.5 × 106 cells in 900 μl mKRB buffer + Annexin V-FITC and 7-AAD and aliquot samples into three different cytofluorimeter-suitable tubes (300 μl sample containing 5 × 105 cells in each tube). These three tubes are technical replicates.
To record staining under basal conditions in all tubes, analyze the green and far-red fluorescence using the cytofluorimeter set-up.
Depending on the cell type, add 1-5 μM cl-HK2pep (and/or your compound of interest; use cl-SCRpep as a control peptide) and gently mix the sample after each addition. Cl-HK2pep is not efficient at killing the non-cancerous cell lines C2C12 or RAW.241, as described in (Ciscato et al., 2020). However, since 5 μM cl-HK2pep can kill 75-96% of cancer cells in less than 15 min (Ciscato et al., 2020), please consider pre-incubating other compounds before cl-HK2pep if they take longer to be effective in intact cells.
Incubate for 15 min at room temperature and reanalyze all tubes in the same order.
Repeat the cytofluorimeter recordings every 15-30 min to perform a kinetic analysis.
Data analysis: Evaluate the percentage of live cells (double-negative for Annexin V-FITC and 7-AAD staining) at each time point and average the technical replicates. Apply the appropriate statistical tests.
Data analysis
Methods of data analysis are specifically described for each technical approach.
Recipes
Lysis Buffer
150 mM NaCl
20 mM Tris
5 mM EDTA
1% Triton X-100
10% glycerolReaction Buffer
50 mM Tris
10 mM MgCl2
4 mM ATP
2 mM glucose0.1 U/ml glucose 6-phosphate-dehydrogenase (G6PDH)
1mM NADP, pH 7.4mKRB Buffer
135 mM NaCl
5 mM KCl
0.4 mM KH2PO4
1 mM MgCl2
1 mM CaCl2
20 mM HEPES
10 mM glucose, pH 7.4 at room temperatureQuenching Solution
0.24% NH4Cl in PBSPermeabilization Solution
0.1% Triton X-100 in PBSBlocking Solution
2% BSA
10% goat serum
0.2% gelatin in PBS
Acknowledgments
This work was supported by grants from the University of Padova, Neurofibromatosis Therapeutic Acceleration Program, Associazione Italiana Ricerca Cancro (AIRC grant IG 2017/20749 to A.R.), Piano for Life Onlus, and Linfa OdV. Protocols were derived from Ciscato et al., 2020.
Competing interests
A patent application (Italian patent No. IT102019000002321, priority: 18/02/2019; PCT: PCT/IB2020/051329) has been filed by the University of Padova for the use of the HK2-targeting peptide described in this manuscript as an anti-neoplastic tool in in vitro and in vivo experiments.
References
- Akins, N. S., Nielson, T. C. and Le, H. V. (2018). Inhibition of Glycolysis and Glutaminolysis: An Emerging Drug Discovery Approach to Combat Cancer. Curr Top Med Chem 18(6): 494-504.
- Boroughs, L. K. and DeBerardinis, R. J. (2015). Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 17(4): 351-359.
- Cannino, G., Ciscato, F., Masgras, I., Sanchez-Martin, C. and Rasola, A. (2018). Metabolic Plasticity of Tumor Cell Mitochondria. Front Oncol 8: 333.
- Chiara, F., Castellaro, D., Marin, O., Petronilli, V., Brusilow, W. S., Juhaszova, M., Sollott, S. J., Forte, M., Bernardi, P. and Rasola, A. (2008). Hexokinase II detachment from mitochondria triggers apoptosis through the permeability transition pore independent of voltage-dependent anion channels. PloS One 3(3): e1852.
- Cieri, D., Vicario, M., Giacomello, M., Vallese, F., Filadi, R., Wagner, T., Pozzan, T., Pizzo, P., Scorrano, L., Brini, M. and Cali, T. (2018). SPLICS: a split green fluorescent protein-based contact site sensor for narrow and wide heterotypic organelle juxtaposition. Cell Death Differ 25(6): 1131-1145.
- Ciscato, F., Filadi, R., Masgras, I., Pizzi, M., Marin, O., Damiano, N., Pizzo, P., Gori, A., Frezzato, F., Chiara, F., Trentin, L., Bernardi, P. and Rasola, A. (2020). Hexokinase 2 displacement from mitochondria-associated membranes prompts Ca2+ -dependent death of cancer cells. EMBO Rep 21(7): e49117.
- Faubert, B., Solmonson, A. and DeBerardinis, R. J. (2020). Metabolic reprogramming and cancer progression. Science 368(6487): eaaw5473.
- Filadi, R., Leal, N. S., Schreiner, B., Rossi, A., Dentoni, G., Pinho, C. M., Wiehager, B., Cieri, D., Cali, T., Pizzo, P. and Ankarcrona, M. (2018). TOM70 Sustains Cell Bioenergetics by Promoting IP3R3-Mediated ER to Mitochondria Ca2+ Transfer. Curr Biol 28(3): 369-382 e366.
- Hay, N. (2016). Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat Rev Cancer 16(10): 635-649.
- Masgras, I., Rasola, A. and Bernardi, P. (2012). Induction of the permeability transition pore in cells depleted of mitochondrial DNA. Biochim Biophys Acta 1817: 1860-1866.
- Mathupala, S. P., Ko, Y. H. and Pedersen, P. L. (2010). The pivotal roles of mitochondria in cancer: Warburg and beyond and encouraging prospects for effective therapies. Biochim Biophys Acta 1797(6-7): 1225-1230.
- Miyamoto, S., Murphy, A. N. and Brown, J. H. (2008). Akt mediates mitochondrial protection in cardiomyocytes through phosphorylation of mitochondrial hexokinase-II. Cell Death Differ 15(3): 521-529.
- Nakazawa, M. S., Keith, B. and Simon, M. C. (2016). Oxygen availability and metabolic adaptations. Nat Rev Cancer 16(10): 663-673.
- Pantic, B., Trevisan, E., Citta, A., Rigobello, M. P., Marin, O., Bernardi, P., Salvatori, S., and Rasola, A. (2013). Myotonic dystrophy protein kinase (DMPK) prevents ROS-induced cell death by assembling a hexokinase II-Src complex on the mitochondrial surface. Cell Death Dis 4: e858.
- Patra, K. C., Wang, Q., Bhaskar, P. T., Miller, L., Wang, Z., Wheaton, W., Chandel, N., Laakso, M., Muller, W. J., Allen, E. L., Jha, A. K., Smolen, G. A., Clasquin, M. F., Robey, B. and Hay, N. (2013). Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 24(2): 213-228.
- Rasola, A. and Bernardi, P. (2011). Mitochondrial permeability transition in Ca2+-dependent apoptosis and necrosis. Cell Calcium 50: 222-233.
- Roberts, D.J. and Miyamoto, S. (2015). Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ 22(2): 248-257.
- Semenza, G. L. (2013). HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest 123(9): 3664-3671.
- Vander Heiden, M. G. and DeBerardinis, R. J. (2017). Understanding the Intersections between Metabolism and Cancer Biology. Cell 168(4): 657-669.
- Wilson, J. E. (2003). Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol 206(Pt 12): 2049-2057.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ciscato, F., Chiara, F., Filadi, R. and Rasola, A. (2021). Analysis of the Effects of Hexokinase 2 Detachment From Mitochondria-Associated Membranes with the Highly Selective Peptide HK2pep. Bio-protocol 11(14): e4087. DOI: 10.21769/BioProtoc.4087.
Category
Cell Biology > Cell viability > Cell death
Cancer Biology > Cell death > Cancer therapy
Biochemistry > Protein
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








