- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Electron Tomography to Study the Three-dimensional Structure of the Reovirus Egress Pathway in Mammalian Cells
Published: Vol 11, Iss 13, Jul 5, 2021 DOI: 10.21769/BioProtoc.4080 Views: 4393
Reviewed by: Juan Facundo Rodriguez AyalaKristin L. ShinglerAnca Flavia Savulescu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Acutely Modifying Phosphatidylinositol Phosphates on Endolysosomes Using Chemically Inducible Dimerization Systems
Wei Sheng Yap [...] Maxime Boutry
Oct 5, 2024 1621 Views

Protein Turnover Dynamics Analysis With Subcellular Spatial Resolution
Lorena Alamillo [...] Edward Lau
Aug 5, 2025 2397 Views

Generating ER-TRG and CA-ER-TRG Knock-in Mice and Quantitative in vivo Imaging of ER-phagy
Mengyuan Zhang [...] Qiming Sun
Jan 5, 2026 190 Views
Abstract
Mammalian orthoreoviruses (reoviruses) are nonenveloped, double-stranded RNA viruses that replicate and assemble in cytoplasmic membranous organelles called viral inclusions (VIs). To define the cellular compartments involved in nonlytic reovirus egress, we imaged viral egress in infected, nonpolarized human brain microvascular endothelial cells (HBMECs). Electron and confocal microscopy showed that reovirus mature virions are recruited from VIs to modified lysosomes termed sorting organelles (SOs). Later in infection, membranous carriers (MCs) emerge from SOs and transport new virions to the plasma membrane for nonlytic egress. Transmission electron microscopy (TEM) combined with electron tomography (ET) and three-dimensional (3D) reconstruction revealed that these compartments are connected and form the exit pathway. Connections are established by channels through which mature virions are transported from VIs to MCs. In the last step, MCs travel across the cytoplasm and fuse with the plasma membrane, which facilitates reovirus egress. This bio-protocol describes the combination of imaging approaches (TEM, ET, and 3D reconstruction) to analyze reovirus egress zones. The spatial information present in the 3D reconstructions, along with the higher resolution relative to 2D projections, allowed us to identify components of a new nonlytic viral egress pathway.
Keywords: ReovirusBackground
Viruses recruit and transform cellular compartments to build membranous viral factories or inclusions, where genome replication and particle assembly take place (Miller and Krijnse-Locker, 2008; Sachse et al., 2019). Following genome packaging and assembly, progeny virions leave the host cell. Both enveloped and nonenveloped viruses can induce cell death by lysis, resulting in the release of viral particles. However, viral egress pathways can also be mediated by intracellular organelles that transport new virions to the plasma membrane, allowing them to exit cells without lysis (Roth et al., 2020). Therefore, many viruses use cellular membranous organelles for replication and assembly, as well as for egress and cell-to-cell transmission (Altan-Bonnet, 2017; Bird and Kirkegaard, 2015).
Reoviruses belong to the Reoviridae family, which includes several pathogens of plants, animals, and humans. Reoviruses are nonenveloped, double-stranded RNA viruses that assemble membranous cytoplasmic structures termed VIs. Early in infection, reovirus nonstructural proteins σNS and μNS induce the remodeling of ER membranes to form VIs (Tenorio et al., 2018). Viral genome replication, secondary rounds of transcription, and capsid assembly occur in VIs and, as a result, mature virions as well as empty capsids gather in this network of membranes. Reovirus uses different types of egress mechanisms depending on the cell type and culture conditions. In some types of cultured cells, reovirus infection leads to NF-κB activation, inducing apoptotic signaling and eventually lytic cell death (Danthi et al., 2013); however, reovirus can undergo nonlytic egress in other cell types such as HBMECs (Lai et al., 2013). In nonpolarized HBMECs, progeny virions exit from discrete areas at the basal surface. The detailed study of these regions is difficult because of their low frequency and location at the cell base. Due to these constraints, late steps of reovirus infection, including nonlytic egress, are not well understood.
The use of several imaging techniques in combination allowed us to discover that the reovirus egress pathway is composed of two different membranous elements called SOs and MCs. SOs are modified lysosomes that are recruited to the periphery of VIs during late phases of infection. Only mature genome-containing virions are collected in SOs, whereas empty capsids are absent. The smaller MCs are formed by budding from SOs and transport viral progeny to the plasma membrane (Fernandez de Castro et al., 2020). ET and 3D reconstruction were essential in defining the spatial relationships of the different compartments and elucidating the intracellular reovirus egress pathway (a summary of the protocol is shown in Figure 1).
ET can be used to visualize the structure of viruses, cells, and their constituents in three dimensions. The chosen approach defines the volume that can be analyzed as well as the possible resolution that can be obtained. In TEM-ET, micrographs of the sample are obtained from various orientations by tilting the specimen, and the resulting micrographs are computationally combined to form a 3D volume (Ercius et al., 2015, Hoppe 1974). Developments in hardware and software during recent years have allowed automatization of the image acquisition process. These innovations enable the production of tomograms, which allows appreciation of cellular organelle ultrastructure and inter-organelle connections in unprecedented detail in 3D. The 3D volumes are useful for studies to determine how cellular compartments interact since they display the spatial organization of structures within the volume, whereas a single 2D image provides only a projection of this information. Interestingly, 3D-ET data showed connections between VIs, SOs, and MCs, the three main components of the reovirus egress pathway. Furthermore, we were able to determine the size of the channels that connect the egress compartments (around 90-100 nm in diameter), which could not be established by analysis of ultra-thin sections (50-70 nm). We were also able to observe that viral egress from MCs to the extracellular medium is mediated by membrane fusion events at the plasma membrane. These events were visualized using double-tilt ET, which orients the specimen around two orthogonal axes, resulting in two independent tomograms computed from each tilt series. The two tomograms were aligned with each other and combined to obtain a single tomogram. As a result, a dual-axis tomogram shows better resolution at any orientation in the plane of the specimen than a single-axis series, in which the specimen is tilted about only one axis (Mastronarde 1997).
In this Bio-protocol, we describe the use of 3D-ET to study the reovirus nonlytic egress pathway and define the morphology and connections between the cellular organelles that compose the exit route. This methodology provides great potential to characterize many other cellular pathways in which different organelles are involved and connected as well as the complexity of pathogen-cell interactions.
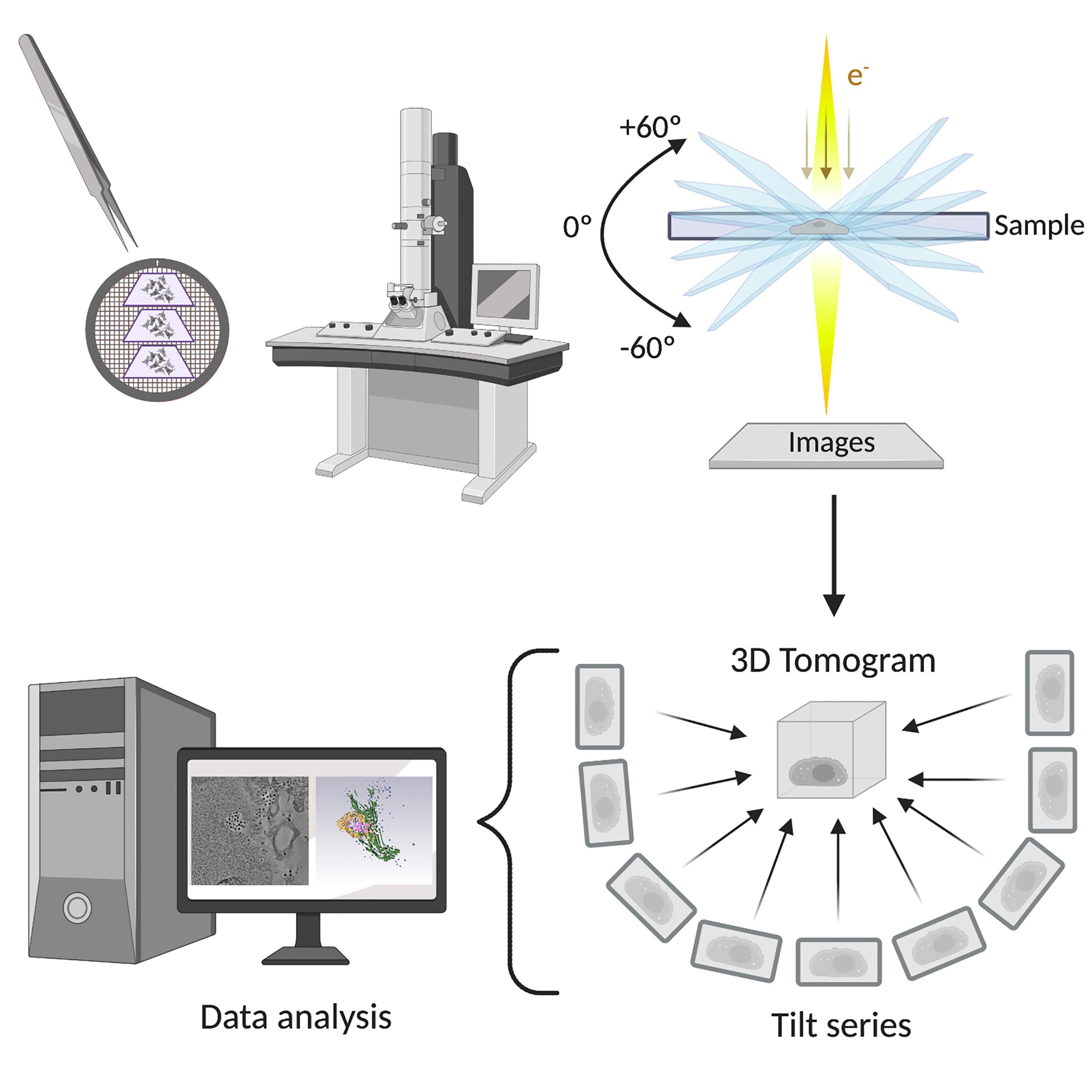
Figure 1. Electron tomography of the reovirus egress pathway workflow. Sections of reovirus-infected cells were imaged by transmission electron microscopy to discern reovirus exit zones. The structures of interest were captured, and tilt series were obtained. After image processing and reconstruction, the 3D tomogram can be analyzed. Created with BioRender.com. © 2020 Fernández de Castro et al., originally published in J Cell Biol https://doi.org/10.1083/jcb.201910131.
Materials and Reagents
Human brain microvascular endothelial cells (HBMECs) (Stins et al., 1997; Stins et al., 2001)
Reovirus strain T1L M1-P208S was recovered from plasmids by reverse genetics as described previously (Kobayashi et al., 2007 and 2009). This strain was engineered by the Terence Dermody Lab.
Roswell Park Memorial Institute (RPMI)-1640 Medium (Merck, catalog number: R8758)
Trypsin-ethylenediaminetetraacetic acid (EDTA) solution 10× (Merck, catalog number: 9002-07-7)
Fetal bovine serum (FBS) (BI Biological Industries, catalog number: 04-007-1A)
Corning® Nu-SerumTM IV Growth Medium Supplement (Corning®, Life Sciences, catalog number: 55004)
Minimum Essential Medium (MEM) vitamin solution 100× (ThermoFisher Scientific, GibcoTM, catalog number: 11120052)
Sodium pyruvate solution (Merck, Sigma-Aldrich, catalog number: 113-24-6)
MEM non-essential amino acid Solution 100× (Merck, catalog number: M7145)
L-glutamine (Merck, catalog number: 56-85-9)
Penicillin-streptomycin (Merck, Sigma-Aldrich, catalog number: P4333)
Amphotericin B solution (Merck, Sigma-Aldrich, catalog number: 1397-89-3)
4-(2-hydroxyethyl) piperazine-1-ethanesulfonic acid, N-(2-Hydroxyethyl) piperazine-N’-(2-ethanesulfonic acid) HEPES (Merck, Sigma-Aldrich, catalog number: 7365-45-9)
Distilled 50% glutaraldehyde (TAAB Laboratories, catalog number: G015)
Sapphire disks Ø1.4 mm (Martin Wohlwend, catalog number: 454)
NuncTM cell culture-treated multidishes (ThermoFisher Scientific, catalog number: 140675)
Bovine serum albumin (BSA) (Merck, Sigma-Aldrich, catalog number: 9048-46-8)
Phosphate-buffered saline (PBS) (Merck, Sigma-Aldrich, catalog number: P3813)
Ultrapure Milli-Q water
Dumont Tweezers type 4 (TAAB Laboratories, catalog number: T052)
Dumont Tweezers type 7 (TAAB Laboratories, catalog number: T050)
Osmium Tetroxide Electron Microscopy (EM) solution (TAAB Laboratories, catalog number: O015)
Uranyl acetate (Electron Microscopy Sciences, catalog number: 22400)
Methanol dried (Merck, catalog number: 67-56-1)
Acetone dried (Merck, catalog number: 1-00299-0500)
812 resin (TAAB Laboratories, catalog number: T026)
Dodecenyl succinic anhydride (DDSA) (TAAB Laboratories, catalog number: D027)
Methyl nadic anhydride (MNA) (TAAB Laboratories, catalog number: MG12)
Benzyldimethylamine (BDMA) (TAAB Laboratories, catalog number: B022)
BEEM® embedding capsule 00 (TED PELLA, INC., catalog number: 130)
GEM® stainless-steel blade (TED PELLA, INC., catalog number: 62-0179)
Perfect loop (Electron Microscopy Sciences, catalog number: 70945)
Ultra-diamond knife 45° dry (DIATOME, catalog number: DU4530)
QUANTIFOIL® R 3.5/1 Holey Carbon Film Grids Cu 300 mesh (QUANTIFOIL®, catalog number: Q11394)
Protein A Gold 10 nm (PAG10). The source is the Cell Microscopy Core (Department of Cell Biology, University Medical Center Utrecht)
Ethanol absolute (100% ethanol) (Merck, catalog number: 64-17-5)
Liquid nitrogen
Equipment
Water purification system (Merck, Milli-Q®, model: Advantage A10)
pH meter Basic 20 (HACH LANGE SPAIN, Crison, model: Basic 20)
Carbon coating system (Leica Microsystems, model: Leica EM MED020)
Cell incubator (BINDER, model: CB 170)
High-pressure vitrification system (Leica Microsystems, model: Leica EM PACT2)
Automatic cryosubstitution system (Leica Microsystems, model: Leica EM AFS2)
Fume hood (Flow-Tronic)
Ultramicrotome (Leica Microsystems, model: Leica EM UC6)
Field Electron and Ion Company (FEI) Tecnai electron microscope (FEI, model: G2 F20 (200 kV))
Charge-couple device (CCD) camera (FEI, model: Eagle 4k×4k)
Software
Tomography 3 (https://www.thermofisher.com/) (EM software, ThermoFisher Scientific). This software was used for Tecnai imaging and tilt series acquisitions.
IMOD (Kremer et al., 1996) (https://bio3d.colorado.edu/imod/) was used for the alignment of the raw tilt series and tomogram generation.
Amira (https://www.thermofisher.com/) (2D-5D visualization and analysis software, ThermoFisher Scientific). Amira was used for tomogram segmentation, reconstruction, and visualization.
Procedure
Note: This protocol involves the manipulation of dangerous compounds, namely osmium tetroxide, glutaraldehyde, epoxy resin, and uranyl acetate. Osmium tetroxide, glutaraldehyde, and epoxy resin are a contact and inhalation hazard. Prepare and use these toxic compounds in a chemical fume hood and wear gloves. Uranyl acetate is a uranium salt that does not represent a significant external radiation hazard, as the alpha particles do not penetrate the external layer of skin. There are beta and gamma emitters, but the betas also do not have enough energy to penetrate the skin, and the amount of gamma radiation is minimal. Despite this, uranyl acetate has associated chemical and radiological hazards, and some basic safety precautions must be adopted, with emphasis on avoiding the possibility of inhalation or ingestion of the material. Uranyl acetate has to be stored and disposed of as a low-level radioactive compound. Please read the official guideline of each compound carefully before use and follow the local safety regulations.
Culturing cells on sapphire disks and viral infection
Before plating cells, the 50-µm-thick sapphire disks (1.4 mm in diameter) must be carefully cleaned with 100% ethanol and covered with a carbon layer of 4-7-µm thickness using a coating device. Most cell lines propagate most efficiently using this substrate. To know on which side of the disk the cells are, it is useful to scratch an asymmetric letter or a number in the carbon.
The carbon coating must be stabilized to avoid detachment from the disks. To stabilize before use, place the carbon-coated disks in a glass dish and bake overnight at 120°C. If the disks will not be used right away, they can be stored in a humid atmosphere in a refrigerator at 4°C for 2-4 weeks. After storage, the disks should be glow-discharged before use as a cell substrate. Finally, the disks should be sterilized prior to culturing cells (e.g., using ultraviolet radiation).
Note: The glow-discharge treatment of TEM grids makes the carbon film surface negatively charged, which allows aqueous solutions to spread easily and facilitates attachment of cells and particles.
Place the sapphire disks in plastic dishes. HBMECs (5 × 105 cells per well) are added and maintained in 2 ml complete RPMI-1640 medium (see Recipes) supplemented with 10% heat-inactivated fetal bovine serum (FBS).
Cells are incubated at 37°C and 5% CO2 for 24 h and adsorbed with reovirus strain T1LM1 P208S at a multiplicity of infection (MOI) of 1 plaque-forming unit (PFU)/cell (stock diluted in culture medium) at 37°C for 1 h.
Note: The maintenance medium must be aspirated prior to reovirus absorption.
After the inoculum is removed, fresh RPMI medium supplemented with 2% FBS is added, and cells are incubated for 18 h post-infection (hpi).
For biosafety reasons, HBMECs are fixed with 1 ml 1% glutaraldehyde in HEPES buffer (see Recipes) at room temperature (RT) for 1 h.
Note: Prior to fixation, the culture medium must be aspirated.
Video 1. Coating of the sapphire disks using the Leica EM MED020
High pressure freezing (HPF)Note: HPF was accomplished using the Leica (EM PACT2) HPF instrument. For other HPF instruments, different sized sapphire discs may be required, and the loading protocol may differ.
Remove the disks from the culture dish and dip them into 20% BSA in PBS.
Place sapphire disks with cells facing into the cavity of the flat specimen holders filled with 20% BSA (see Figure 2A, Verkade, 2008). The BSA functions as a cryoprotectant. Other cryoprotectants can be used, including FBS and normal goat serum [NGS]. Ensure that there are no air bubbles in the liquid present in the sample holder.
The flat specimen holders are placed in a sample holder pod and tightly sealed by pressing them against a surface made of black diamond (see Figure 2).
Load the sample holder in the Leica EM PACT2 with the loading stage (see Figure 2).
To freeze the sample, on the touch screen press “start,” and the system starts the high pressure freezing. After freezing is complete, the pod loaded with the sample falls automatically into a liquid nitrogen bath.
Check the cooling rate and the pressure rise on the screen.
Place the pod holder in the socket bath of the liquid nitrogen and unscrew the pod holder, leaving frozen samples in the bottom of the bath.
Following freezing, the flat specimen holders are transferred under liquid nitrogen from the EM PACT2 instrument to a Leica AFS2 freeze substitution system pre-cooled to -90°C.
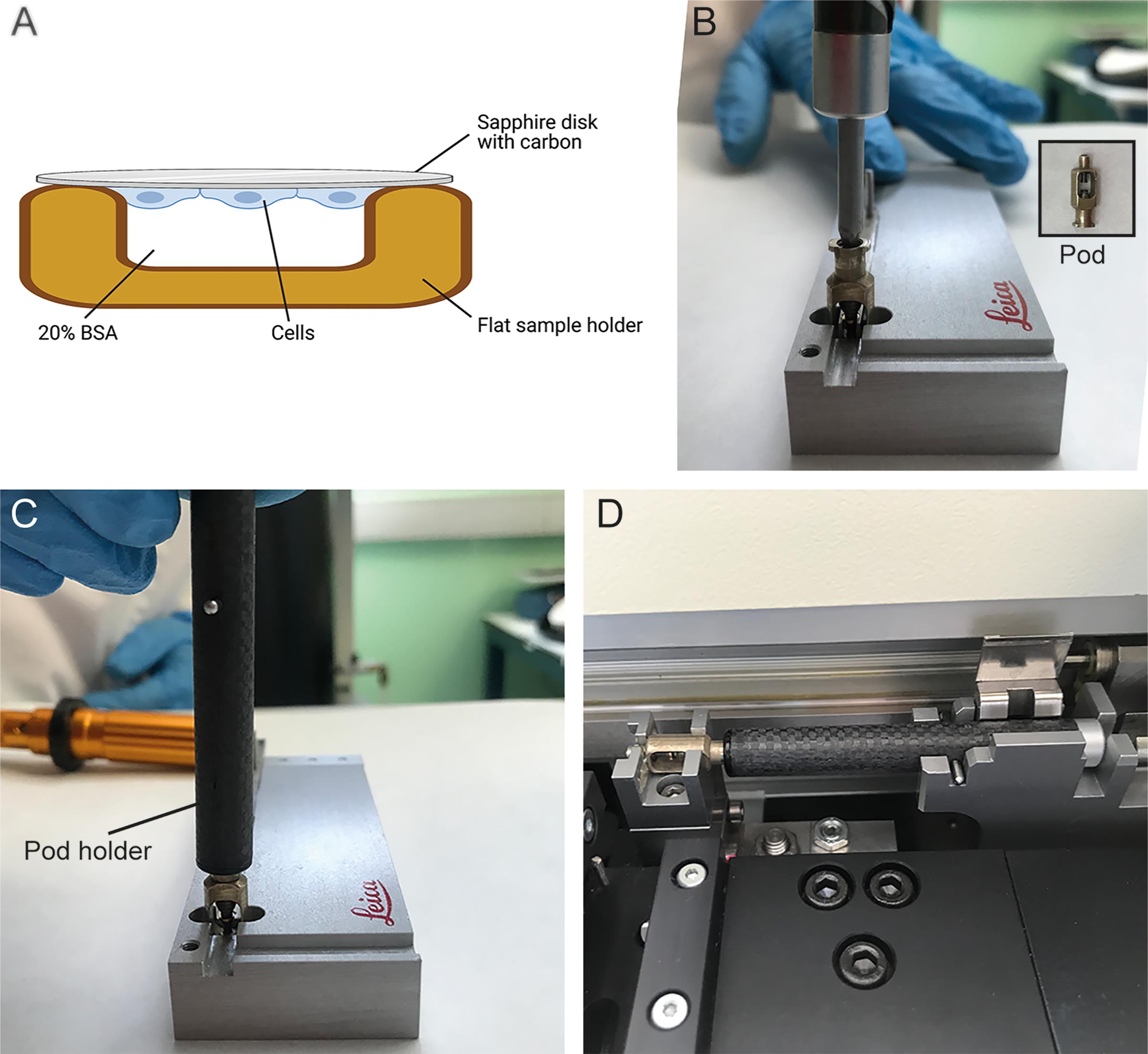
Figure 2. Loading of sapphire disks for high pressure freezing. (A) Diagram showing the sandwich of the sapphire disk with the flat sample holder for HPF (Step B2). Prepared with BioRender.com. (B) A torque screwdriver was used to tighten the flat sample holder containing the sapphire disk in the pod (Step B3). (C) A pod holder was used to pick up and introduce the pod with the sample to the Leica EM PACT2 (Step B3). (D) The pod holder was charged in the loading stage of the Leica EM PACT2 (Step B4).
Freeze substitution and embedding in epoxy resin
Note: Always maintain the frozen samples in liquid nitrogen before loading into the freeze substitution liquid. All instruments (e.g., forceps to transfer sample to the tube containing the substitution liquid) and solutions should be precooled before use.
Samples are freeze-substituted at -90°C for 24 h in a mixture of 1% osmium tetroxide, 0.1% uranyl acetate, 5% water, and 2% methanol in dry acetone.
The temperature is increased to -25°C at a rate of 2°C/h, and finally to 0°C in increments of 5°C/h.
Following completion of the -90°C to 0°C transition, the machine can be held at 0°C for 1-2 h. This procedure increases contrast of the sample. If the duration is longer than 2 h, membranes will not be as well-preserved.
All of the following steps are conducted at RT. Wash samples three times for 20 min with 1 ml dried-pure acetone.
The samples were gradually infiltrated with mixtures of acetone-epoxy resin (see Recipes), increasing the concentration of resin in steps of 30%, 50%, 70%, and 90% for 1 h each.
For infiltration in pure epoxy resin, the sapphire disks are separated from the flat sample holders using tweezers and placed into flat-bottomed beam capsules.
Capsules are filled with freshly prepared epoxy resin for polymerization at 60°C for 2 days.
Ultramicrotomy
Beam capsules are removed using a stainless-steel blade, and the sapphire disks are separated from the resin blocks with thermal shocks. The samples are loaded into liquid nitrogen for a few seconds and then returned to room temperature. Repeat these two steps until the sapphire disks are detached from the resin blocks.
Samples are cut into small pieces (~1 × 1 mm) using a stainless-steel blade and affixed to a resin block as a holder (see Figure 3).
The sample surface is hand-trimmed to obtain a trapezoidal shape using a new stainless-steel blade.
Collect semi-thick sections (~ 350-400 nm) using a ultramicrotome UC6 with a perfect loop and place the sections in Quantifoil® cooper grids.
Note: Other types of grid can be used for ET.
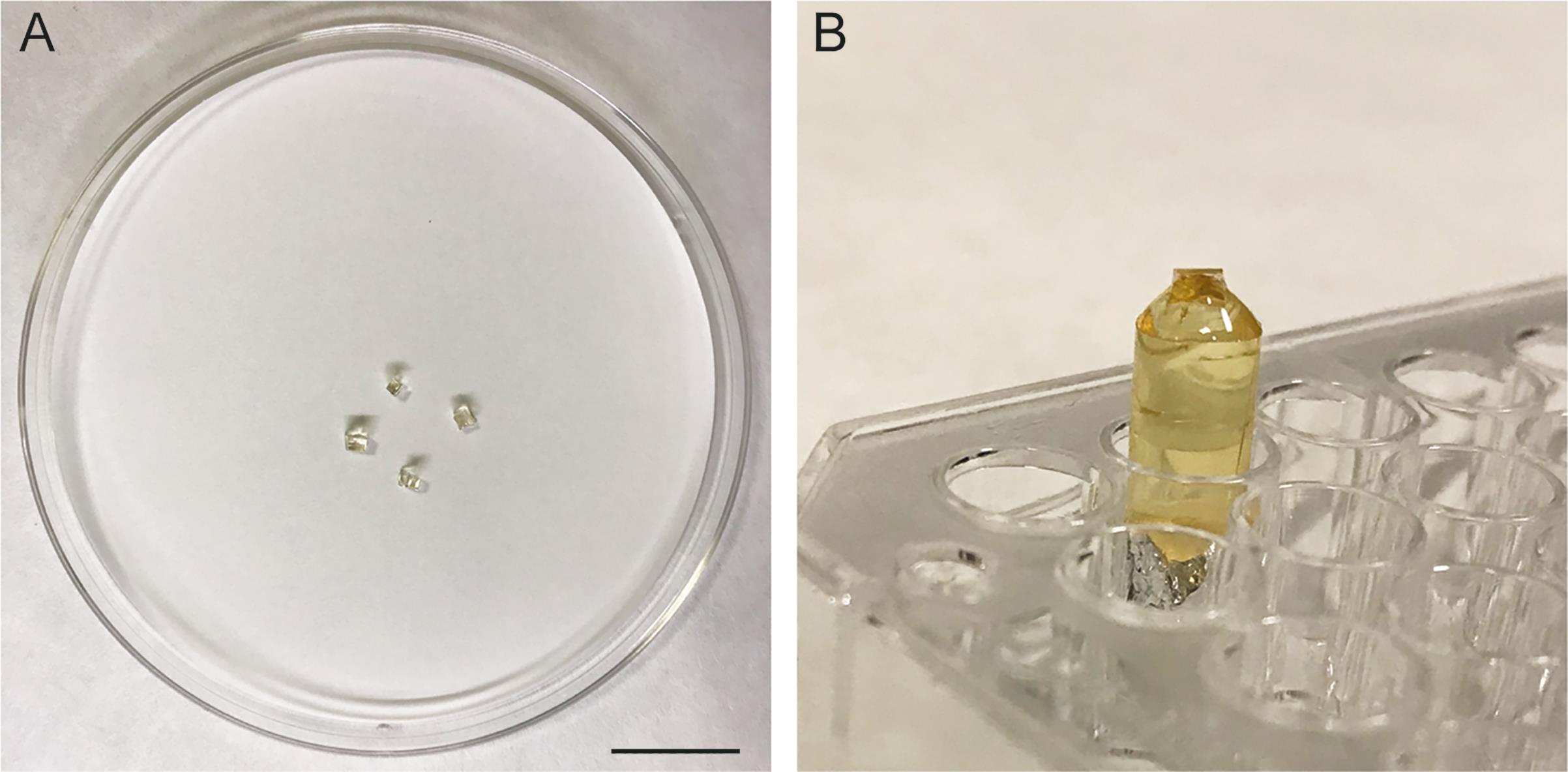
Figure 3. Mounting the samples for ultramicrotomy. (A) After detachment of the sapphire disc, the resin with embedded cell monolayers is cut into small pieces of about 1-mm in size. (B) The small pieces of sample are glued onto cylindrical resin blocks, which allows mounting of the sample in the sample holder of the microtome. Scale bar = 10 mm.
Coating grids with fiducial markers (protein A gold spheres [10 nm diameter], PAG10) on both sides of the section for better alignment and staining with uranyl acetate. The protocol is summarized in Figure 4.
Note: The fiducial markers are gold particles of 10 nm diameter required for the alignment of the tilt images during tomogram processing prior to image reconstruction.
On a parafilm sheet, place one drop per sample of the protein A gold solution (see Recipes) and three drops per sample of water.
Incubate the grids on the gold drops for 10 min.
Sequentially lay the grids for a few seconds on the three water drops to remove excess gold solution.
Absorb the excess liquid with absorbent filter paper.
Repeat Steps E1-E4, incubating the other side of the grid.
Let the grids dry for at least 2 h.
Note: To increase the contrast of the images, stain the sections with uranyl acetate.
On a parafilm sheet, place one drop of 2% uranyl acetate (see Recipes) and incubate the grids for 20 min.
Wash the grids with three drops of water for a few seconds to remove the uranyl acetate.
Absorb the excess liquid with filter paper.
Allow the grids to dry for at least 2 h prior to imaging with an electron microscope.
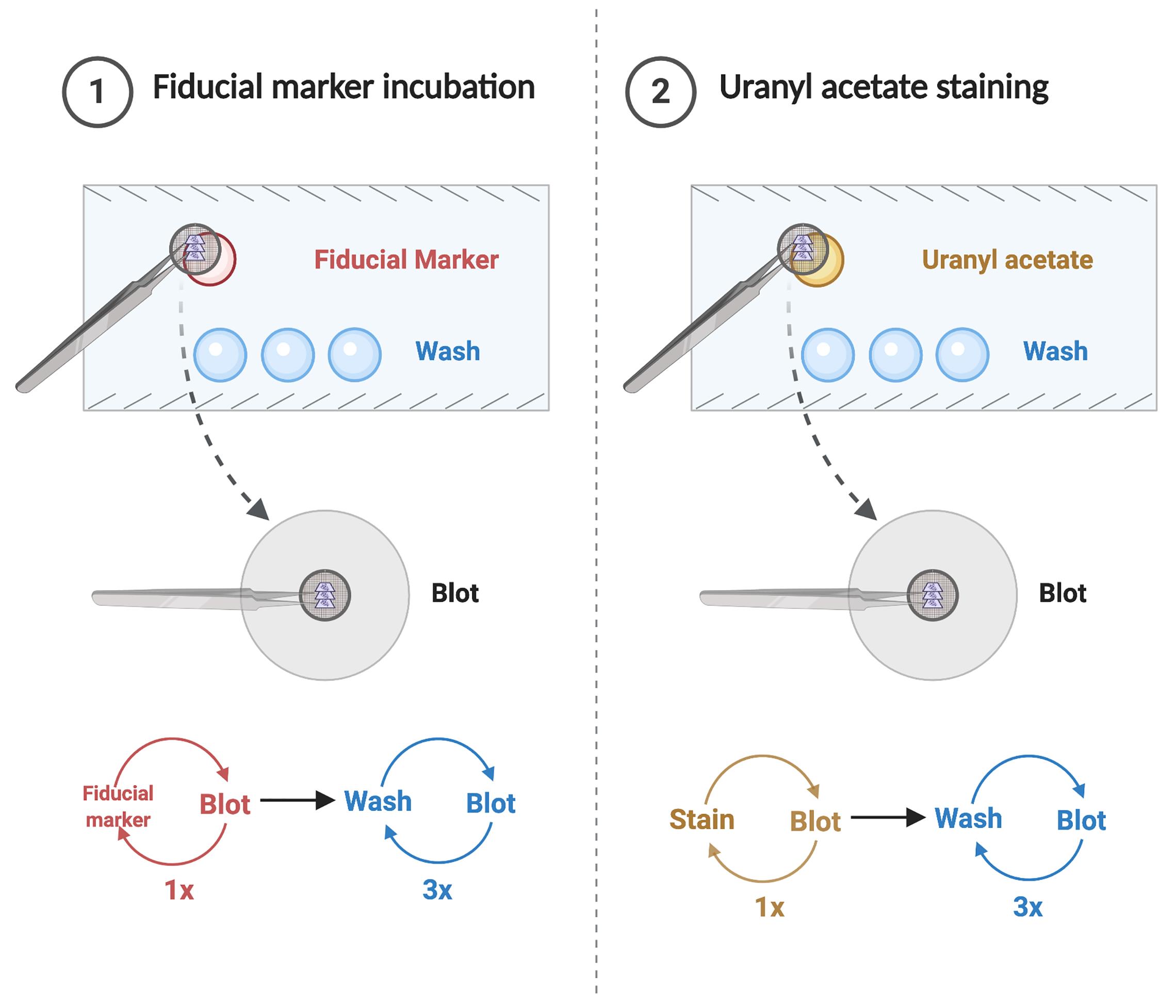
Figure 4. Schematic of grid preparation prior to electron tomography. Created with BioRender.com.
Electron tomography
Examine the sections using an electron microscope for reovirus egress zones, selecting and saving the regions of interest (ROIs) for the tilt series.
Pre-irradiate the ROIs prior to collection of the tilt series. The electron beam produces damage in resin-embedded samples, resulting in specimen shrinkage, especially along the Z-axis (Luther et al., 1988). To avoid this problem during tomogram acquisition, it is important to pre-irradiate the ROIs, including a larger area than the ROI since outside areas could later enter in the field.
Notes:
About 2,000 electrons per square Angstrom at a low magnification (2,000×) of EM-irradiation is enough to allow the ROIs to assume sufficient size and stability.
The ROIs should not be close to the grid bars, as the grid bars will block acquisition at high tilt angles.
Single- and dual-axis ET is conducted using a Tecnai G2 F20 transmission electron microscope (FEI) operated at 200 kV.
The specimen is tilted within the electron microscope to produce transmission electron microscopy images from many different angles, resulting in a tilt series.
Acquire tilt series automatically at 1° increments over an angule range of −60° to 60°. The camera parameters should be set to allow optimum resolution and contrast for the sample. In our studies, we acquired the tilt series with a nominal magnification of ×11,500, resulting in a pixel size of 1.01 nm.
Images are recorded using an Eagle 4k×4k slow-scan charge-coupled device (FEI) and Tomography 3 software.
To conduct dual-axis ET, collect a second tilt series after rotation of the specimen by 90° around an axis perpendicular to its support plane.
Note: Dual-axis or double-tilt ET increases the information collected and reduces artifacts of acquisition and reconstruction (Mastronarde, 1997).
Tomogram processing
Note: The different perspectives obtained by ET are used to reconstruct a 3D view, which resolves the ultrastructure of the reovirus egress organelles in three dimensions.
Download and install the IMOD software to align tilt series and obtain tomographic reconstructions.
Start eTomo, providing the entries in the Setup Panel. The important parameters regarding the tilt series are:
Axis type: single or dual axis
Frame type
Pixel size and fiducial diameter
Tilt angles
Note: In this Setup Panel, it is possible to remove excluded views (images that are suboptimal for analysis).
Once the Setup Panel is complete, activate Create Com Scripts.
Preprocessing. Run Ccderaser to erase X-rays and other artifacts from the images. The X-ray model creates a fixed stack that can be saved and used.
Coarse Alignment. Calculate the cross-correlation with Tiltxcorr to determine the X and Y translations required to align each image with the previous one, generating the coarse aligned stack.
Fiducial Model Generation. There are several ways to generate a fiducial model. We selected well-distributed fiducials or gold beads on the zero-degree view and used Beadtrack to find their locations in the rest of the tilt series. Finally, a Seed Model was created. The seed points can be marked manually or automatically. An overview of the alignment process is shown in Figure 5.
Fine Alignment. Perform the alignment using Tiltalign and examine the results in the log file after the process is complete. It is possible to view a model of the Residual Vectors, which gives an idea of the alignment quality (see Figure 5). The smaller the residual vectors, the better the alignment. A typical mean residual value of a good alignment is around 1 pixel.
Tomogram Positioning. Prepare a simple model with horizontal lines across the top and bottom faces of the section at three locations in Y. It is important to delineate the thickness of the sections. After defining the edge of the section, select Find boundary model automatically, creating the Boundary Model (see Figure 5).
Final Aligned Stack. Use Newstack to obtain the aligned stack. As an option, it is possible in this step to correct the microscope CTF (contrast transfer function) and filter the aligned stack.
Tomogram Generation. Use Tilt to generate the tomogram.Note: In double-tilt tomography, both tomograms, called A and B, are processed separately. Tomogram A is first aligned, indicating that it is dual axis. Tomogram B is aligned with reference to tomogram A using its fiducials (matching the tomogram B to A). As a result, a combined tomogram is prepared.
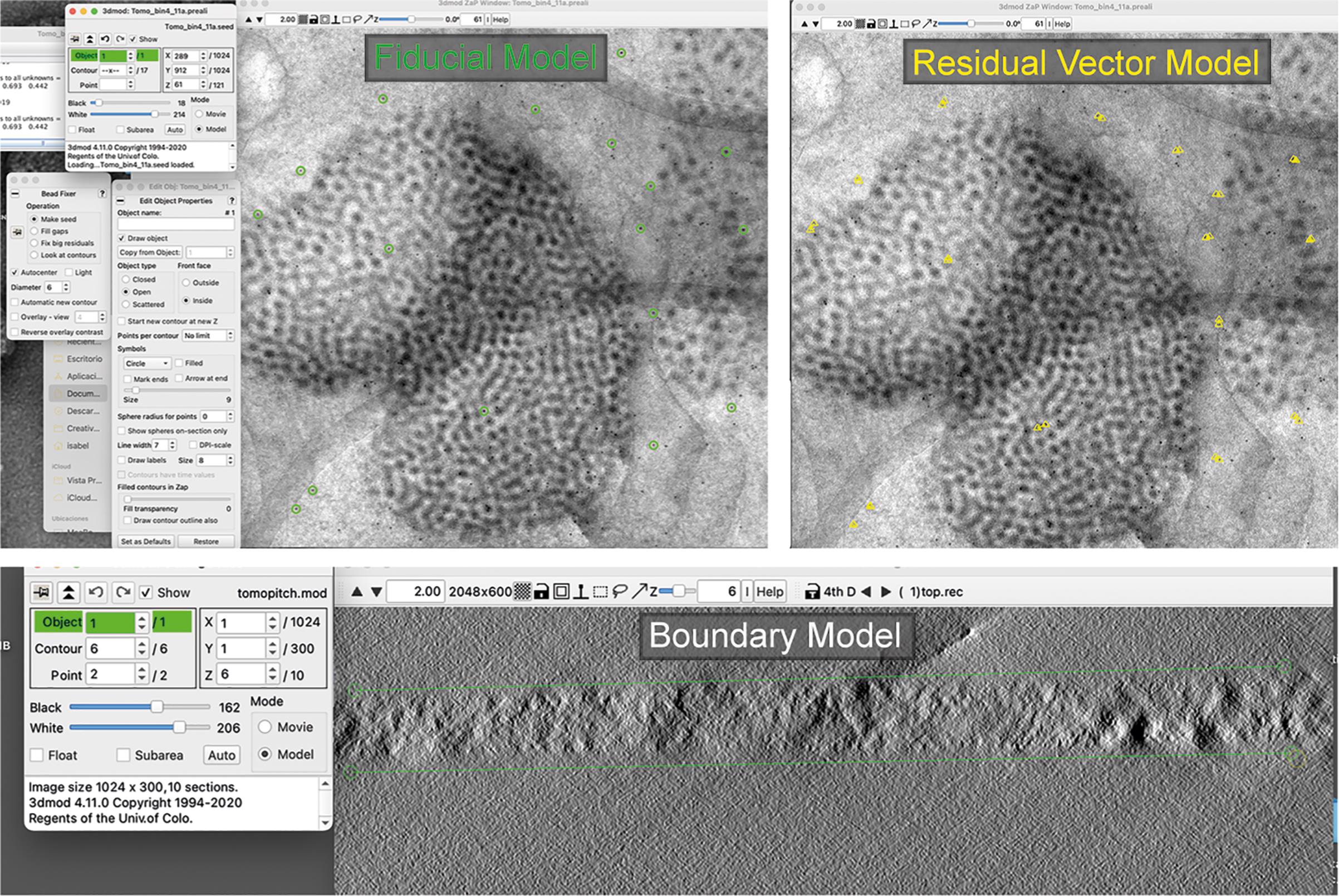
Figure 5. Screen captures of the IMOD software. Representative models constructed using tomogram alignment are shown.
Tomogram segmentation and 3D visualization
The tomogram segmentation and 3D visualization were conducted with the Amira Software.
Load the tomogram (.rec) in the Amira interface and apply a Gaussian filter (3D type) for noise reduction.
An Orthoslice can be made to visualize and compare the tomogram with the filtered tomogram.
The filtered tomogram was segmented with the function LabelField, which allows the area of the different structures to be marked in the images. Labeling a structure in all images of the tomogram is tedious work, which can be simplified with the Interpolate function. The Interpolate function allows the segmentation to be done automatically after marking the images.
Note: It is important to verify that the Interpolate function is working well, as occasionally manual corrections of the automatic segmentation must be made to some of the labeled images or structures.
The functions Arithmetic and Isosurface were used to visualize the structures in 3D.
Note: The threshold values of labeled structures are obtained using histograms for each structure. Adjusting the threshold value using Isosurface improves visualization of the reconstructions.
Data analysis
We used 3D-ET to analyze the reovirus egress pathway. The tomogram shown in Figure 6 was obtained using a semi-thick section (350 nm), which allowed us to identify the spatial relationship between the different structures. The analysis of the VI-SO-MC interfaces by ET revealed the presence of physical connections between these structures (Figure 6). The computational slices of the tomogram in combination with the 3D visualization revealed that there are points of membrane continuity between VIs and SOs (dashed box in Figure 6A) and SOs and MCs (dashed box in Figure 6B), respectively. These interactions are restricted to a few computational slices, suggesting that the association between these compartments occurs through discrete channels observed in the higher-magnification images shown in Figure 6A and B.
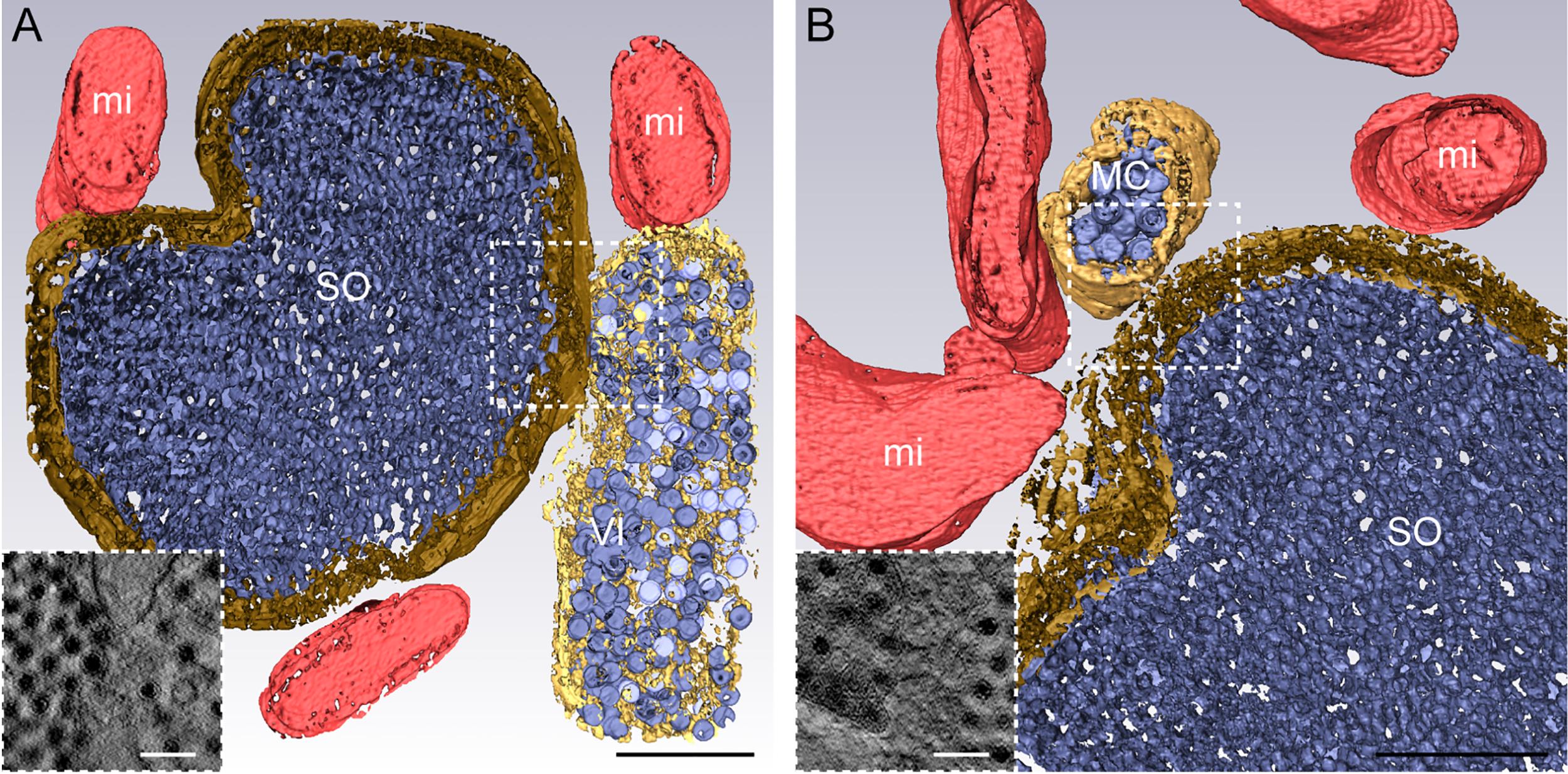
Figure 6. Electron tomography of the reovirus egress machinery. (A and B) HBMECs were adsorbed with reovirus T1L M1-P208S at an MOI of 1 PFU/cell and processed at 18 hpi by high-pressure freezing, freeze substitution, and semi-thick sectioning. Single-tilt tomography and 3D reconstruction show the connection between the components of the reovirus egress machinery (dashed boxes). Higher magnifications of the specific membrane fusion points between a sorting organelle (SO) and a viral inclusion (VI) in (A) or a membranous carrier (MC) adherent to an SO through a channel in (B). mi, mitochondrion. Scale bars = 500 nm A and B; 100 nm in the higher magnifications. © 2020 Fernández de Castro et al. originally published in J Cell Biol https://doi.org/10.1083/jcb.201910131.
We used double-tilt ET to study the final steps of the reovirus egress pathway (Figure 7). Examination of the MCs at the plasma membrane by ET and 3D reconstruction revealed details of the carrier morphology before egress and the virion exit mechanism. Mature virions are distributed along arrays of membranes or channels inside the MCs (Figure 7A), which may increase the carrying capacity of the MC or the efficiency of viral transport. The 3D volume depicts the total content of the MC and its association with the plasma membrane (Figures 7B and C). Therefore, progeny reovirus particles exit infected cells through distinct egress zones after virion-filled MCs contact the plasma membrane (Fernandez de Castro et al., 2020).
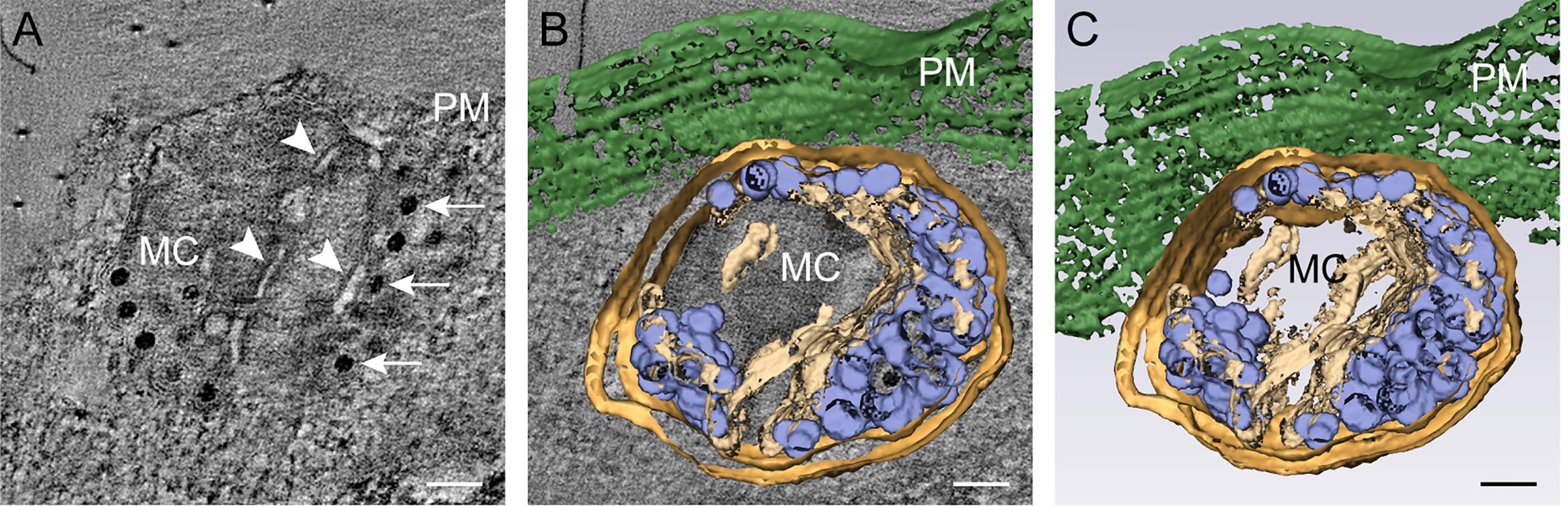
Figure 7. Electron tomography of a membranous carrier. HBMECs were adsorbed with reovirus T1L M1-P208S at an MOI of 1 PFU/cell and processed at 18 hpi by high-pressure freezing, freeze substitution, and semi-thick sectioning. (A) Computational tomography slice showing a membranous carrier (MC) with membranous channels (white arrowheads) and virions (white arrows) close to the plasma membrane (PM). (B) A 3D reconstruction of the MC overlapping with a 2D computational slice. (C) One view of the 3D volume showing the total content of the MC after segmentation. Mature virions, blue; channels, beige; membranous carrier, light brown; plasma membrane, green. Scale bars = 200 nm. © 2020 Fernández de Castro et al. originally published in J Cell Biol https://doi.org/10.1083/jcb.201910131.
The ET experiments were conducted with at least two biological replicates. Eight tomograms showing different reovirus egress zones were recorded, and the VI-SO-MC interface was analyzed from nine tomograms. Statistical significance was determined using a two-sample unequal variance t-test with two-tailed distribution.
Notes
The reovirus strain T1L M1-P208S is identical to the prototype T1L strain except for a proline-to-serine substitution at position 208 of the μ2 protein (encoded by the M1 gene). This mutation changes inclusion morphology from filamentous to globular (Parker et al., 2002).
In the tilt series alignment, the most critical part is the distribution of the fiducial markers in the sections. Only those fiducials maintained in every section should be marked at the same Z-plane, and they should be well-distributed throughout the sections.
Recipes
Complete RPMI-1640 medium
10% Nu serum
1% MEM vitamins
1% sodium pyruvate
1% MEM nonessential amino acids
1% L-glutamine
1% penicillin/streptomycin
0.1% amphotericin
Store at 4°C
HEPES 0.4 M
9.54 g HEPES
Add Milli-Q water to 50 ml and mix well until HEPES is dissolved entirely
Adjust pH with 1N NaOH solution to 7.2-7.4
Add Milli-Q water to 100 ml
Store at 4°C
Epoxy resin
Use plastic Pasteur pipettes to weigh the epoxy resin components.
Mix the resin components by inverting the tube several times.
The accelerator (BDMA) has to be added at the moment of use.
Centrifugate the mixture at 14,100 × g for 10 min to eliminate small bubbles.
19.2 g 812 resin
7.6 g DDSA
13.2 g NMA
0.8 g (or 20 drops) BDMA
Protein A Gold or fiducial marker solution
Mix colloidal gold solution in water (1:50 ratio)
Prepare this solution fresh every time
Note: The dilution (and size) of the fiducial marker depends on the magnification of the tilt series acquisition. The higher the magnification, the denser the fiducials and the smaller the gold size.
Acknowledgments
This work was supported in part by Public Health Service awards AI032539 (T.S.D. and C.R.). Special thanks to Dr. Martin Sachse for expert technical advice and support throughout this work and for critical review of the manuscript, Dr. Francisco Javier Chichón from the CNB-CIB (CSIC) cryoEM facility for technical advice with ET, and Cristina Patiño, Beatriz Martín, and Pablo Solá from the CNB (CSIC) EM facility. This protocol was adapted from previous work: Fernández de Castro et al., originally published in J Cell Biol https://doi.org/10.1083/jcb.201910131.
References
- Altan-Bonnet, N. (2017). Lipid Tales of Viral Replication and Transmission. Trends Cell Biol 27(3): 201-213.
- Bird, S. W. and Kirkegaard, K. (2015). Escape of non-enveloped virus from intact cells. Virology 479-480: 444-449.
- Danthi, P., Holm, G. H., Stehle, T. and Dermody, T. S. (2013). Reovirus receptors, cell entry, and proapoptotic signaling. Adv Exp Med Biol 790: 42-71.
- Ercius, P., Alaidi, O., Rames, M. J. and Ren, G. (2015). Electron Tomography: A Three-Dimensional Analytic Tool for Hard and Soft Materials Research. Adv Mater 27(38): 5638-5663.
- Fernandez de Castro, I., Tenorio, R., Ortega-Gonzalez, P., Knowlton, J. J., Zamora, P. F., Lee, C. H., Fernandez, J. J., Dermody, T. S. and Risco, C. (2020). A modified lysosomal organelle mediates nonlytic egress of reovirus. J Cell Biol 219(7).
- Hoppe W. (1974). Towards three-dimensional "electron microscopy" at atomic resolution. Naturwissenschaften 61(6):239-49.
- Kobayashi, T., Antar, A. A., Boehme, K. W., Danthi, P., Eby, E. A., Guglielmi, K. M., Holm, G. H., Johnson, E. M., Maginnis, M. S., Naik, S., Skelton, W. B., Wetzel, J. D., Wilson, G. J., Chappell, J. D. and Dermody, T. S. (2007). A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1(2): 147-157.
- Kobayashi, T., Ooms, L. S., Chappell, J. D. and Dermody, T. S. (2009). Identification of functional domains in reovirus replication proteins muNS and mu2. J Virol 83(7): 2892-2906.
- Kremer, J. R., Mastronarde, D. N. and McIntosh, J. R. (1996). Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116(1): 71-76.
- Lai, C. M., Mainou, B. A., Kim, K. S. and Dermody, T. S. (2013). Directional release of reovirus from the apical surface of polarized endothelial cells. mBio 4(2): e00049-00013.
- Luther, P. K., Lawrence, M. C. and Crowther, R. A. (1988). A method for monitoring the collapse of plastic sections as a function of electron dose. Ultramicroscopy 24(1): 7-18.
- Mastronarde, D. N. (1997). Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol 120(3): 343-352.
- Miller, S. and Krijnse-Locker, J. (2008). Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6(5): 363-374.
- Parker, J. S., Broering, T. J., Kim, J., Higgins, D. E. and Nibert, M. L. (2002). Reovirus core protein mu2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J Virol 76(9): 4483-4496.
- Roth, A. N., Aravamudhan, P., Fernandez de Castro, I., Tenorio, R., Risco, C. and Dermody, T. S. (2021). Ins and Outs of Reovirus: Vesicular Trafficking in Viral Entry and Egress. Trends Microbiol 29(4): 363-375.
- Sachse, M., Fernandez de Castro, I., Tenorio, R. and Risco, C. (2019). The viral replication organelles within cells studied by electron microscopy. Adv Virus Res 105: 1-33.
- Stins, M. F., Badger, J. and Sik Kim, K. (2001). Bacterial invasion and transcytosis in transfected human brain microvascular endothelial cells. Microb Pathog 30(1): 19-28.
- Stins, M. F., Gilles, F. and Kim, K. S. (1997). Selective expression of adhesion molecules on human brain microvascular endothelial cells. J Neuroimmunol 76(1-2): 81-90.
- Tenorio, R., Fernandez de Castro, I., Knowlton, J. J., Zamora, P. F., Lee, C. H., Mainou, B. A., Dermody, T. S. and Risco, C. (2018). Reovirus sigmaNS and muNS Proteins Remodel the Endoplasmic Reticulum to Build Replication Neo-Organelles. mBio 9(4).
- Verkade, P. (2008). Moving EM: the Rapid Transfer System as a new tool for correlative light and electron microscopy and high throughput for high-pressure freezing. J Microsc 230(Pt 2): 317-328.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Fernandez de Castro, I., Fernández, J. J., Dermody, T. S. and Risco, C. (2021). Electron Tomography to Study the Three-dimensional Structure of the Reovirus Egress Pathway in Mammalian Cells. Bio-protocol 11(13): e4080. DOI: 10.21769/BioProtoc.4080.
- Fernandez de Castro, I., Tenorio, R., Ortega-Gonzalez, P., Knowlton, J. J., Zamora, P. F., Lee, C. H., Fernandez, J. J., Dermody, T. S. and Risco, C. (2020). A modified lysosomal organelle mediates nonlytic egress of reovirus. J Cell Biol 219(7).
Category
Cell Biology > Cell structure > Cell organelle
Biophysics > Microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link