- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fluorescence-based Heme Quantitation in Toxoplasma Gondii
Published: Vol 11, Iss 12, Jun 20, 2021 DOI: 10.21769/BioProtoc.4063 Views: 4022
Reviewed by: Alexandros AlexandratosMarcelo S. da SilvaSneha Ray

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Femtoliter Injection of ESCRT-III Proteins into Adhered Giant Unilamellar Vesicles
Vasil N. Georgiev [...] Rumiana Dimova
Feb 20, 2022 3732 Views

Artificial Metalloenzymes in Artificial Sanctuaries Through Liquid–Liquid Phase Separation
Kaixin Wang [...] Tong Wu
Oct 5, 2025 1585 Views

Dynamic Mapping of RNA-Binding Proteins During Bacillus subtilis Sporulation Using Orthogonal Organic Phase Separation
Thomas Kaboré and Clémentine Delan-Forino
Mar 5, 2026 133 Views
Abstract
Toxoplasma gondii is a highly prevalent protozoan pathogen throughout the world. As a eukaryotic intracellular pathogen, Toxoplasma ingests nutrients from host cells to support its intracellular growth. The parasites also encode full or partial metabolic pathways for the biosynthesis of certain nutrients, such as heme. Heme is an essential nutrient in virtually all living organisms, acting as a co-factor for mitochondrial respiration complexes. Free heme is toxic to cells; therefore, it gets conjugated to proteins or other metabolites to form a “labile heme pool,” which is readily available for the biosynthesis of hemoproteins. Previous literature has shown that Toxoplasma gondii carries a fully functional de novo heme biosynthesis pathway and principally depends on this pathway for intracellular survival. Our recent findings also showed that the parasite’s intracellular replication is proportional to the total abundance of heme within the cells. Moreover, heme abundance is linked to mitochondrial oxygen consumption for ATP production in these parasites; thus, they may need to regulate their cellular heme levels for optimal infection when present in different environments. Therefore, quantitative measurement of heme abundance within Toxoplasma will help us to understand the roles of heme in subcellular activities such as mitochondrial respiration and other events related to energy metabolism.
Keywords: Toxoplasma gondiiBackground
Heme, an organic molecule, plays a vital role in virtually all living organisms. For example, heme binds to hemoglobin and myoglobin for oxygen transport and serves as a co-factor for several enzymes within the electron transport chain for mitochondrial respiration (Ponka, 1999). Previous literature has shown that Toxoplasma has a fully functional de novo heme biosynthesis pathway (Bergmann et al., 2020). Genetic deletion of Toxoplasma heme biosynthetic enzymes results in decreased replication in vitro (Bergmann et al., 2020; Krishnan et al., 2020; Tjhin et al., 2020) and the loss of acute virulence in a murine model (Bergmann et al., 2020); therefore, inhibition of de novo heme production sheds light on the development of novel therapeutic strategies for managing Toxoplasma infections.
Generally, there are four methods for the measurement of heme abundance in cells: 1) Pyridine hemochrome-based heme quantitation (Sinclair et al., 2001). This strategy replaces the nitrogen ligands of protein-bound heme with pyridine under alkaline conditions. The resulting hemochrome is further reduced and oxidized before spectrophotometric quantitation; 2) Reversed-phase HPLC-based quantitation of heme and its intermediates (Sinclair et al., 2001). An acetone/HCl/water solution is used to extract heme and its biosynthetic intermediates from intact cells or cell homogenates, followed by separation on a C18 HPLC column. The standards of pure heme and its intermediates are run on the column before sample measurement to help recognize their peaks and quantitate their abundances; 3) Protoporphyrin IX (PPIX)-based fluorescence assay (Sinclair et al., 2001). PPIX is produced by the penultimate enzyme within the classic de novo heme biosynthesis pathway (Phillips, 2019). PPIX is further conjugated with a ferrous iron group to form a functional heme molecule. The non-conjugated PPIX molecule displays strong fluorescence, whereas heme is non-fluorescent; therefore, stripping ferrous ion from heme generates fluorescent PPIX, which can be quantitated by a fluorometer and represents heme abundance; 4) Biosensor-based heme quantitation by live-cell imaging or flow cytometry (Song et al., 2015; Hanna et al., 2016; Yuan et al., 2016). Genetically encoded hemoproteins, such as horseradish peroxidase (HRP)- or ascorbate peroxidase (APX)-based biosensors, can be expressed in different organelles to detect their labile heme content (Yuan et al., 2016). Additionally, the heme-binding proteins are genetically fused to heme-sensitive fluorescent proteins to quantitate labile heme within live cells by ratiometric fluorescence quantitation or fluorescence resonance energy transfer (FRET) (Song et al., 2015; Hanna et al., 2016).
The pyridine hemochrome-based strategy is not very sensitive and requires large amounts of parasites as initial material. The HPLC-derived quantitation requires expensive equipment and columns. Additionally, it only can quantitate one sample per run. The biosensor-based method demands the implementation of biosensors in target organisms. Overexpression of biosensors may disturb labile heme pools in the cells and further impair their health status. The PPIX-based fluorescence heme quantitation is economical and sensitive. Moreover, it can be performed in 96-well plate format as a semi high-throughput strategy for multiple samples simultaneously.
In the procedure for the PPIX-based heme quantitation assay, the heme-bound ferrous ion can be chemically removed by oxalic acid via boiling before measurement. During the assay, non-boiled parallel samples must be included to detect background fluorescence signals within parasite strains, which will be subtracted from the corresponding boiled samples. Here, we modify the previously published PPIX-based fluorescence heme quantitation assay (Sinclair et al., 2001). In this protocol, the parasites are mechanically liberated from host cells, filter-purified, and ruptured by sonication prior to heme quantitation. As an example, we measured and compared heme abundances in wildtype parasites and a heme-deficient transgenic Toxoplasma strain. This method employs a plate reader to quantitate heme in a 96-well plate format; it shows high sensitivity and only requires 5-10 × 107Toxoplasma parasites for measurement, which can be harvested from 1-2 T25 flasks. This strategy can be modified for measuring heme abundances in other apicomplexan parasites.
Materials and Reagents
Nucleopore track-etched hydrophilic membrane filter, 3.0-µm pore size (VWR, catalog number:28158-624) with filter holder (Fisher Scientific, catalog number: SX0002500)
15 ml polystyrene conical tubes (VWR, catalog number: 10026-084)
1.5 ml Eppendorf microcentrifuge tubes (VWR, catalog number: 89000-028)
Black 1.5 ml microcentrifuge tubes (VWR, catalog number: 47751-688)
10 ml syringes (VWR, catalog number: BD309646)
Blunt-end needles (McMaster-Carr, catalog numbers: 75165A761 [20G]; 76165A759 [25G])
Cellstar® black 96-well plates without lids (VWR, catalog number: 82050-732)
1,000 ml glass beaker (VWR, catalog number: 10754-960)
5 ml and 10 ml serological pipettes (VWR, catalog numbers: 89130-896, 89130-898)
20 µl, 200 µl, and 1,000 µl micropipette tips (VWR, catalog numbers: 76322-134, 76322-150, 76322-138)
Toxoplasma gondii RH∆ku80∆hxg strain lacking the ku80 and hypoxanthine-xanthine-guanine phosphoribosyl transferase genes. This strain is provided by the Carruthers lab at the University of Michigan Medical School. Lack of the ku80 gene boosts homology-dependent recombination in the parasites (Huynh and Carruthers, 2009). Loss of hypoxanthine-xanthine-guanine phosphoribosyl transferase (HXG) allows this strain to be genetically modified by exogenous DNA vectors carrying the HXG gene. This strain is widely used in the Toxoplasma research field as a wildtype parental strain.
RH∆ku80∆hxg∆cpox strain (abbreviated to ∆cpox). A heme-deficient parasite strain was generated in a previous study (Bergmann et al., 2020). This strain lacks coproporphyrinogen-III oxidase (TgCPOX) that catalyzes the antepenultimate reaction within the parasite’s heme biosynthesis pathway.
RH∆ku80∆hxg∆cpoxCPOX strain (abbreviated to ∆cpoxCPOX). A TgCPOX complementation strain was generated in a previous study (Bergmann et al., 2020). Heme deficit is restored in this strain.
Human Foreskin Fibroblasts (HFFs) (American Type Culture Collection (ATCC), catalog number: SCRC-1041)
Corning® Dulbecco’s Modified Eagle Medium (DMEM) (VWR, catalog number: 45000-304)
100× Penicillin-Streptomycin (VWR, catalog number: 45000-652)
200 mM L-glutamine (VWR, catalog number: 45000-676)
1 M HEPES Free Acid (VWR, catalog number: 45000-696)
Hyclone® Cosmic Calf Serum (Cytiva, catalog number: SH30087.03)
HyClone® Dulbecco’s Phosphate-Buffered Saline (DPBS) powder, without calcium and magnesium (Powder) (Cytiva, catalog number: SH30013.04)
Pig hemin powder (VWR, catalog number: BT138155-1G)
Oxalic acid dihydrate (VWR, catalog number: 97064-978)
Isopropanol (VWR, catalog number: BDH11744)
DPBS (see Recipes)
D10 medium (see Recipes)
1 mM heme solution (see Recipes)
2 M oxalic acid solution (see Recipes)
Equipment
Eppendorf CellXpert® C170 cell culture incubator (Eppendorf)
Eppendorf refrigerated centrifuge (Eppendorf, model: 5810R)
Branson Analog Sonifier ultrasonic cell disrupter S-250A (VWR, catalog number: 33995-309) with a tapered 1/8-inch microtip (VWR, catalog number: 33996-163)
Biotek Synergy H1 Hybrid Multi-Mode microplate reader (Biotek Instruments)
Micropipette set (P-1000, P-200, and P-20) (VWR, catalog number: 75788-456)
Hemocytometer (VWR, catalog number: 15170-168). Instructions can be found in http://www.hausserscientific.com/products/reichert_bright_line.html
Hot plate (VWR, Advanced Hot Plate, catalog number: 97042-642)
Software
BioTek® Gen5 Data Analysis Software
Microsoft Excel
GraphPad Prism (Version 9)
Procedure
Please see Figure 1 for a schematic description of the entire procedure.
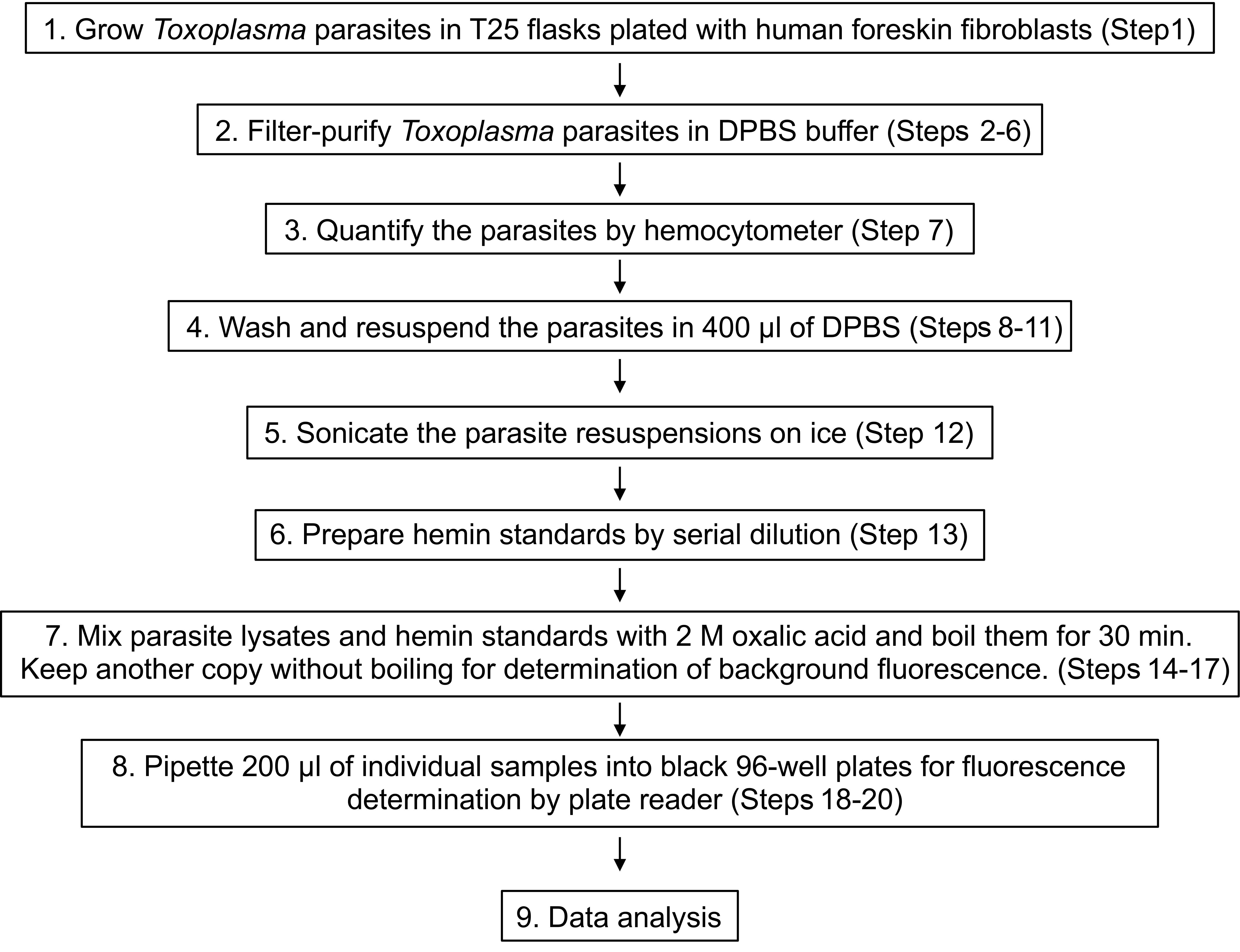
Figure 1. A schematic outline of the fluorescence-based heme quantitation in Toxoplasma gondii. Please refer to the text for more details.
In this protocol, wildtype (RH∆ku80∆hxg), ∆cpox (a heme-deficient strain), and ∆cpoxCPOX (a TgCPOX complementation strain) strains are included as examples. If parasite strains are stored at -80°C, they must be thawed at 37°C and immediately passed to a T25 tissue culture flask containing fully confluent human foreskin fibroblast (HFF) cells. After the parasites lyse the host cells, ~300 µl fully lysed wildtype and ∆cpoxCPOX parasites were passed to one T25 tissue culture flask coated with HFF cells for a 2-day passage. For the ∆cpox strain, the inoculum was increased to 2 ml to compensate for its severe growth defects. 1-2 T25 flasks of parasites per strain are needed depending on their growth phenotype. Since the ∆cpox showed severe growth defects, two flasks of ∆cpox parasites were used in this assay. The wildtype and ∆cpoxCPOX strains were passed into one T25 flask.
Pre-cool DPBS on ice and keep the infected cells on ice. Scrape the infected HFFs and syringe them three times using a 20 G needle, followed by three times using a 25 G needle. The lysates were passed through a 3.0-µm filter to remove intact host cells and large host cell debris. Rinse the flasks with 5 ml ice-cold DPBS and pass through the filter to wash.
Fill a 1,000-ml glass beaker with water and keep it boiling on a hot plate for sample heating. The boiling water will be used in Step 16 to heat the samples.
Centrifuge parasites at 1,000 × g for 10 min at 4°C. After centrifugation, a parasite pellet is expected to be seen at the bottom of the centrifuge tube.
Carefully aspirate the supernatant and resuspend the parasite pellet in 10 ml ice-cold DPBS. Repeat Step 4. Check for the pellet before proceeding.
Aspirate the supernatant carefully and resuspend the pellet in 10 ml ice-cold DPBS.
Quantitate the yield of purified parasites using a hemocytometer following the vendor’s instructions.
Pellet the parasites at 1,000 × g for 10 min at 4°C.
Aspirate the supernatant and resuspend the parasites in 1 ml ice-cold DPBS and transfer to a 1.5-ml microcentrifuge tube.
Spin down the parasites at 5,000 × g at 4°C for 5 min.
Aspirate the supernatant and resuspend the remaining pellet in 400 µl ice-cold DPBS.
Sonicate each purified parasite strain inside a biosafety cabinet using a Branson Analog Sonifier ultrasonic cell disrupter S-250A with a tapered 1/8-inch microtip, using the following settings: output intensity = 3 and Duty% = 20%. Sonicate each purified parasite strain for 10 s and repeat 4 times. Wait for 30 s between each repeat and keep samples on ice to avoid overheating.
A hemin standard curve is needed for the measurement of absolute heme abundance per parasite. The hemin stock is initially diluted in DPBS to 1,200 nM, followed by a 3-fold serial dilution to generate 5 additional concentrations at 400, 133.3, 44.4, 14.8, and 4.9 nM. DPBS alone is also included as a blank for 0 nM hemin.
Add 100 µl each sample or hemin standard to black 1.5-ml centrifuge tubes. Two sets for each strain or hemin standard are prepared; one will be boiled and another one will remain at room temperature.
Add 900 µl 2 M oxalic acid to each sample and vortex for complete mixing.
Keep one set of samples and hemin dilutions in a rack at room temperature and place another set in a tube holder to boil for 30 min.
Place the boiled samples on ice for 5 min and then leave them at room temperature for 10 min. Mix all samples by vortexing.
Pipette 200 µl each parasite stain or hemin standard into each well of a black 96-well plate in triplicate.
Read the samples using a BioTek Synergy H1 Hybrid Multi-Mode Microplate Reader with the following settings: Excitation: 400 nm, Emission: 608 nm, Optics: Top, Gain: 135, Read Speed: Normal, Delay: 100 msec, Measurements/Data Point: 10, and Read Height: 7 mm. Export the acquired data to an Excel spreadsheet.
Repeat the assay in at least three biological replicates for statistical significance comparison.
Data analysis
Calculation of the normalized heme abundance in Toxoplasma parasites (Figure 2):
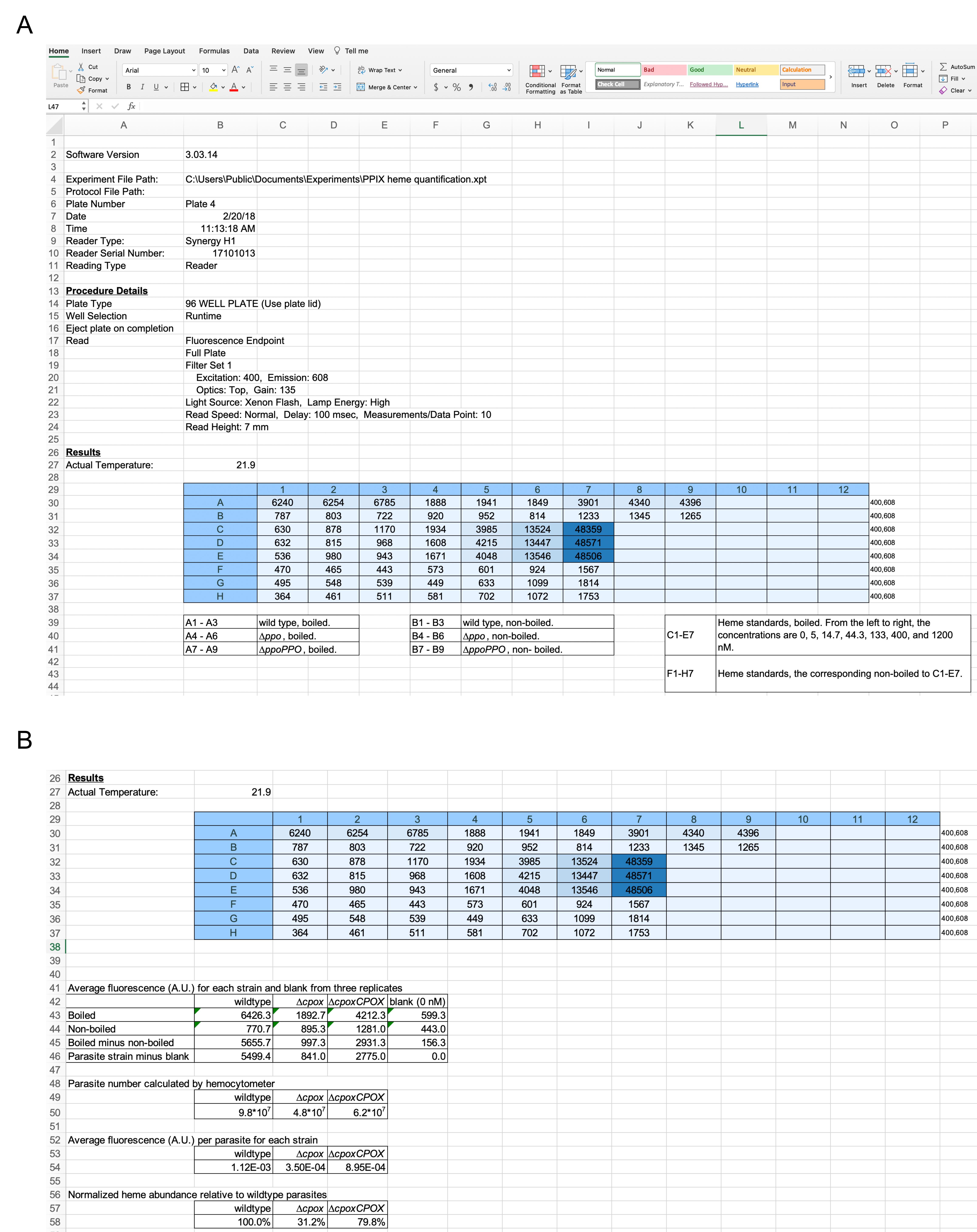
Figure 2. Example data and analysis for PPIX-based heme quantitation in Toxoplasma parasites. A. Excel table of the raw data from one biological replicate of heme quantitation in wildtype, ∆cpox, and ∆cpoxCPOX parasites. B. Data analysis of normalized heme abundances in Toxoplasma parasites. The heme abundance in wildtype parasites was set as 100% for normalization of heme amounts in other strains.Calculate the average readings for each boiled and non-boiled sample.
Subtract the fluorescence of every non-boiled sample from the signal of the corresponding boiled sample.
Subtract the blank value (0 nM hemin sample) from each parasite strain.
Divide the mean values for the individual samples by the number of parasites calculated by the hemocytometer counts.
Note: One-twentieth of the purified parasites are included in each well for quantitation of heme abundance (A quarter of the total purified parasites are mixed with oxalic acid before boiling, and one-fifth of the boiled mixture is pipetted into each well of a 96-well plate for fluorescence measurement).
Divide the average fluorescence value per parasite for each sample by that from the wildtype strain for normalization. The normalized value for wildtype parasites is set as 100% for comparison with other strains.
Calculation of the absolute heme abundance in Toxoplasma parasites (Figure 3):
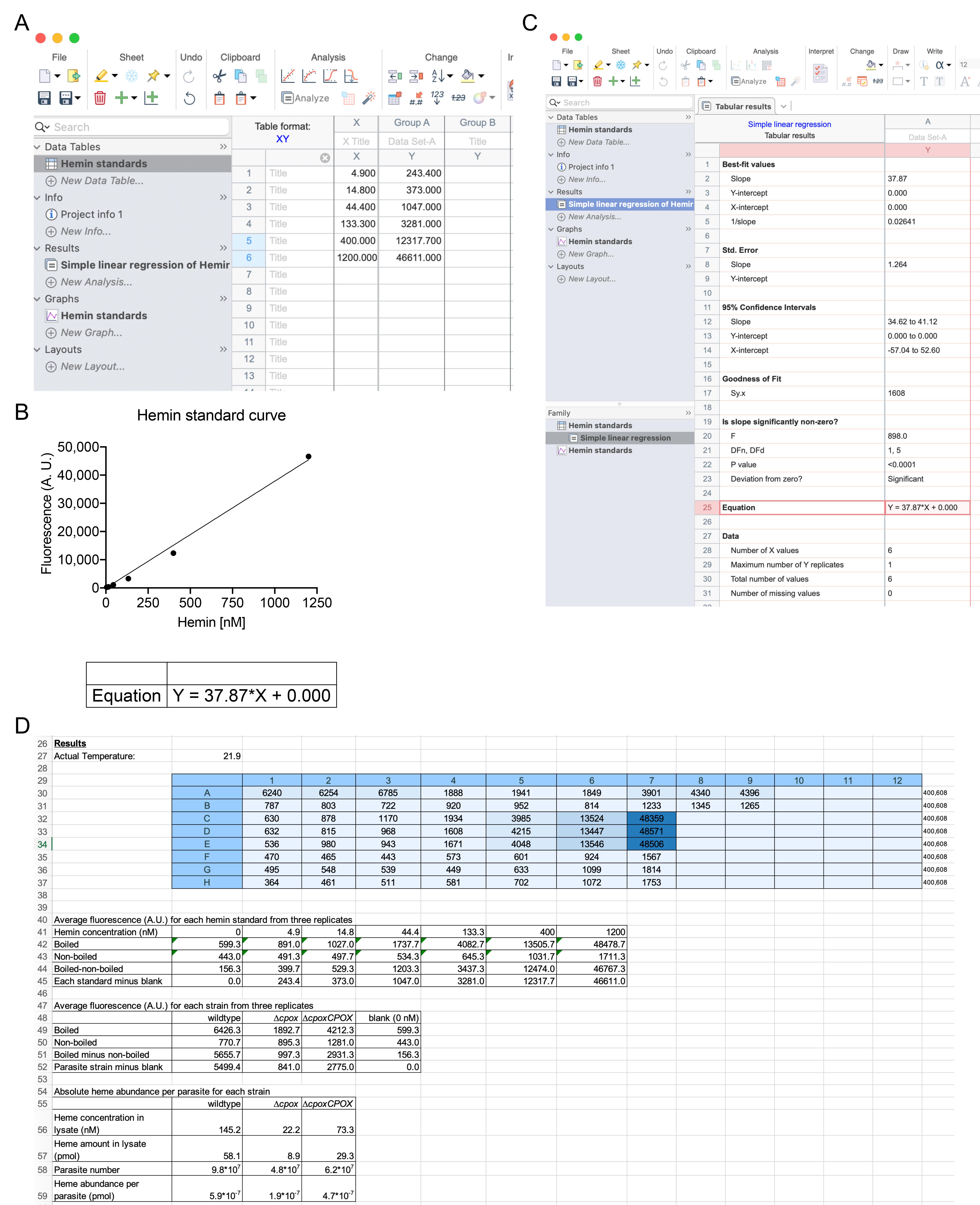
Figure 3. Calculation of the absolute heme abundance in Toxoplasma parasites. A. Table of fluorescence (A.U.) signals under each concentration of hemin standard. B. Graph of a linear regression of the plotted hemin standard curve. C. Linear regression analysis to generate the equation for calculating heme concentrations in lysates of purified parasites. D. Step-by-step data analysis for calculating heme amounts per parasite.If the absolute heme abundance per parasite is desired, the standard heme curve is required to determine the heme concentration in sonicated parasite lysates by substituting fluorescence values from each strain in the heme standard curve equation. The total amount of heme in each parasite lysate is determined by multiplying the heme concentration of each sample by the volume (400 µl) and dividing by the total number of parasites to yield the absolute heme abundance per parasite.
To generate the hemin standard curve:
Average the fluorescence intensities of boiled and non-boiled samples for each concentration of hemin standard.
Subtract the average fluorescence values for the individual non-boiled samples from the corresponding boiled samples.
Subtract the blank value (0 nM hemin) from the individual hemin concentrations.
Plot the subtracted fluorescence values against the individual hemin concentrations at 4.9, 14.8, 44.4, 133.3, 400, and 1,200 nM.
Perform linear regression on the plotted data points for curve fitting to produce the heme standard curve equation, which will be used to calculate heme abundances in the tested parasite lysates.
Recipes
DPBS
Add 9.6 g HyClone Dulbecco's phosphate-buffered saline powder in 1 L deionized water. The solution is autoclaved before use.
D10 medium
Dulbecco’s Modified Eagle Medium (DMEM) is supplemented with 10 mM HEPES, 10% (v/v) Cosmic calf serum, 2 mM L-glutamine, 100 IU/ml penicillin, and 100 µg/ml streptomycin.
1 mM heme solution
Dissolve 32.6 mg hemin in 50 ml 20 mM NaOH.
2 M oxalic acid solution
Add 126.07 g oxalic acid dihydrate to 500 ml deionized water. Heat water to ~50°C for solution preparation. The solution is oversaturated, and the solute will precipitate when the solution cools to room temperature. Use the supernatant in the assay.
Acknowledgments
The authors would like to thank Dr. Carruthers for sharing the RH∆ku80∆hxg Toxoplasma strain for this study. This work was supported by the Clemson Startup fund (to Z.D.), Knights Templar Eye Foundation Pediatric Ophthalmology Career-Starter Research Grant (to Z.D.), a pilot grant of an NIH COBRE grant P20GM109094 (to Z.D.), and NIH R01AI143707 (to Z.D.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This protocol was modified from the previously published PPIX-based heme quantitation assay (Sinclair et al., 2001) used for heme quantification in mammalian cells.
Competing interests
The authors declare no conflicts or competing interests.
References
- Bergmann, A., Floyd, K., Key, M., Dameron, C., Rees, K. C., Thornton, L. B., Whitehead, D. C., Hamza, I. and Dou, Z. (2020). Toxoplasma gondii requires its plant-like heme biosynthesis pathway for infection. PLoS Pathog 16(5): e1008499.
- Hanna, D. A., Harvey, R. M., Martinez-Guzman, O., Yuan, X., Chandrasekharan, B., Raju, G., Outten, F. W., Hamza, I. and Reddi, A. R. (2016). Heme dynamics and trafficking factors revealed by genetically encoded fluorescent heme sensors.Proc Natl Acad Sci U S A 113(27): 7539-7544.
- Huynh, M. H. and Carruthers, V. B. (2009). Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot Cell 8(4): 530-539.
- Krishnan, A., Kloehn, J., Lunghi, M., Chiappino-Pepe, A., Waldman, B. S., Nicolas, D., Varesio, E., Hehl, A., Lourido, S., Hatzimanikatis, V. and Soldati-Favre, D. (2020). Functional and Computational Genomics Reveal Unprecedented Flexibility in Stage-Specific Toxoplasma Metabolism. Cell Host Microbe 27(2): 290-306 e211.
- Phillips, J. D. (2019). Heme biosynthesis and the porphyrias. Mol Genet Metab 128(3): 164-177.
- Ponka, P. (1999). Cell biology of heme. Am J Med Sci 318(4): 241-256.
- Sinclair, P. R., Gorman, N. and Jacobs, J. M. (2001). Measurement of heme concentration. Curr Protoc Toxicol Chapter 8: Unit 8 3.
- Song, Y., Yang, M., Wegner, S. V., Zhao, J., Zhu, R., Wu, Y., He, C. and Chen, P. R. (2015). A Genetically Encoded FRET Sensor for Intracellular Heme. ACS Chem Biol 10(7): 1610-1615.
- Tjhin, E. T., Hayward, J. A., McFadden, G. I. and van Dooren, G. G. (2020). Characterization of the apicoplast-localized enzyme TgUroD in Toxoplasma gondii reveals a key role of the apicoplast in heme biosynthesis. J Biol Chem 295(6): 1539-1550.
- Yuan, X., Rietzschel, N., Kwon, H., Walter Nuno, A. B., Hanna, D. A., Phillips, J. D., Raven, E. L., Reddi, A. R. and Hamza, I. (2016). Regulation of intracellular heme trafficking revealed by subcellular reporters. Proc Natl Acad Sci U S A 113(35): E5144-5152.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Bergmann, A. and Dou, Z. (2021). Fluorescence-based Heme Quantitation in Toxoplasma Gondii. Bio-protocol 11(12): e4063. DOI: 10.21769/BioProtoc.4063.
Category
Microbiology > Microbial physiology
Biochemistry > Other compound > Heme
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









