- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Peptide-mediated Targeting of Nanoparticles with Chemical Cargoes to Chloroplasts in Arabidopsis Plants
Published: Vol 11, Iss 12, Jun 20, 2021 DOI: 10.21769/BioProtoc.4060 Views: 4795
Reviewed by: Zinan ZhouLusheng FanDemosthenis Chronis

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Mobilization of Plasmids from Bacteria into Diatoms by Conjugation Technique
Federico Berdun [...] Eduardo Zabaleta
Mar 5, 2024 1917 Views

A Step-by-step Protocol for Crossing and Marker-Assisted Breeding of Asian and African Rice Varieties
Yugander Arra [...] Wolf B. Frommer
Sep 20, 2024 2333 Views

A Detailed Guide to Recording and Analyzing Arabidopsis thaliana Leaf Surface Potential Dynamics Elicited by Mechanical Wounding
Fatiha Atanjaoui [...] Michael M. Wudick
Apr 5, 2025 1444 Views
Abstract
Plant nanobiotechnology is a flourishing field that uses nanomaterials to study and engineer plant function. Applications of nanotechnology in plants have great potential as tools for improving crop yield, tolerance to disease and environmental stress, agrochemical delivery of pesticides and fertilizers, and genetic modification and transformation of crop plants. Previous studies have used nanomaterials functionalized with chemicals, including biocompatible polymers with charged, neutral, or hydrophobic functional groups, to improve nanomaterial uptake and localization in plant cells. Recently, the use of biorecognition motifs such as peptides has been demonstrated to enable the targeted delivery of nanoparticles in plants (Santana et al., 2020). Herein, we describe a bio-protocol to target nanoparticles with chemical cargoes to chloroplasts in plant leaves and assess targeting efficiency using advanced analytical tools, including confocal microscopy and elemental analysis. We also describe the use of isothermal titration calorimetry to determine the affinity of nanomaterials for their chemical cargoes. Nanotechnology-based methods for targeted delivery guided by conserved plant molecular recognition mechanisms will provide more robust plant bioengineering tools across diverse plant species.
Graphic abstract:
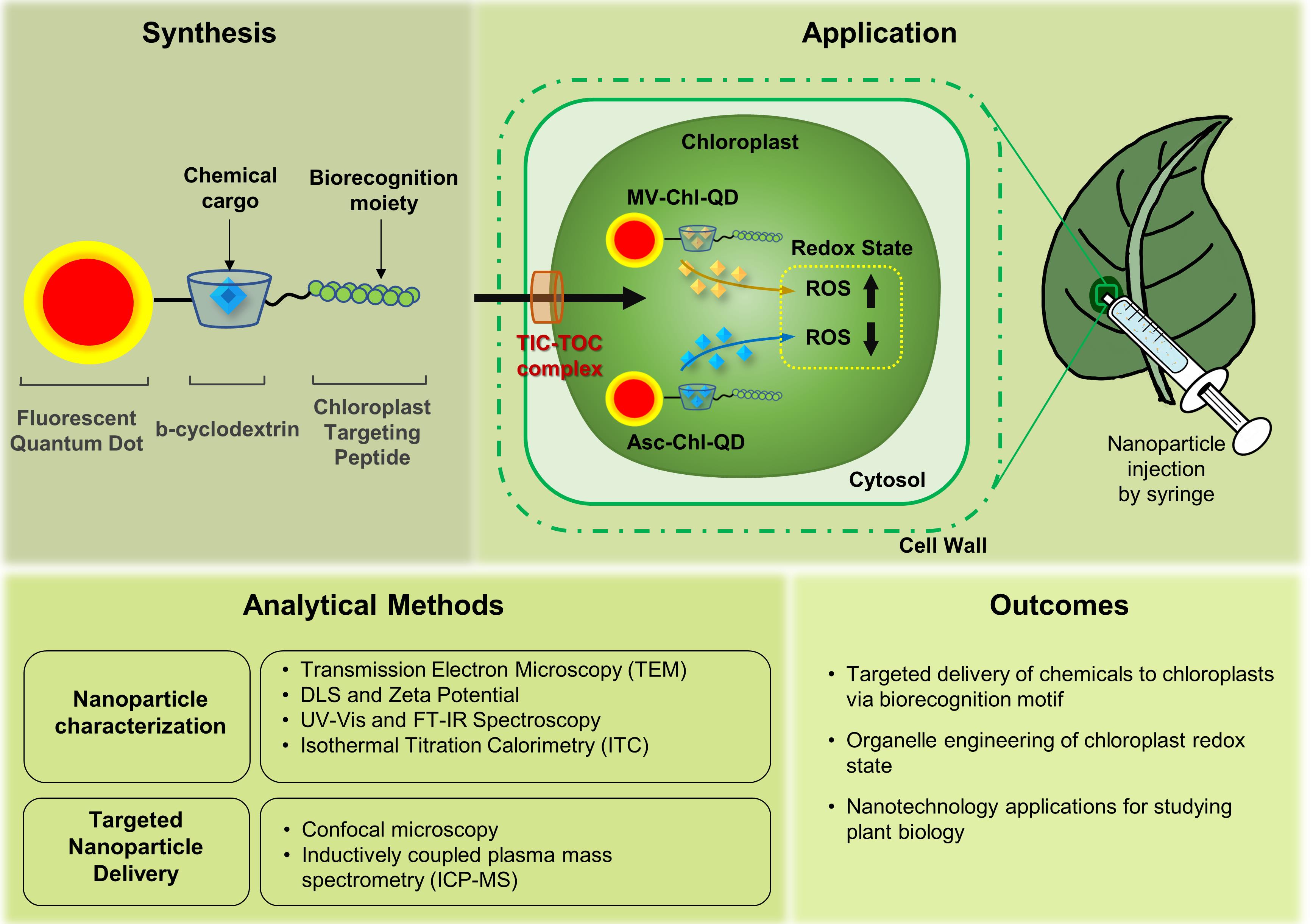
Targeted delivery of nanomaterials with chemical cargoes to chloroplasts enabled by plant biorecognition
Background
Nanomaterials have enabled improved diagnostic tools, drug delivery, bioengineering, and tissue regeneration platforms for mammalian systems (Das et al., 2014; Li et al., 2016; Patra et al., 2018). Applications of nanotechnology in plant bioengineering and nano-enabled agriculture have recently emerged (Kah et al., 2019; Lowry et al., 2019). The use of plant nanobiotechnology has great potential in developing valuable diagnostic and therapeutic tools for improving crop management, resistance to diseases and environmental stresses, targeting the delivery of agrochemicals, and genetic bioengineering tools (Wang et al., 2016; Yin et al., 2018; Giraldo et al., 2019; Wang et al., 2019; Santana et al., 2020).
Currently, the delivery of chemicals in plants leads to unintended alterations in plant function and environmental pollution from chemical leaching (Nagajyoti et al., 2010; Smith and Gilbertson, 2018; Lowry et al., 2019). Nanotechnology approaches have relied on size, surface charge, and hydrophobicity modifications to tune their distribution in plant cells (Asati et al., 2010; Wong et al., 2016; Wu et al., 2017; Demirer et al., 2019; Hu et al., 2020); however, these approaches based on chemical coatings cannot target specific plant subcellular compartments with high precision. Nanoparticle functionalization with targeting peptide recognition motifs enables plant molecular machinery to guide the nanomaterials to plant organelles in vivo with high specificity (Santana et al., 2020).
Herein, we present a protocol for synthesizing, characterizing, and detecting a nanomaterial platform that targets the delivery of chemicals to chloroplasts in wild-type Arabidopsis thaliana (Col-0) using a peptide recognition motif that is relatively conserved in dicotyledonous plants. We describe techniques for the synthesis and characterization of targeted quantum dots using UV-vis spectroscopy, Fourier-transform infrared spectroscopy (FTIR), dynamic light scattering (DLS), and transmission electron microscopy (TEM). We outline methods to image and quantitate the nanoparticles in plant chloroplasts using advanced analytical tools with high resolution, including confocal microscopy and inductively coupled plasma mass spectrometry (ICP-MS). These nanomaterials target the delivery of chemicals to the chloroplast, allowing tuning of their function, e.g., redox state, with higher specificity and efficiency than chemicals alone.
The use of quantum dots coated with biorecognition moieties to target the delivery of chemicals to chloroplasts can be extended to sustainable nanomaterials for the targeted delivery of genetic elements, nanosensors, nutrients, or pesticides across multiple plant species.
Materials and Reagents
Centrifugal filter (Merck Millipore, Amicon Ultra 15, molecular weight cut-off, 10,000 Da)
Arabidopsis thaliana seeds (ecotype Columbia seed stock source CS60000)
NaBH4, purum p.a., ≥96% (gas-volumetric) (Sigma-Aldrich, catalog number: 71320-25G, CAS Number 16940-66-2)
Succinimidyl-[(N-maleimidopropionamido)-tetraethyleneglycol] ester (NHS-PEG4-MAL linker, Thermo Fisher Scientific, U.S.A.)
Perfluorodecalin (Acros Organics, 25 g, 90% mixture of cis and trans, CAS 306-94-5)
Tellurium powder, Te; 99.8%, (Sigma-Aldrich, catalog number: 266418-25G)
Ethanol 200 proof (Fisher Scientific, Acros organics 61509-0040 4L)
Molecular grade H2O (Corning, catalog number: 46-000-CM)
Cadmium chloride hydrate (Sigma-Aldrich catalog number: 529575)
Mercaptopropionic acid (Sigma-Aldrich, catalog number: M5801-100G)
Sodium hydroxide solution (50% w/w certified; Fisher Scientific, catalog number: SS254-1, 1L, CAS number 1310-73-2)
1 ml NORM-JECT® (4010-2000V0)
1-Ethyl-3-[3-dimethylaminopropyl] carboamide hydrochloride, EDC, 5 g (G-Biosciences, catalog number: BC 25-5)
N-Hydroxysuccinimide, NHS, 25 g (Thermo Scientific, catalog number: 24500)
TES buffer (Sigma Life Science, catalog number: T1375-25G)
3-Aminophenylboronic acid hydrochloride (Sigma-Aldrich, catalog number: 410705-1G)
L-Ascorbic acid (Fisher Chemical, catalog number: A61-100)
Methyl viologen (Acros organics, catalog number: A227320010)
Centrifugal filter (Merck Millipore, Amicon Ultra 15, molecular weight cut-off, 10,000 Da, catalog number: UFC901024)
NaOH 1% solution
HCl 1% solution
Mono-(6-ethanediamine-6-deoxy)-beta-cyclodextrin (PONOCO enterprise, CAVCON, catalog number: 60984-63-6) (store at -4°C)
NHS-PEG4-Maleimide, SM-PEG pegylated crosslinker (NHS-PEG4-MAL) (Thermo Scientific, catalog number: 22107) (store at -20°C)
RbcS targeting peptide was designed from Rubisco small subunit transit peptide sequence (GenBank: OAP15425): MASSMLSSATMVGGC (1432.72 g/mol) (Genscript) (store at -20°C)
Carolina microscope observation gel (Corning, 2984-75x25)
Cheesecloth grade 50 (VWR International, catalog number: 470150-438 (PK))
1× chilled sucrose buffer (pH 7.3) (see Recipes)
Equipment
Erlenmeyer flasks (200 ml)
Fume hood
-20°C freezer
Plastic plant growth inserts (T.O. plastics st-10804)
Hot plate/stir plate (IKA RCT basic safety control magnetic stirrer, RCT BS001)
Bath sonicator (Elmasonic p, P-30H, #101-3737)
UV-vis spectrophotometer (Shimadzu 2600, UV-2600 EN)
Malvern 1600 zetasizer (Nano S, ZEN1600)
Folded capillary zeta cell cuvette (Malvern Panalytics, DTS1070)
Disposable cuvettes (Spectrum Laboratory Products, 330-10304P5)
Quartz open top cuvette 10 mm (Starna Cells Inc, 18-Q-10)
FTIR spectrometer (Bruker Alpha I)
TEM (Philips FEI Technai 12 microscope)
Leica SP5 Confocal Microscope
Malvern isothermal titration calorimeter (G.E. Healthcare, MicroCal ITC200 instrument)
ICP-MS (Agilent 7700x)
Grinder or macerator (Intertek KWG-100A)
Software
ImageJ
Procedure
Plant growth
Germinate Arabidopsis thaliana seeds (ecotype Columbia seed stock source CS60000) in (2.5” × 2.5” × 3”) pots filled with soil containing 1% marathon (OHP, Inc., Marathon 1% Granular, 5 lbs., 985490.0) and 1% osmocote (Classic 3-4 Month 14-14-4 Fertilizer 50 lb., E90550).
Grow plants in Adaptis A1000 growth chambers (Conviron MODEL No. A1000, SERIAL No. 150031) set to 200 μmol m−2 s−1 photosynthetic active radiation (PAR), 24 ± 1°C, 60% humidity, and a 14 h/10 h day/night regime.
Water the plants once every three days.
For all experiments, use Arabidopsis thaliana Col-0 plants that are 3 weeks old (Figure 1).

Figure 1. Leaves of 3-week-old Arabidopsis thaliana plants (Col-0) were used for nanoparticle infiltration
Synthesis of quantum dots
To synthesize quantum dots (QDs), prepare a colloidal solution of 0.01 g CdCl2 and 40 µl mercaptopropionic acid in 50 ml molecular grade water.
Label this solution A, add a stir bar, and stir the solution at 500 rpm and room temperature.
Next, adjust the pH of solution A to 11.4 with 1 M NaOH added dropwise. Place solution A on a hot plate set to 100°C and allow to reflux. Stir solution A at 700 rpm.
Meanwhile, label a 20-ml glass vial with a cap as solution B.
Add 0.05 g NaBH4 and 0.02 g tellurium powder into the 20-ml vial labeled solution B.
Add 600 µl 50% ethanol into the 20-ml vial and add a stir bar. Make sure to dispense the ethanol gently into the glass vial labeled solution B.
Keep the solution lightly capped to avoid air entering the reaction (Figure 2A).
Place solution B onto a hot plate set to 70°C and stir at 300 rpm.
Allow solution B to react for 5-10 min. The solution will turn from a dark black color to purple-blue, producing NaHTe for later use (Figure 2A-2B).
Immediately after the color change (Figure 2B), use a pipet tip to collect 150 µl freshly prepared solution B and quickly dispense directly into solution A.
Allow the mixture to react for 5 min under reflux conditions with vigorous stirring (700 rpm) (Figure 2C).
An increase in fluorescence of the mixture can be monitored when excited under U.V. light (375 nm) (Figure 2D).
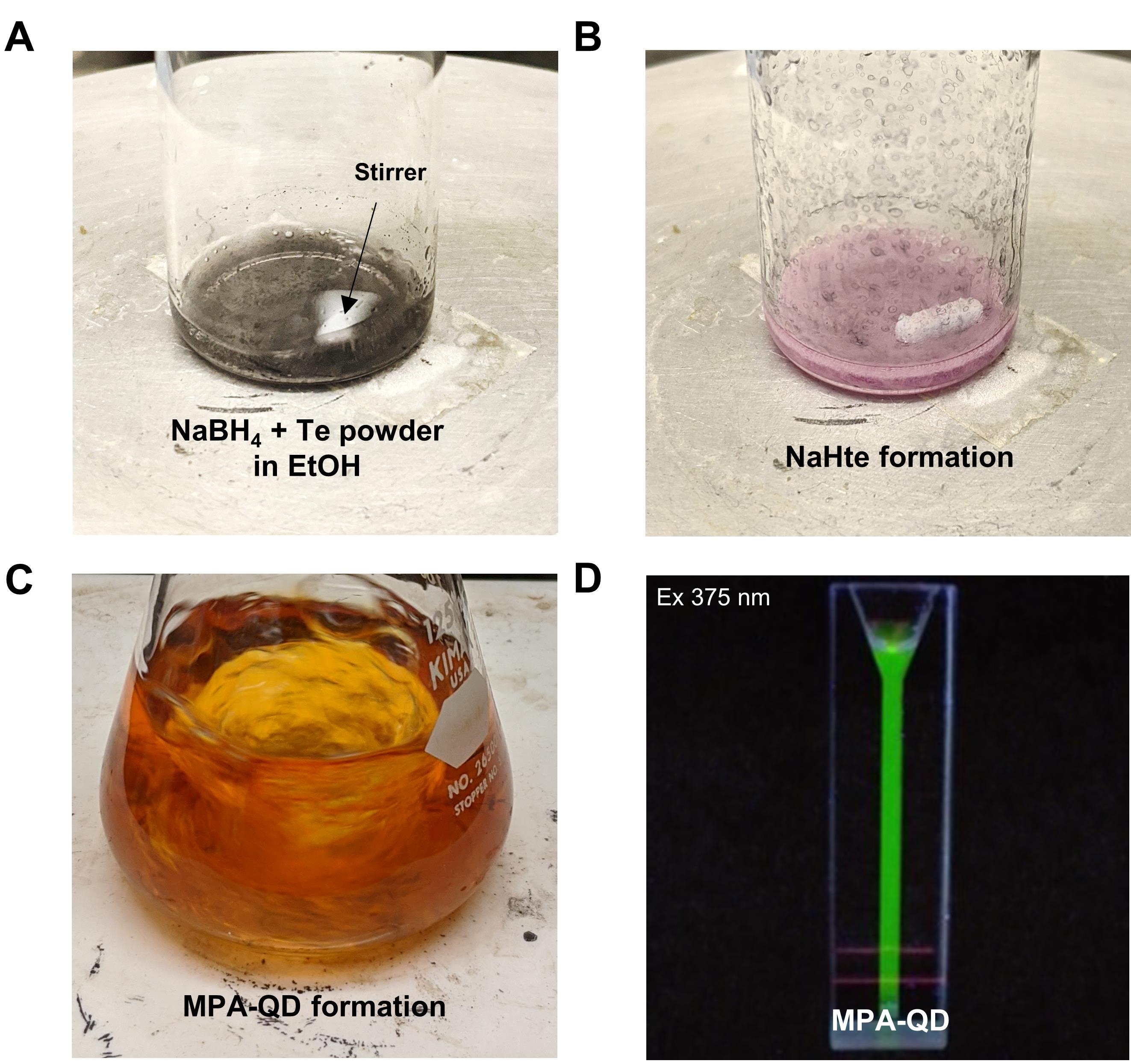
Figure 2. Synthesis steps for mercaptopropionic-coated quantum dots. A. Image of a 20-ml vial labeled solution B filled with NaBH4 and tellurium powder in 50% ethanol. B. Final product NaHTe after the reaction. C. Formation of MPA-QD crystals in an Erlenmeyer flask containing solutions A and B under reflux. D. Image of fluorescent MPA-QDs under U.V. light excitation (375 nm).Stop the reaction by removing it from the hot plate after 5 minutes and cooling it to room temperature. The emission of QD could be tuned to a specific wavelength by adjusting the reaction time (Table 1).
Table 1. QD synthesis reaction time versus emission peak wavelength
Reaction time (min) Emission peak wavelength (nm) 1 524 3 537 5 552 The resulting solution contains fluorescent Cd/Te-Cd/S core quantum dots functionalized with mercaptopropionic acid with terminal carboxyl groups on the outer shell. Label this solution as MPA-QD.
Formation of 3-aminophenylboronic acid (APBA)-capped QDs (APBA-QD)
Prepare APBA-QDs by reacting the MPA-QD terminal carboxyl group with 3-aminophenylboronic acid using the 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysuccinimide (NHS) activated reaction method.
First, determine the concentration of QDs (see section F-M).
Bath sonicate MPA-QDs for 30 min at 80% power and 37 Hz.
Dilute the MPA-QD solution to 1 µM in 10 mM TES buffer at pH 7.0.
Add NHS (2000 nmol) dissolved in molecular grade H2O to 1 nmol MPA-QD in 10 mM TES buffer at pH 7.0.
Next, add EDC/HCl (2000 nmol) dissolved in molecular grade H2O into 1 nmol MPA-QD in 10 mM TES buffer at pH 7.0.
Gently stir the mixture (500 rpm) for 15 min at room temperature.
Add 80 μl 25 mM APBA dissolved in molecular grade H2O to the activated MPA-QD solution to generate aminophenylboronic acid-functionalized quantum dots (APBA-QD). Allow the reaction to stir (500 rpm) for 3 h at room temperature.
Wash the APBA-QD solution twice with molecular grade H2O to remove the excess APBA using a centrifugal filter (Merck millipore, Amicon Ultra 15, molecular weight cut-off, 10,000 Da). Set the centrifuge to 2,360 × g for 10 min.
Bath sonicate the APBA-QD solution for 30 min at 80% power at 37 Hz to break down any agglomerated particles.
Synthesis of β-cyclodextrin-capped QD
Suspend the resulting APBA-QD in 10 ml TES buffer 10 mM pH 10.4.
Add 1 μmol β-cyclodextrin (β-CD, Cavcon) dissolved in molecular grade H2O (0.5 ml) to the APBA-QD solution.
Allow the mixture to react overnight at room temperature with stirring set at 500 rpm.
Remove the excess of β-cyclodextrin by washing through a centrifugal filter (Merck millipore, Amicon Ultra 15, molecular weight cut-off, 10,000 Da). Centrifuge at 2,360 × g for 10 min.
Bath sonicate the resulting solution for 30 min at 80% power and 37 Hz.
Suspend the β-cyclodextrin-coated quantum dots (CD-QD) in 10 ml 10 mM TES pH 7.5.
Confirm CD-QD formation by collecting the FTIR spectrum (see section K).
Preparation of peptide-conjugated β-CD-capped QD
To conjugate the RbcS targeting peptide to CD-QD nanoparticles, first dissolve succinimidyl-[(N-maleimidopropionamido)-tetraethyleneglycol] ester (NHS-PEG4-MAL linker, Thermo Fisher Scientific, U.S.A.) in DMSO to make a 200 mM stock solution.
Next, add 5 μl (1 μmol) NHS-PEG4-MAL stock solution to 1 μM CD-QDs in 10 ml final volume.
Incubate the mixture at room temperature for 1 h and stir at 500 rpm to yield MAL-PEG4-QD.
Remove excess NHS-PEG4-MAL by washing the mixture through a centrifugal filter (Merck Millipore, Amicon Ultra 15, molecular weight cut-off, 10,000 Da) with molecular grade H2O.
Resuspend the MAL-PEG4-QD in 10 ml 10 mM TES pH 8.0.
A peptide sequence from the Rubisco small subunit (RbcS) was used for targeting nanomaterials to the chloroplast. The peptide was synthesized by Genscript containing the amino acid sequence MASSMLSSATMVGGC.
RbcS chloroplast-targeting peptide was dissolved in 5% DMSO diluted with TES buffer pH 8.0 to 10 mg·ml-1 (equivalent to 7 mM).
Finally, add 0.143 ml (1 μmol) RbcS chloroplast-targeting peptide to the resulting MAL-PEG4-QD.
React for 1 h at room temperature and stir at 500 rpm to form chloroplast-targeting peptide-functionalized QD (Chl-QD).
Remove excess peptide and reactants by washing at least twice through a centrifugal filter (Merck Millipore, Amicon Ultra 15, molecular weight cut-off, 10,000 Da) with molecular grade H2O loaded onto a centrifuge set to 2,360 × g for 10 min.
Notes:
Make sure not to completely wash away the buffer from the centrifugal filter. Keep Chl-QD in solution during washing.
The resulting Chl-QD can be stored for up to one week without significant aggregation.
Bath sonicate the Chl-QD solution for 30 min at 80% power and 37 Hz to break down any agglomerated particles.
Characterize the Chl-QD using the methods described below (see section F).
Measure the hydrodynamic size, zeta potential, and fluorescence.
Confirm the formation of Chl-QD by collecting the FTIR spectrum.
Characterization of quantum dots (MPA-QD)
The resulting nanomaterial absorbance, size, zeta potential, and fluorescence emission (under 405 nm excitation) can be characterized accordingly.
UV-vis spectroscopy
Measure the UV-vis absorption spectrum of QDs on a Shimadzu UV-2600 spectrophotometer (Figure 3A).
The spectrophotometer settings are scanning range from 200 to 700 nm, interval of 0.5 nm, and integration time of 0.1 s.
Collect a background spectrum using molecular grade H2O in a 1-ml quartz spectrophotometer cuvette (10 mm × 4 mm).
Take 10 µl prepared MPA-QD and dilute it into 990 µl molecular grade H2O.
Place the sample in a quartz cuvette to record the UV-vis absorbance from 200 to 700 nm.
Record the peak absorbance value. The typical absorption peak of green fluorescent QD is 500-510 nm (Figure 3A).
Fluorescence emission
Measure the fluorescence of MPA-QD with the PTI QuantaMaster 600 fluorometer (Figure 3B).
Prepare the MPA-QD solution (200 nM) in molecular grade H2O.
Fill 3 ml solution into a quartz fluorescence cuvette and insert the cuvette into the holder of the PTI QuantaMaster 600 fluorometer.
The fluorometer settings are 5 nm slit size, 1 nm step size, and 0.1 s integration time.
Collect the emission spectra of MPA-QD following excitation at 405 nm (Figure 3B).
Determination of nanomaterial concentration
Determine the concentration of the solution using the following equations according to the peak absorbance and size.
c=Abs/(L×ϵ)
ε=10043×d2.12 (Yu et al., 2003)
where,
c is the QD concentration in M,
Abs is the peak absorbance at 500-510 nm,
L is the path length (1 cm),
𝜖 is the extinction coefficient in L mol-1 cm-1,
d is the diameter of QD 10-9 meters.
Hydrodynamic size
Measure the hydrodynamic size using a Malvern Zetasizer Nano S (Figure 2C) (model 1600).
The zetasizer settings we set for water as the solvent, a temperature of 20°C, material refractive index of 1.350, and a material absorbance of 1.000. Repeat the measurement 3 times.
Place 1 ml diluted MPA-QD in a 4-ml disposable cuvette. Insert the cuvette into the zetasizer for size measurements.
Measure the particle size distribution and take the average volume-based particle size distribution. A summary table will be displayed alongside the graph, which can help to determine the peak distribution of particles and the average from each measurement (Figure 3C).
FTIR spectroscopy
To collect the FTIR spectrum of nanomaterials, take a 2-ml centrifuge tube, add 500 µl 3 µM MPA-QD, and suspend in 1.5 ml ethanol at a 3:1 (v/v) ratio mixture.
Centrifuge at a max speed of 847 × g to precipitate MPA-QD into a pellet.
Remove the supernatant and allow it to air dry in a fume hood overnight.
A small dry pellet should form at the bottom of the centrifuge tube, which is used for subsequent FTIR spectroscopy using a Bruker Alpha II FTIR spectrophotometer or equivalent.
Zeta potential
Measure the zeta potential of MPA-QD with a Malvern Zetasizer (Nano Z.S.) (Figure 2D).
Prepare the MPA-QD solution (200 nM) in molecular grade H2O.
Fill about 0.7 ml solution into a folded capillary zeta cell cuvette (Malvern Panalytics, DTS1070) with a 1-ml syringe and insert the cuvette into the zetasizer (Nano Z.S.).
The zetasizer (Nano Z.S.) settings are water as the solvent, a temperature of 20°C, material refractive index of 1.350, material absorbance of 1.000, Hückel approximation, and a measurement repeat of 5.
Record the zeta potential and calculate the average and standard deviation (Figure 3D).
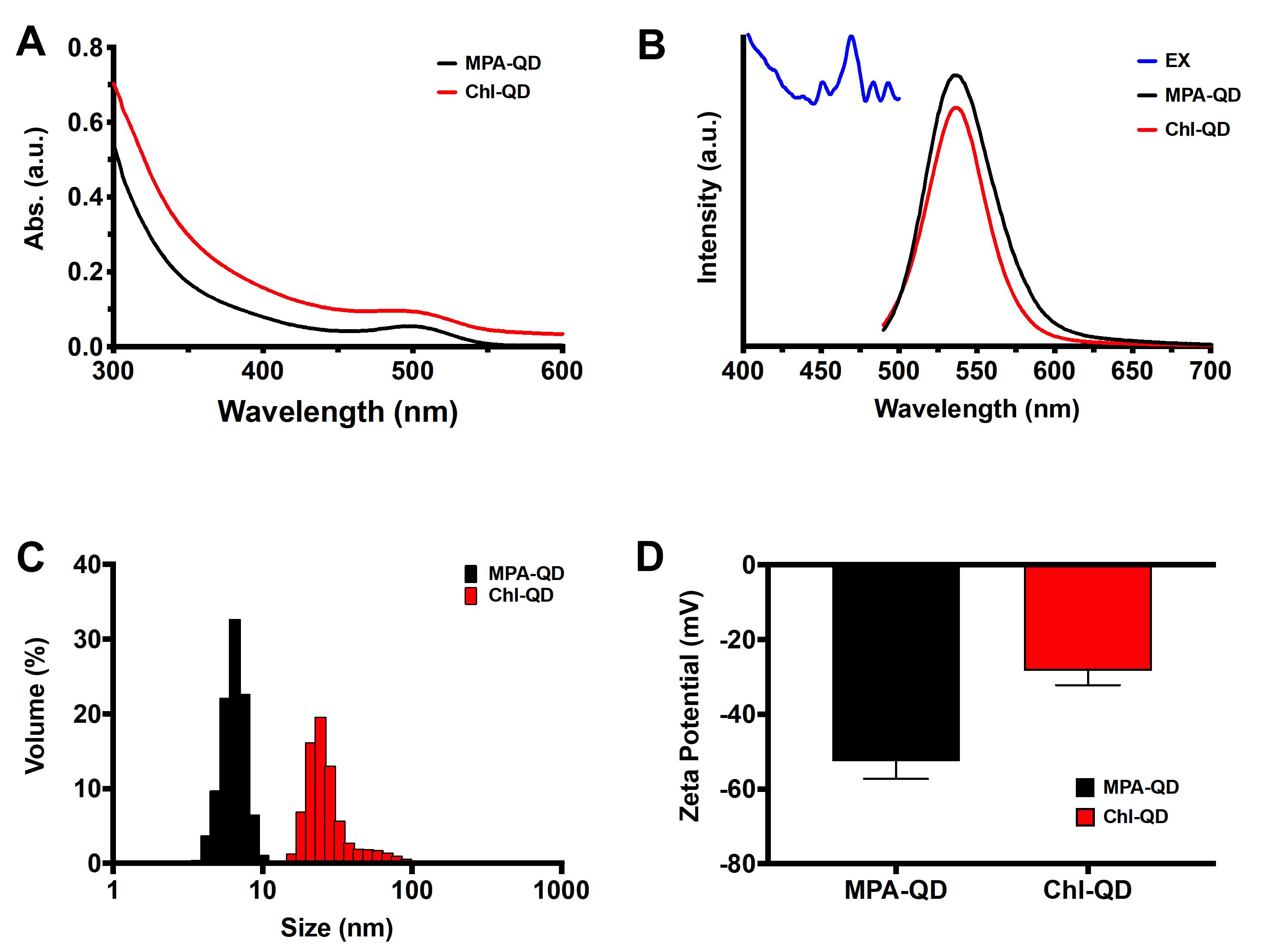
Figure 3. Characterization of MPA-QD and Chl-QD. A. UV-vis absorption spectra. B. Fluorescence excitation and emission spectra. C. Hydrodynamic size distribution. D. Zeta potential.
Transmission electron microscopy (TEM) of MPA-QDs
Load one drop (about 2 μl) MPA-QD onto the TEM grid (ultrathin carbon film on lacey carbon support film, 400 mesh, Cu, Ted Pella).
Allow the droplet to air dry.
Image the MPA-QD on the prepared grid with a Philips FEI Tecnai 12 microscope operated at an accelerating voltage of 120 kV.
Analyze the images obtained by TEM using ImageJ by measuring the particle diameter using the line segment tool (Figure 4A). Outline over 100 particles in each image and calculate the average particle diameter. To obtain lattice spacing of nanoparticles using the line segment tool (Figures 4A-4B), select one single particle clearly showing the lattice fringe, measure the distance between two planes that are separated by (at least) 10 lattice spacings, and calculate the lattice spacing by dividing the measured distance by 10.
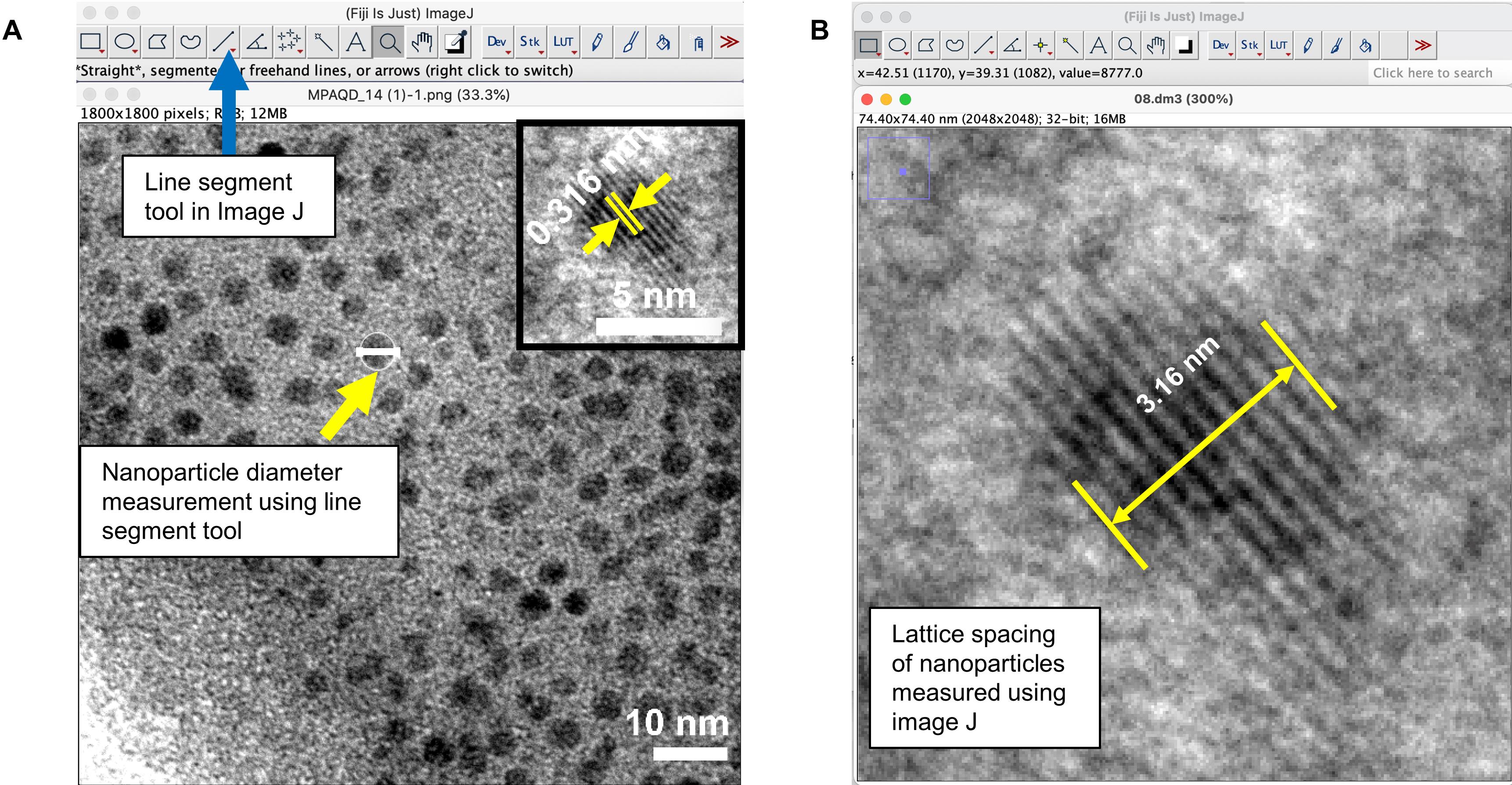
Figure 4. The ImageJ line segment tool was used to measure particle diameter and lattice spacing in TEM imagesCalculate the average size of MPA-QD with statistical analysis on more than 100 particles and measure the lattice spacing (Figure 4B).
Loading chemicals into Chl-QD
To load chemicals into Chl-QD, make a 1 ml stock solution of 0.1 mM methyl viologen or ascorbic acid in molecular grade H2O.
Add 100 µl 0.1 mM methyl viologen or ascorbic acid to 1 ml 200 nM Chl-QD in TES buffer pH 7.0.
Allow the mixture to incubate for 30 min and wash once through a centrifugal filter (Merck millipore, Amicon Ultra 15, molecular weight cut-off, 10,000 Da) with molecular grade H2O to remove excess molecules.
Determine the Chl-QD nanoparticle concentration loaded with chemicals, methyl viologen or ascorbic acid (MV-Chl-QD or Asc-Chl-QD, respectively), using the Beer-Lambert law, which allows determination of the unknown concentration of sample mixture to be calculated by measuring its absorbance.
Make a set of standards in triplicate using a fixed concentration of Chl-QD (200 nM) and a gradient concentration of methyl viologen or ascorbic acid ranging from 0 to 100 µM.
Record the max absorbance value of each standard mixture between 260 and 265.5 nm.
Use the known concentration of the standards and their respective absorbance values to generate a standard curve and determine the dilution factor (slope of the line).
Using the Beer–Lambert law, determine the unknown concentration of chemicals loaded into the 200 nM Chl-QDs.
Next, measure and record the initial absorbance of the Chl-QD sample without chemicals and the mixed chemical cargo-Chl-QD complex (from Step 3) after washing.
Use the Beer-Lambert law to determine the concentration of the chemicals mixed with the Chl-QD samples.
The final dosage of chemicals infiltrated into plants with 200 nM Chl-QD should be approximately 60 µM methyl viologen or 60 µM ascorbic acid in 1 ml TES buffer pH 7.0.
Measurement of the affinity of QDs (MPA-QD and CD-QD) for the chemicals (methyl viologen and ascorbic acid) using isothermal titration calorimetry (ITC)
Isothermal titration calorimetry (ITC) was performed using a MicroCal ITC200 instrument (G.E. Healthcare) and MPA-QD and cyclodextrin-coated QDs (CD-QD).
Prepare a 10 mM TES buffer pH 7.3.
Prepare CD-QD (0.5 μM) and methyl viologen (25 mM) stock solutions with TES buffer 10 mM pH 7.3.
Thoroughly clean the reference cell, sample cell, and the injection syringe with molecular grade H2O.
Load 0.3 ml CD-QD in 10 mM TES buffer pH 7.3 solution into the reference and sample cells.
Load methyl viologen solution into the calorimeter injection syringe and ensure that no bubbles are present.
Set up the instrument as follows: Temperature of 25°C, injection volume of 2 μl, 21 injections, time intervals between two consecutive injections of 180 s, reference compensation power of 5 μcal/s.
Run the measurement to obtain the thermogram and plot the thermal power against time.
Display the thermal power peaks corresponding to each chemical injection.
Use Origin (MicroCal) to integrate the thermal power peaks and normalize according to the amount of injected methyl viologen in moles to obtain the enthalpy changes, and plot the enthalpy changes against the molar ratios of injected methyl viologen to CD-QD in the sample cell to obtain the binding isotherm.
Fit the binding isotherm raw data using a one-set-of-sites binding model to generate the best-fit curve and change in enthalpy of CD-QD interacting with methyl viologen (Figure 5A-5B).
Record the thermodynamic parameters of methyl viologen binding to CD-QD, including the number of binding sites on CD-QD (n), association constant (Ka, M-1), dissociation constant (Kd, M), enthalpy change (ΔH, cal mol-1), and entropy change (ΔS, cal mol-1 K-1) (Figure 5A-5B).
Use the same method to determine the thermodynamic parameters for the binding between methyl viologen and MPA-QD, ascorbic acid and CD-QD, and ascorbic acid and MPA-QD, respectively.
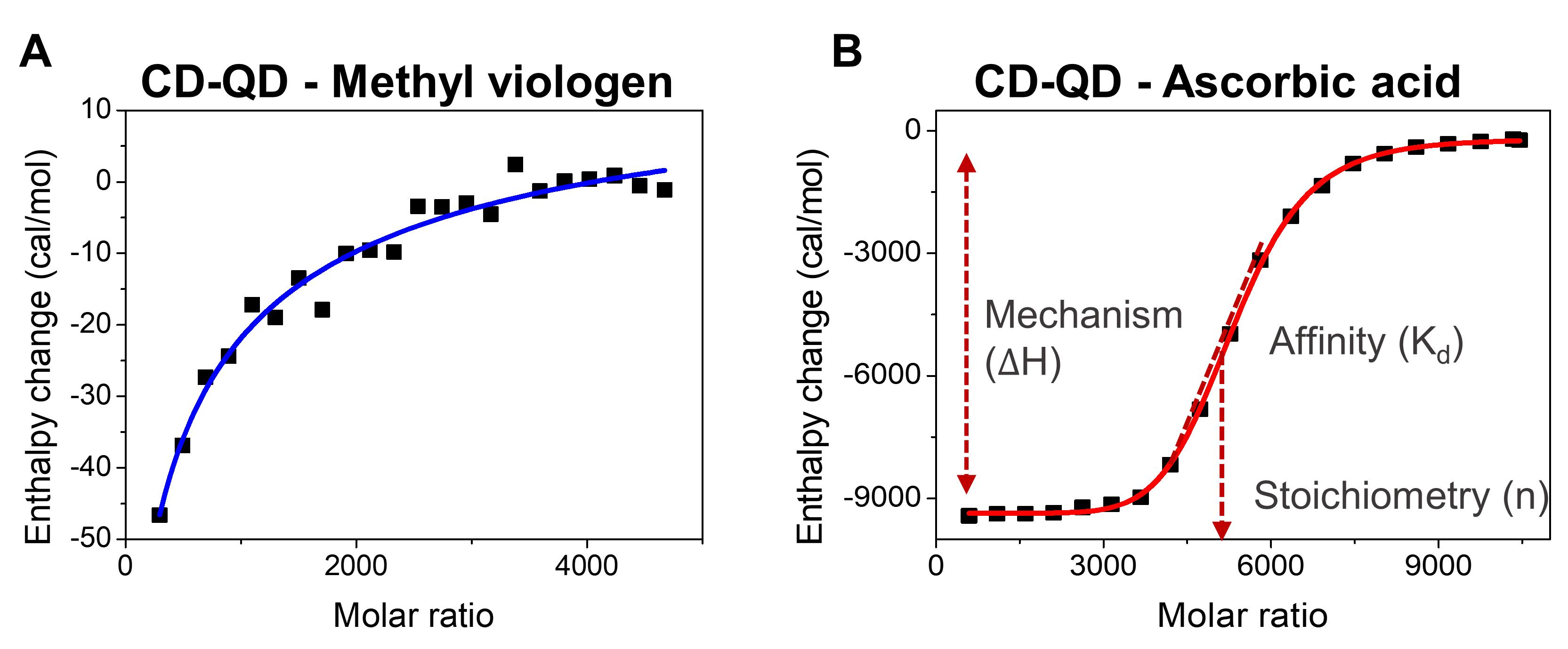
Figure 5. Isothermal titration calorimetry (ITC) of cyclodextrin-coated quantum dots (CD-QD) with chemical cargoes. A. The change in enthalpy of CD-QD interacting with methyl viologen and ascorbic acid. B. Thermal parameters that can be extrapolated from ITC, including enthalpy change (ΔH, cal mol-1), stoichiometry (n), and binding affinity (Kd).
Determination of nanoparticle bound and unbound chemicals in solution
Use the thermodynamic parameters, such as the number of binding sites (n), dissociation constant (Kd), and concentration of chemical ligand acquired from the ITC data analysis, to determine the bound and unbound fractions of chemicals in cyclodextrin-coated nanoparticles.
Use the following equation to determine the bound and unbound fractions:
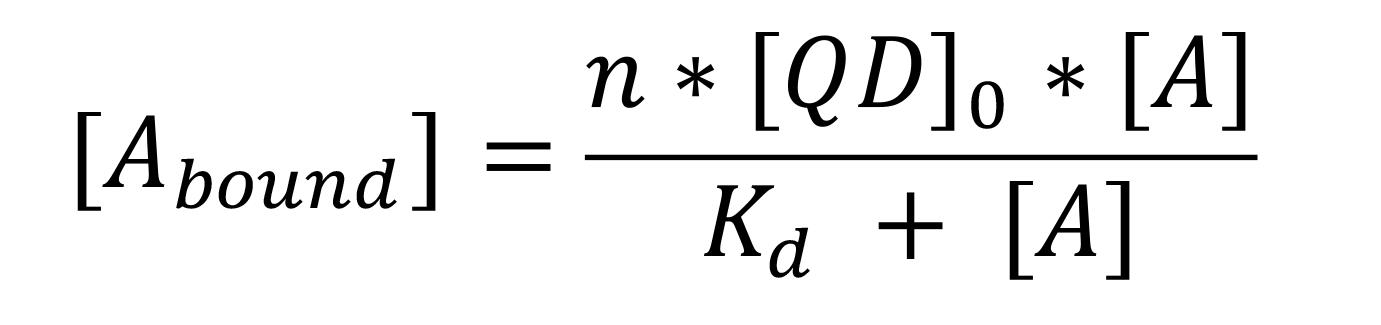
Where [Abound] and [A] is the concentration of bound and unbound chemicals in solution, respectively; n is the number of binding sites on QD; [QD]0 is the initial QD concentration; and Kd is the dissociation constant between QD and the chemicals.
Nanoparticle and chemical delivery into plant leaves
Dilute Chl-QD to 200 nM (0.17 mg ml-1) in 10 mM TES buffer pH 7.0.
Load Chl-QDs with 60 µM methyl viologen or ascorbic acid (see section N).
Take a 1-ml NORM-JECT needleless syringe and fill with 100 μl Chl-QD solution loaded with methyl viologen or ascorbic acid.
Gently press the tip of the syringe up against the abaxial side of the leaf and hold the index finger up against the adaxial side for support (Figure 6A).
Slowly perfuse each plant leaf by gently depressing the plunger. Gently remove any excess solution with Kimwipes (Figure 6B).
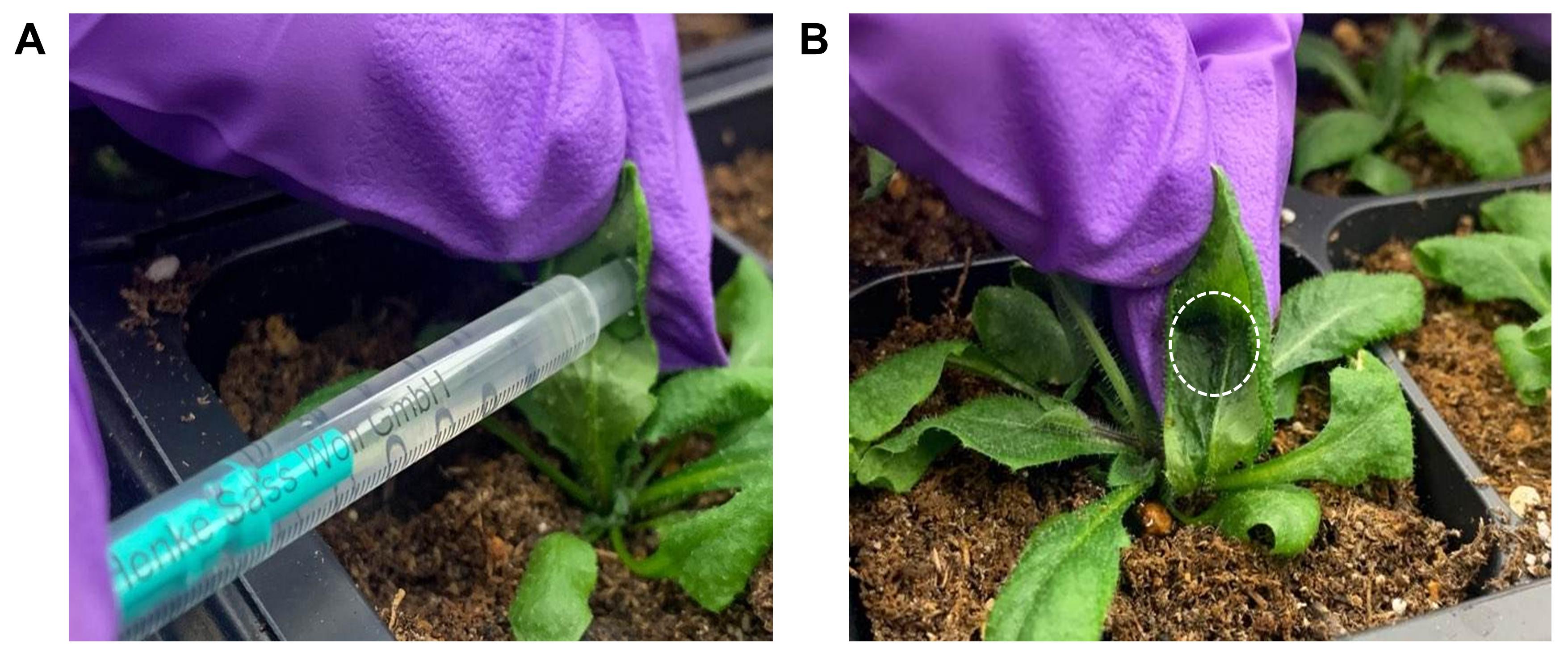
Figure 6. Nanoparticle and chemical cargo delivery into plant leaves. Nanomaterials are delivered through the leaf lamina using a needleless syringe. A. Gently press the bottom of the syringe up against the abaxial side of the leaf and hold the index finger up against the adaxial side. B. After infiltration with nanoparticles, the leaf appears darker as the particle solution fills the mesophyll space (dashed circle).Place the plant on a lab bench and incubate for 15 min before placing the plant back into the growth chamber.
Confocal fluorescence microscopy sample preparation
Take a pea-sized amount of Carolina observation gel, make a thin film (~1 mm in thickness) on a microscope slide (Corning 2984-75x25), and create a chamber using a cork border (diameter 8 mm) to remove a circular section from the film center (Figure 7A).
Take a leaf punch from the treated leaf with a cork border (diameter 6 mm) and incubate the leaf disk in 0.5 ml 10 µM DHE (Thermo Fisher Scientific, U.S.A.) in 10 mM TES buffer pH 7.0 for 30 minutes.
Immerse the DHE-incubated leaf disk in perfluorodecalin (Acros Organics, 25 g, 90% mixture of cis and trans, CAS 306-94-5) (Figure 7A-7B) filled in the gel chamber and seal it with a coverslip. The sample is ready for confocal fluorescence microscopy imaging.
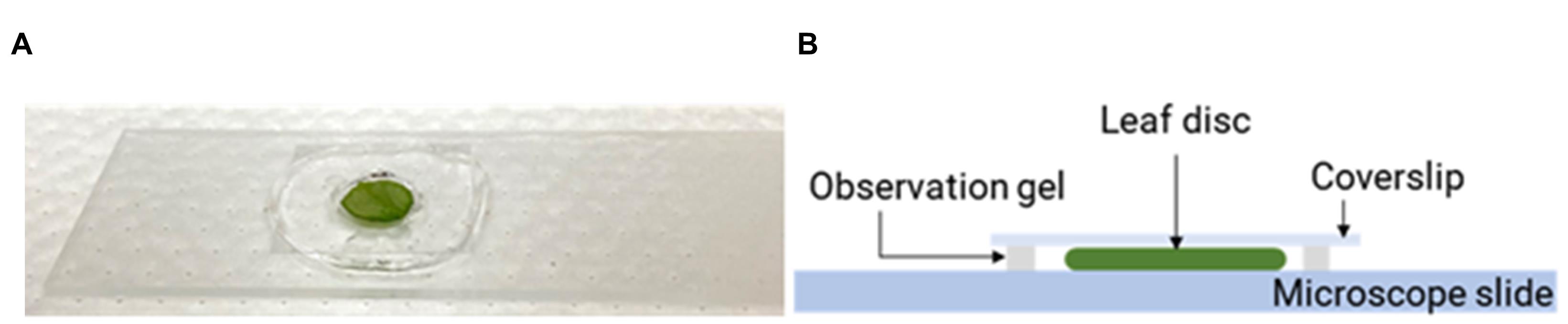
Figure 7. Preparation of a microscopy slide for confocal imaging of nanoparticles in leaf samples. A. Leaf disk samples were placed inside a gel chamber and mounted on glass slides for confocal imaging. B. The gel chamber containing a leaf disk was filled with perfluorodecalin.
Confocal fluorescence microscopy
Load the leaf sample on the Leica laser scanning confocal microscope TCS SP5 (Leica Microsystems, Germany).
Set up the microscope as follows: 40× wet objective (HCX PL APO CS 40.0 × 1.10 WATER UV, Leica Microsystems, Germany); 405 nm laser excitation for QDs; 514 nm for DHE; z-stack section thickness of 2 µm; line average of 4; PMT detection range of 500-550 nm for QDs, 580-615 nm for DHE, and 720-780 nm for chloroplast autofluorescence.
Scan to locate a flat-leaf surface region of interest and image the sample.
Collect the QD and DHE signals separately to avoid overlap between the excitation of the DHE dye and the emission detection range of QDs.
Image three or more leaf discs for each plant and take images of two different regions per leaf disc for confocal analysis (Figure 8).
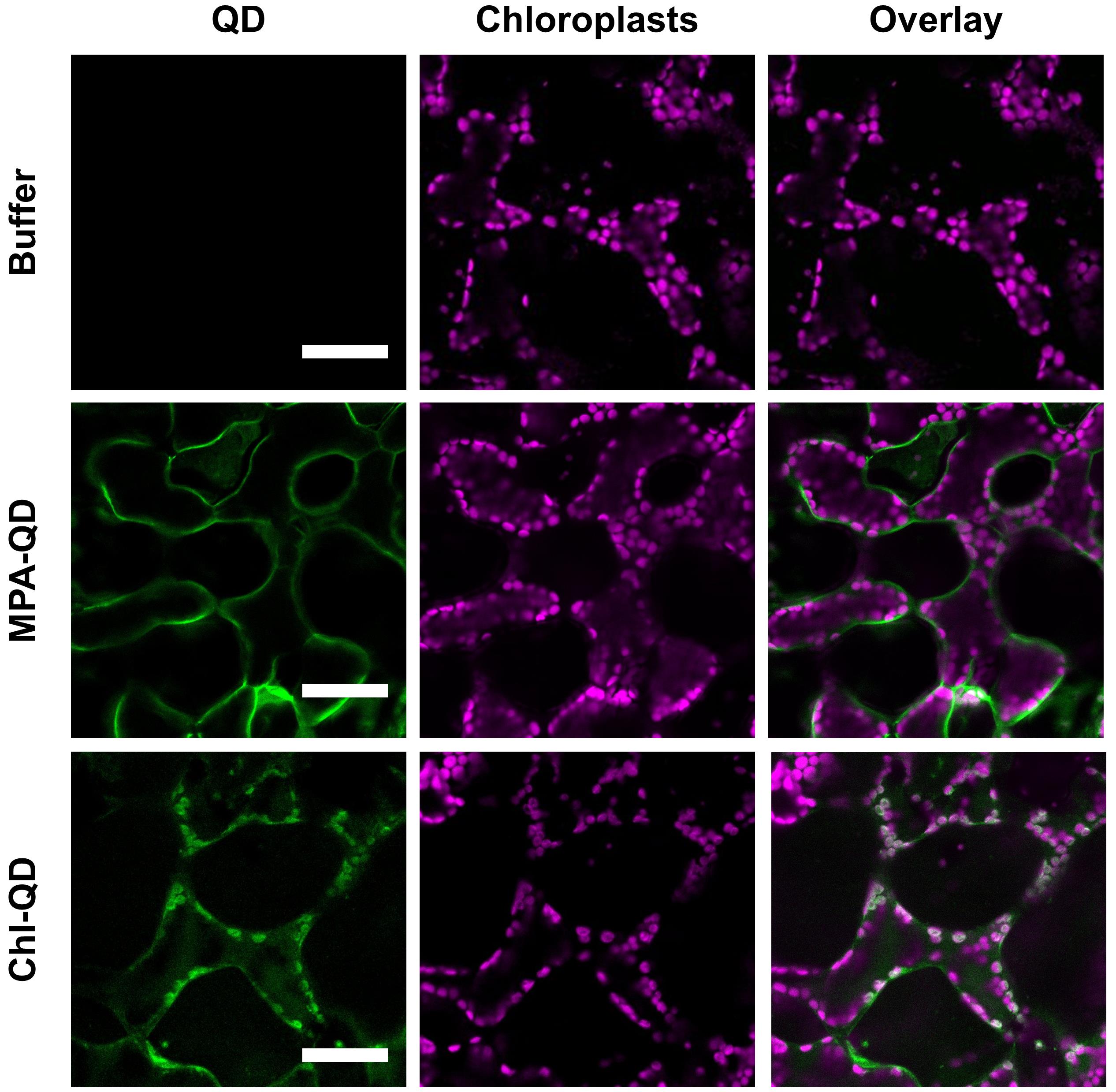
Figure 8. Confocal images of nanoparticles in plant leaves. Confocal images of mesophyll cells from leaves infiltrated with 10 mM TES buffer pH 7.0, MPA-QD, and Chl-QD. Images show the degree of overlap of the QD fluorescence signal with chloroplast autofluorescence. Scale bars: 50 µm.
Chloroplast isolation for nanoparticle detection by confocal microscopy and ICP-MS
Infiltrate the plant leaf tissue with approximately 100 µl 500 nM Chl-QD in TES buffer 10 mM pH 7.3.
Allow the plant leaves to incubate for 6 h at room temperature.
Collect approximately 8 g leaf tissue from 3-week-old Arabidopsis thaliana plants treated with buffer control or Chl-QD from 5-6 plants per treatment. Macerate the leaf tissue in a grinder or macerator with 1× chilled sucrose buffer (Recipe 1).
Grind the tissue by pulsing (3-4 pulses) for 5-s intervals until the leaf tissue is homogenized into a slurry (Figure 9A).
Strain the slurry through 4 layers of cheesecloth into a glass beaker on ice. Place the filtered flowthrough into a 50-ml centrifuge tube and centrifuge twice with 1× sucrose buffer at 3,082 × g for 10 min (Figure 9B).
Remove the supernatant each time and refill with a new sucrose buffer solution.
A pellet should form at the bottom of the centrifuge tube (Figure 9B).
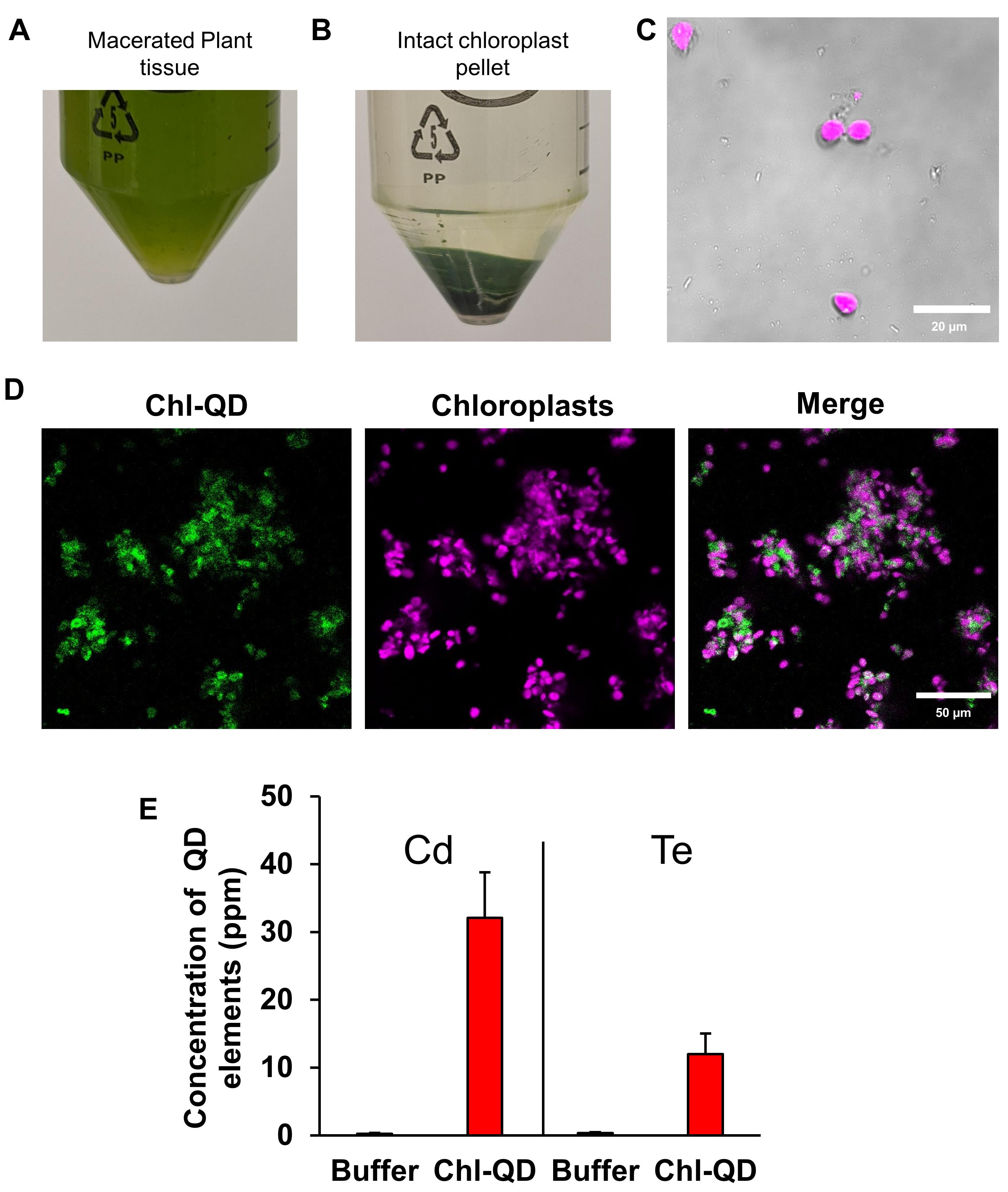
Figure 9. Chloroplast isolation schematic. Chloroplasts were isolated from 3-week-old Arabidopsis thaliana plants. A. Centrifuge tube filled with macerated leaf homogenate in a sucrose buffer. B. Pellet of intact chloroplasts. C. Confirmation of the colocalized Chl-QDs in isolated chloroplasts by confocal microscopy. The QD signal is visualized in green and the chloroplast autofluorescence in magenta. Scale bar: 20 µm. D. Detection and quantitation of QD elements (Cd and Te) detected in chloroplasts by ICP-MS analysis.Following chloroplast isolation, a small stab sample of chloroplasts was placed on a glass slide to detect quantum dot fluorescence within extracted chloroplasts using confocal microscopy (see section S for settings) (Figure 9C-9D).
Remove the supernatant (sucrose buffer and damaged chloroplasts) from the tube. Allow pelleted chloroplasts to air dry in a fume hood for more than 24 h. A small green sticky pellet will form.
Once the pellet is dry, place the tube in a -20°C freezer for 1 h until the pellet solidifies to allow easy removal from the centrifuge tube and preparation for ICP-MS analysis.
Inductively coupled plasma mass spectrometry (ICP-MS)
Following chloroplast isolation, dry the sample pellets (~0.1 g) in air for 48 h.
Place the air-dried samples in 50-ml polypropylene digestion tubes and digest with a solution of 5% HNO3/1% HCl/1% H2O2 v/v. Digest the samples in 1 ml HNO3/0.4 ml HCl while heating at 115°C for 5 min using a heat block (DigiPREP System; S.C.P. Science, Champlain, NY). Add 0.4 ml H2O2 and incubate for an additional 10 min.
Dilute the solution and analyze the samples by ICP-MS (Agilent 7700x ICP-MS) to quantitate the Cd and Te content. Report individual element concentrations in μg g-1 as in Figure 8D (element mass in μg per g dry chloroplast).
Data analysis
Confocal fluorescence microscopy image analysis with Fiji (ImageJ)
To analyze the images, open the image in the Fiji (ImageJ) software (Figure 10A-10C).
Using the line tool, draw six transect lines on an image set to be analyzed evenly. Save the lines to the region of interest (ROI) manager (Figure 10A).
Measure the corresponding fluorescence intensity profiles for the QD and DHE fluorescence and chloroplast autofluorescence channels across each of the six ROI line sections. Using the Fiji software, select Analyze > Plot profile (Figure 10B).
Count the number of overlapping peaks between the QD and DHE channels over chloroplast fluorescence, and calculate the colocalization percentage (Figure 10B-10C).
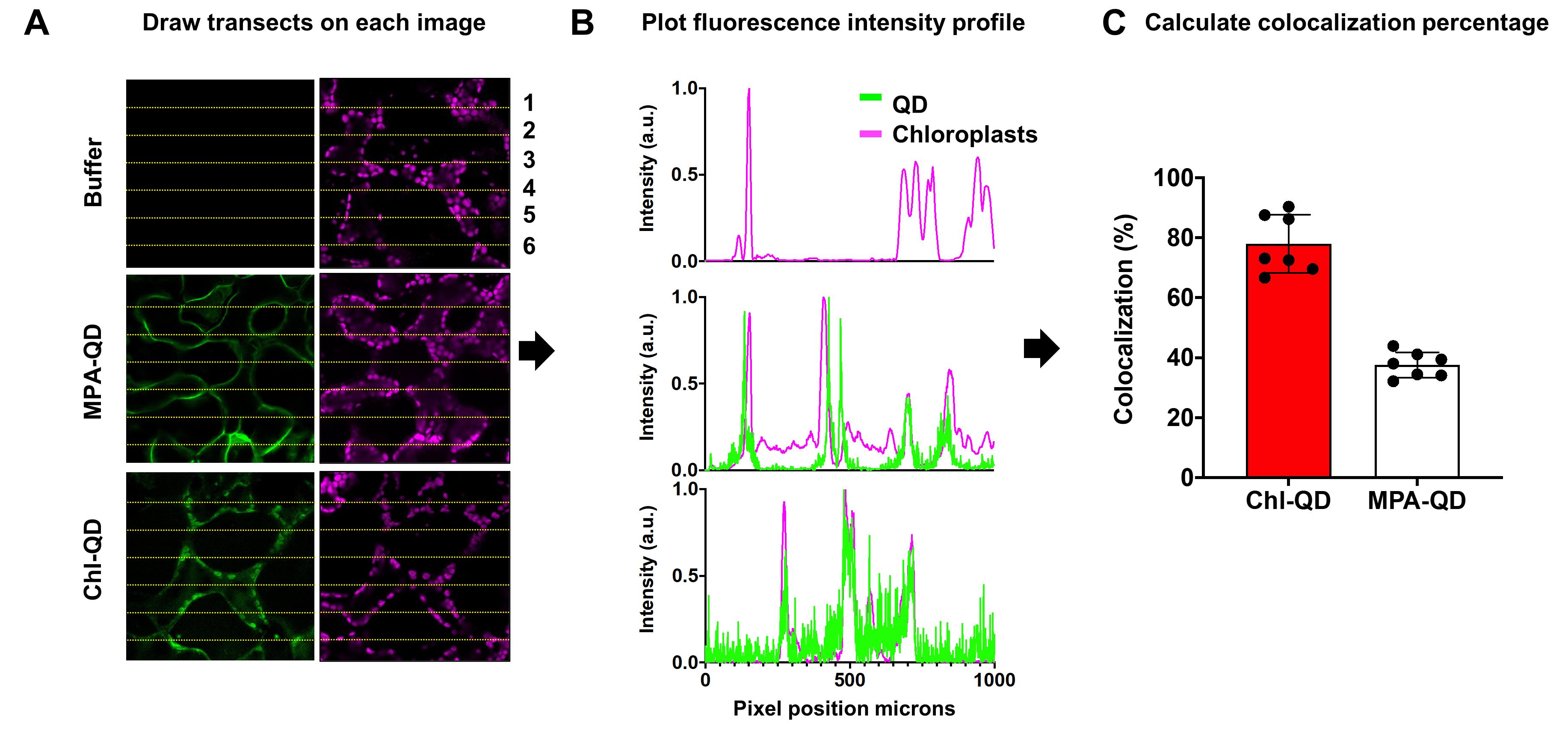
Figure 10. Stepwise schematic for colocalization analysis between targeted nanomaterials (Chl-QD) and chloroplasts in leaf mesophyll cells. A. Confocal fluorescence microscopy image analysis using the Fiji (ImageJ) software. B. Plots of normalized fluorescence intensity profiles collected by confocal microscopy and processed in Fiji (ImageJ). C. Calculated colocalization percentage of overlapping fluorescence intensity peaks.
Notes
If air enters, the reaction solution will turn from blue to black, thereby oxidizing the catalyst and causing the inefficient formation of quantum dot crystals.
Solution B should exhibit a color change as the reaction progresses, turning black-blue to pink-purple (Figure 2B).
EDC is hygroscopic and will degrade in molecular grade H2O; make sure EDC is prepared fresh.
QD solution can be filtered through a 20-nm filter to remove agglomeration.
QDs tend to aggregate at a pH lower than 6.5.
Make sure not to completely wash away the buffer from the 10 K filter column. Keep APBA-QD in solution during washing.
Recipes
1× chilled sucrose buffer (pH 7.3)
28 mM Na2HPO4
22 mM KH2PO4
2.5 mM MgCl2
400 mM sucrose
10 mM KCl
pH 7.3
Acknowledgments
This material is based upon work supported by the National Science Foundation under Grant No. 1817363 to J.P.G. Students C.C. and H.T., and Postdoc P.H. were supported by the National Science Foundation under Grant No. CHE-2001611, the NSF Center for Sustainable Nanotechnology. The CSN is part of the Centers for Chemical Innovation Program. This protocol was based on our previous publication in Nature Communications (Santana et al., 2020).
Competing interests
There is a pending U.S. patent entitled "Compositions and methods for chloroplast genetic and biochemical engineering in plants" (16/218.429) that is based on this work. All authors of this Bio-protocol manuscript, J.P.G., I.S., and P.H., are inventors in this patent, including Gregory M. Newkirk (University of California, Riverside) and Hong Hong Wu (Huazhong Agricultural University). Specific aspects outlined in this protocol are covered in the patent application, including methods for the targeted delivery of nanomaterials to chloroplasts using rationally designed guiding peptides.
References
- Asati, A., Santra, S., Kaittanis, C. and Perez, J. M. (2010). Surface-charge-dependent cell localization and cytotoxicity of cerium oxide nanoparticles. ACS Nano 4(9): 5321-5331.
- Das, S., Chigurupati, S., Dowding, J., Munusamy, P., Baer, D. R., McGinnis, J. F., Mattson, M. P., Self, W. and Seal, S. (2014). Therapeutic potential of nanoceria in regenerative medicine. In: Materials Research Bulletin. 39: 976-983.
- Demirer, G. S., Zhang, H., Matos, J. L., Goh, N. S., Cunningham, F. J., Sung, Y., Chang, R., Aditham, A. J., Chio, L., Cho, M. J., Staskawicz, B. and Landry, M. P. (2019). High aspect ratio nanomaterials enable the delivery of functional genetic material without DNA integration in mature plants. Nat Nanotechnol 14(5): 456-464.
- Giraldo, J. P., Wu, H., Newkirk, G. M. and Kruss, S. (2019). Nanobiotechnology approaches for engineering smart plant sensors. Nat Nanotechnol 14(6): 541-553.
- Hu, P., An, J., Faulkner, M. M., Wu, H., Li, Z., Tian, X. and Giraldo, J. P. (2020). Nanoparticle Charge and Size Control Foliar Delivery Efficiency to Plant Cells and Organelles. ACS Nano 14(7): 7970-7986.
- Kah, M., Tufenkji, N. and White, J. C. (2019). Nano-enabled strategies to enhance crop nutrition and protection. Nat Nanotechnol 14(6): 532-540.
- Li, J., Lee, W. Y., Wu, T., Xu, J., Zhang, K., Li, G., Xia, J. and Bian, L. (2016). Multifunctional Quantum Dot Nanoparticles for Effective Differentiation and Long-Term Tracking of Human Mesenchymal Stem Cells In Vitro and In Vivo. Adv Healthc Mater 5(9): 1049-1057.
- Lowry, G. V., Avellan, A. and Gilbertson, L. M. (2019). Opportunities and challenges for nanotechnology in the agri-tech revolution. Nat Nanotechnol 14(6): 517-522.
- Nagajyoti, P. C., Lee, K. D. and Sreekanth, T. V. M. (2010). Heavy metals, occurrence and toxicity for plants: a review. Environ Chem Lett 8: 199-216.
- Patra, J. K., Das, G., Fraceto, L. F., Campos, E. V. R., Rodriguez-Torres, M. D. P., Acosta-Torres, L. S., Diaz-Torres, L. A., Grillo, R., Swamy, M. K., Sharma, S., Habtemariam, S. and Shin, H. S. (2018). Nano based drug delivery systems: recent developments and future prospects. J Nanobiotechnology 16(1): 71.
- Santana, I., Wu, H., Hu, P. and Giraldo, J. P. (2020). Targeted delivery of nanomaterials with chemical cargoes in plants enabled by a biorecognition motif. Nat Commun 11(1): 2045.
- Smith, A. M. and Gilbertson, L. M. (2018). Rational Ligand Design To Improve Agrochemical Delivery Efficiency and Advance Agriculture Sustainability. ACS Sustainable Chem Eng 6: 13599-13610.
- Wang, J. W., Grandio, E. G., Newkirk, G. M., Demirer, G. S., Butrus, S., Giraldo, J. P. and Landry, M. P. (2019). Nanoparticle-Mediated Genetic Engineering of Plants. Mol Plant 12(8): 1037-1040.
- Wang, P., Lombi, E., Zhao, F. J. and Kopittke, P. M. (2016). Nanotechnology: A New Opportunity in Plant Sciences. Trends Plant Sci 21(8): 699-712.
- Wong, M. H., Misra, R. P., Giraldo, J. P., Kwak, S. Y., Son, Y., Landry, M. P., Swan, J. W., Blankschtein, D. and Strano, M. S. (2016). Lipid Exchange Envelope Penetration (LEEP) of Nanoparticles for Plant Engineering: A Universal Localization Mechanism. Nano Lett 16(2): 1161-1172.
- Wu, H., Tito, N. and Giraldo, J. P. (2017). Anionic Cerium Oxide Nanoparticles Protect Plant Photosynthesis from Abiotic Stress by Scavenging Reactive Oxygen Species. ACS Nano 11(11): 11283-11297.
- Yin, J., Wang, Y. and Gilbertson, L. M. (2018). Opportunities to advance sustainable design of nano-enabled agriculture identified through a literature review. Environmental Science: Nano 5: 11-26.
- Yu, W.W., Qu, L., Guo, W. and Peng, X. (2003) Experimental Determination of the Extinction Coefficient of CdTe, CdSe, and CdS Nanocrystals. Chem Mater 15: 2854-2860.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Santana, I., Hu, P., Jeon, S., Castillo, C., Tu, H. and Giraldo, J. P. (2021). Peptide-mediated Targeting of Nanoparticles with Chemical Cargoes to Chloroplasts in Arabidopsis Plants. Bio-protocol 11(12): e4060. DOI: 10.21769/BioProtoc.4060.
Category
Plant Science > Plant cell biology > Intercellular communication
Molecular Biology > Nanoparticle
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








