- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protein Import Assay into Mitochondria Isolated from Human Cells
(*contributed equally to this work) Published: Vol 11, Iss 12, Jun 20, 2021 DOI: 10.21769/BioProtoc.4057 Views: 6324
Reviewed by: Alexandros AlexandratosIbrahim AlharbiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Modified Acyl-RAC Method of Isolating Retinal Palmitoyl Proteome for Subsequent Detection through LC-MS/MS
Sree I. Motipally [...] Saravanan Kolandaivelu
Apr 20, 2023 2631 Views

Analysis and Quantification of the Mitochondrial–ER Lipidome
Alexis Diaz-Vegas [...] James G. Burchfield
Jul 5, 2024 2580 Views

Preparation of Protein Lysates Using Biorthogonal Chemical Reporters for Click Reaction and in-Gel Fluorescence Analysis
Yaxin Xu and Tao Peng
Nov 20, 2024 2011 Views
Abstract
Mitochondria are essential organelles containing approximately 1,500 proteins. Only approximately 1% of these proteins are synthesized inside mitochondria, whereas the remaining 99% are synthesized as precursors on cytosolic ribosomes and imported into the organelle. Various tools and techniques to analyze the import process have been developed. Among them, in vitro reconstituted import systems are of importance to study these processes in detail. These experiments monitor the import reaction of mitochondrial precursors that were previously radiolabeled in a cell-free environment. However, the methods described have been mostly performed in mitochondria isolated from S. cerevisiae. Here, we describe the adaptation of this powerful assay to import proteins into crude mitochondria isolated from human tissue culture cells.
Graphic abstract:
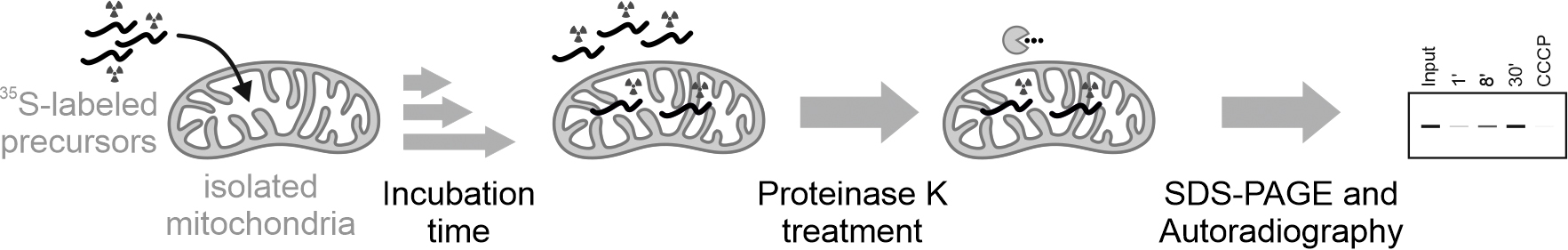
Overview of the assay to monitor protein import into mitochondria isolated from human cells
Background
Mitochondrial proteins perform critical functions in energy conversation, heat production, lipid and iron metabolism, signaling, and cell death pathways (Vafai and Mootha, 2012; Spinelli and Haigis, 2018; Habich et al., 2019b; Pfanner et al., 2019). Human mitochondria contain approximately 1500 different proteins, but only 13 are translated in the matrix on mitochondrial ribosomes. Thus, the vast majority of proteins are synthesized as precursors on cytosolic ribosomes and then imported into mitochondria. Different import pathways have been identified that direct proteins to their correct submitochondrial localization (Hartl et al., 1989; Chacinska et al., 2009; Schmidt et al., 2010; MacPherson and Tokatlidis, 2017; Endo and Tamura, 2018; Hansen and Herrmann, 2019). Most of these pathways have been characterized using a handful of model substrates. However, it becomes increasingly clear that many proteins have different dependencies on import machinery components and that the intrinsic protein properties that drive import can vary. In vitro reconstituted systems using isolated mitochondria are powerful tools that allow rapid analysis of mitochondrial protein import. These assays allow rapid and parallel analysis of protein import and its determinants. In contrast to assays in intact cells, cytosolic processes – including proteasomal degradation, differing protein transcription/translation rates, competition with other precursors, and contributions of specific chaperone systems – can be excluded in import assays using isolated mitochondria.
Most described import experiments have been performed in mitochondria isolated from S. cerevisiae (Mokranjac and Neupert, 2007; Weckbecker and Herrmann, 2013; Tang and Tang, 2015), mainly because yeast genetics allowed detailed mechanistic analysis and insight into import pathways. We adapted the protocol for monitoring protein import into mitochondria enriched from human tissue culture cells. The combination with CRISPR-Cas9 mediated gene editing (knock-outs and knock-ins), siRNA-mediated protein depletion, and stable cell lines overexpressing components of the human import machinery allow a detailed investigation of human mitochondrial protein import. This enables the investigation of the differences between human and yeast mitochondrial import as well as the analysis of human patient mutations.
Materials and Reagents
Cultivation of HEK293 cells
Cell culture dishes 100 × 20 mm (Sarstedt, catalog number: 83.3902)
Cell culture dishes 150 × 20 mm (Sarstedt, catalog number: 83.3903)
Autoclaved disposable glass Pasteur pipets without cotton pad (VWR, catalog number: HECH40567001)
Serological pipettes: Sterile individually packed,
2 ml pipettes (Sarstedt, catalog number: 83.3903)
5 ml pipettes (Sarstedt, catalog number: 86.1252.001)
10 ml pipettes (Sarstedt, catalog number: 86.1254.001)
25 ml pipettes (Sarstedt, catalog number: 86.1685.001)
Flp-In T-REx HEK293 cells (Human embryonic kidney 293, Invitrogen, catalog number: R78007)
Dulbecco’s modified Eagle medium (DMEM) medium-complete (see Recipes)
10% Fetal calf serum (FCS, Sigma-Aldrich, catalog number: F0804) and
1% penicillin/streptomycin (P/S, Sigma-Aldrich, catalog number: P0781-100ML)
DMEM high glucose (Thermo Fisher, catalog number: 41965-062)
Dulbecco's Phosphate Buffered Saline (PBS) (see Recipes)
PBS powder (Sigma-Aldrich, catalog number: D5652)
Trypsin-EDTA solution (see Recipes)
10× Trypsin-EDTA solution (Sigma-Aldrich, catalog number: T4174)
Isolation of mitochondria from HEK293 cells
Cell scraper 16 mm (Sarstedt, catalog number: 831832)
15 ml Falcon tubes (VWR, catalog number: 734-0451)
3 × 150 mm confluent dishes plated with HEK293 cells
Note: Cells are seeded 3-4 days prior to the mitochondrial isolation. Cells are grown to 90% confluency. See Note 2.
CompleteTM Protease Inhibitor Cocktail EDTA-free tablets (Roche, catalog number: 11873580001). Add freshly before use. Aliquots can be stored at -20°C
PBS, store at 4°C before use
Bradford Reagent ROTI® Quant (Roth, catalog number: K015.1)
1× buffer M (see Recipes)
Mannitol (Merck, catalog number: 1.05983)
Sucrose (Sigma-Aldrich, catalog number: S0389)
HEPES-KOH, pH 7.4 (VWR, catalog number: 441487)
EGTA-KOH, pH 7.4 (Roth, catalog number: 3054.1)
Synthesis of [35S]-labeled precursor
TNT® Quick Coupled Transcription/Translation System SP6 Promoter (Promega, catalog number: L2080). See Note 3.
EasyTagTM EXPRESS 35S Protein Labeling Mix, [35S]-, 7 mCi, 1175 Ci/mmol (PerkinElmer, catalog number: NEG772007MC). See Note 4.
DNA template (1 µg ml-1) containing the gene of the mitochondrial precursor inserted downstream of the SP6 promoter. Plasmid vector: pGEM-3/4 (Promega). Store at -20°C.
Nuclease-free double-distilled water
200 mM L-methionine (Merck, catalog number: 1057070100) in nuclease-free double-distilled water. Store at 4°C
50 mM ethylene diamine tetra acidic acid (EDTA) disodium salt 2-hydrate, pH 8 (AppliChem, catalog number: A2937) in nuclease-free double-distilled water
1× sample buffer (see Recipes)
Tris(hydroxymethyl)aminomethane (Tris, VWR, catalog number: 0497)
Glycerol (Sigma-Aldrich, catalog number: G7757)
Bromophenol blue (Roth, catalog number: A512.1)
Dithiothreitol (DTT, AppliChem, catalog number: A1101)
Import of [35S]-labeled precursors into isolated mitochondria
Blotting membrane, nitrocellulose, AmershamTM Protran® (VWR, catalog number: 10600001), 0.2 µm pore size
20 mM Carbonyl cyanide 3-chlorophenylhydrazone (CCCP, Sigma-Aldrich, catalog number: C2759)
200 mM phenylmethylsulfonyl fluoride (PMSF, AppliChem, catalog number: A0999.0100)
Crushed ice
Triton X-100 (AppliChem, catalog number: A1388)
1× buffer M (see Recipes)
20 µg/import reaction mitochondria (see: Recipes)
20 µg ml-1 peqGOLD proteinase K (see Recipes)
1× sample buffer (see Recipes)
SEH buffer (see Recipes)
Equipment
Cultivation of HEK293 cells
Laminar flow hood class II (ENVAIR eco)
CO2 incubator
Vacuum pump
Microscope
Centrifuge (e.g., Eppendorf, model: 5702R)
Equipped cell culture laboratory containing, e.g., a laminar flow hood class II (ENVAIR eco), a CO2 incubator for cell cultivation set at 37°C and 5% CO2, a vacuum pump for removal of medium using sterile glass Pasteur pipets, a microscope, and a cell counter and a cooling centrifuge.
Isolation of mitochondria from HEK293 cells
Electric stirrer (Heidolph RZR 2020)
Potter S homogenizer (15 ml) with tight-fitting PTFE (polytetrafluorethylen) plungers (Sartorius, catalog number: 8542406)
The range of clearance (i.e., the average distance between the pestle and the cylinder walls) optimal for mitochondria isolation is thereby from 0.045-0.065 mm.
Centrifuge (Eppendorf, model: 5702R)
Centrifuge (Eppendorf, model: 5417R)
Vacuum pump (KNF laboport minipump) for removal of medium using sterile glass Pasteur pipets.
[35S]-labeled precursor synthesis
Hypoxic Glove Box and Cabinet (Coy Laboratory Products, “Coy 1 Person O2 Control Glove Box”, catalog number: 031615); not necessary if precursor protein is not prone to air oxidation (e.g., does not contain or contains only few cysteine residues).
Import of [35S]-labeled precursors into isolated mitochondria
ThermoMixer Eppendorf (F1.5, #EP5384000012)
Centrifuge (Eppendorf, model: 5417R)
System for SDS-PAGE electrophoresis and (semidry) blotting (Bio-Rad, MiniPROTEAN Tetra Cell, catalog number: 1658000)
Imaging System Typhoon FLA 9500 (GE Healthcare, catalog number: 9351515)
Image Eraser FLA (GE Healthcare, catalog number: 12999750)
Software
ImageQuant TL 8.1 (GE Healthcare Life Science).
Procedure
An overview of the procedure is depicted in Figure 1.
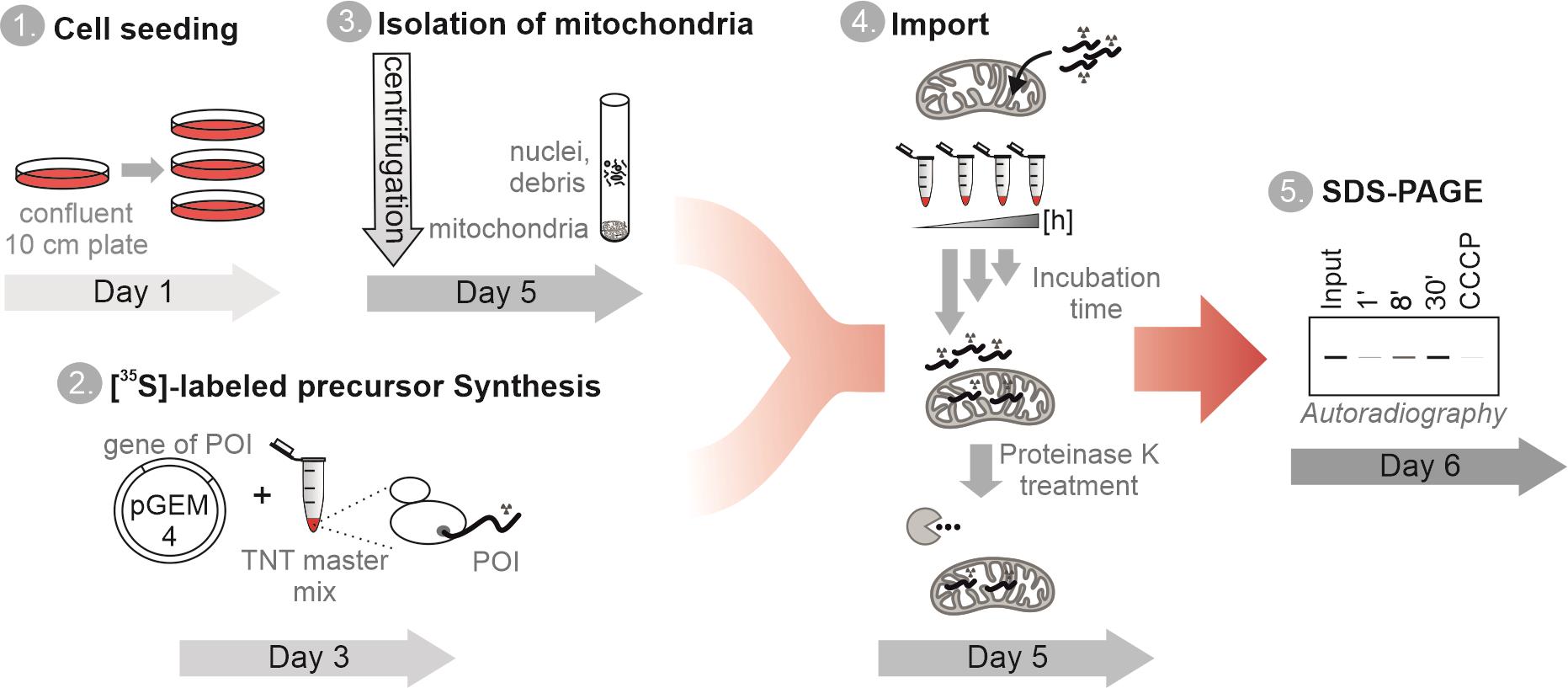
Figure 1. Schematic representation of the steps in the ”in organello import assay” protocol
Cultivation of HEK293 cells
HEK293 cells are cultured in DMEM medium complete containing high glucose, FCS, and a penicillin/streptomycin antibiotic mixture. For passaging, HEK293 cells are cultured on 100 mm dishes in 10 ml DMEM medium complete until 90% confluency. For the described experiment, HEK293 cells are plated on 150 mm dishes in 30 ml DMEM medium complete and cultivated until they reach confluency. Special care must be taken for some cells in which mitochondrial proteins (e.g., of the mitochondrial import machinery) have been either depleted or overexpressed; different cultivating conditions might be required.
Cultivate HEK293 cells on 100 mm dishes in 10 ml DMEM complete medium in an incubator at 37°C and 5% CO2 until they reach 90% confluency.
Transfer the HEK293 dishes into a surface-sterilized laminar flow hood class II.
Transfer the sterile DMEM medium complete, sterile PBS, and sterile Trypsin-EDTA into the laminar flow hood. All solutions should be preheated to 37°C.
Remove the medium using an autoclaved disposable glass Pasteur pipet and a vacuum pump.
Rinse the cells carefully by adding 10 ml of sterile PBS at the edge of the dish using a serological pipette and a pipette boy.
Remove the PBS using an autoclaved disposable glass Pasteur pipet and a vacuum pump.
Add 1 ml of sterile Trypsin-EDTA dropwise onto the cells using a serological pipette and a pipette boy.
Incubate the cells for 5 min at 37°C and 5% CO2 until cells detach from the dish. Flick the dish with your finger.
Transfer the dish back into the surface-sterilized laminar flow hood and add 9 ml of DMEM medium complete.
Singularize the cells using a 10 ml serological pipette and a pipette boy. Press the pipette tip softly onto the dish bottom.
Transfer three times 2 ml of the cell suspension into a 150 mm dish containing 28 ml DMEM medium complete. After this step, you should have three dishes with the same number of cells.
Add 9 ml of DMEM medium complete to the “old” 100 mm dish for further cultivation.
Transfer all plates into the incubator at 37°C and 5% CO2 and cultivate the HEK293 cells plated onto the three 150 mm dishes until they reach confluency.
Isolation of mitochondria from HEK293 cells
This protocol does not yield highly pure mitochondria but a fraction enriched in mitochondria. This is sufficient to monitor protein import into mitochondria. For an efficient isolation procedure, make sure that all buffers, materials, and reagents are precooled at 4°C. Always work on crushed ice. Isolated mitochondria are delicate (the mitochondrial membranes can easily be disrupted); be careful when pipetting solutions containing mitochondria and use cut pipette tips.
Rinse the three confluent 150 mm dishes plated with HEK293 cells (approximately 30 × 106 cells per 150 mm dish) with 10 ml of ice-cold PBS.
Add 10 ml of ice-cold PBS and gently scrape the cells off using a cell scraper. Transfer the cell solutions into 3 × 15 ml Falcon tubes.
Pellet the cells at 500 × g for 5 min at 4°C in 15 ml Falcon tubes using a centrifuge (Eppendorf 5702R). Remove PBS using a vacuum pump.
Resuspend all three cell pellets together in a total of 5 ml 1× M buffer containing 1× CompleteTM Protease Inhibitor Cocktail using a 10 ml serological pipette and a pipette boy. See Note 6.
Transfer cells to precooled Potter S homogenizer cylinder. Homogenize cells using the precooled potter homogenizer on crushed ice (1,000 rpm). Move the cylinder (the pestle/plunger is fixed in the stirrer) up and slowly down (1x up and down counts as one stroke). Perform 15 strokes. In this step, the plasma membrane is opened, and organelles and the cytosol are released. See Note 7.
Cell lysis can be monitored by pipetting 20 µl of suspension onto a glass slide and placing a coverslip on top before looking at cell integrity under the light microscope using Trypan Blue staining.
Transfer the cell homogenate to a 15 ml Falcon tube. Pellet the homogenized cells at 600 × g for 5 min at 4°C using a centrifuge (Eppendorf 5702R).
Collect the supernatant and store at 4°C.
If you suspect that cell lysis might have been not efficient, perform Steps B10-B12; otherwise, proceed with Step B13.
Resuspend the pellet from Step B7, which still might contain intact cells, in 5 ml 1× M buffer and repeat the potter step (15 strokes, 1,000 rpm).
Pellet the homogenized cells at 600 × g for 5 min at 4°C in 15 ml Falcon tubes using a centrifuge (Eppendorf 5702R).
Collect the supernatant and pool with the supernatant from Step B7.
Distribute the supernatant in 2 ml aliquots into 2 ml tubes using cut 1 ml-pipette tips. You should have three completely filled 2 ml tubes at this step. Spin at 600 × g for 5 min at 4°C using a centrifuge (Eppendorf 5417R) to pellet nuclei.
Carefully collect the supernatant from step 13 using cut 1 ml-pipette tips (avoid touching the pellet with the pipet tip) and transfer to fresh 2 ml tubes. Spin supernatant at 8,000 × g for 10 min at 4°C using a centrifuge (Eppendorf 5417R).
Note: The pellet from this step contains the crude mitochondria.
Carefully collect the supernatant and discard it. Avoid touching the pellet with the pipette tip but still remove the supernatant completely.
Following this protocol will yield 3 × 2 ml tubes containing pellet. With a cut 200 µl-pipette tip, gently resuspend these pellets, which contain crude mitochondria in a total of 1 ml ice-cold 1× M buffer (without complete protease inhibitor cocktail). Combine pellets into one 2 ml tube. Add another 1 ml ice-cold 1× M buffer (without the complete protease inhibitor cocktail). See Note 8.
Spin the 2 ml tube at 6,000 × g for 10 min at 4°C using a centrifuge (Eppendorf 5417R) and carefully remove the supernatant.
Note: The pellet from this step contains the mitochondria.
Resuspend the mitochondrial fraction using a cut 200 µl-pipette tip in 400 µl of ice-cold 1× M buffer.
Store on ice until the protein content has been determined. Afterward, proceed immediately with the import experiments.
Quantify the concentration of the isolated mitochondria using the Bradford Reagent ROTI® Quant Assay according to the manufacturer’s instructions. Use a range of different dilutions of the mitochondria solution for protein determination to ensure that at least one sample falls within the standard range. For wild-type HEK293 cells, a good start is protein determination on a 1:10 dilution of the mitochondria solution.
[35S]-labeled precursor synthesis
Cytosolic ribosomes synthesize precursors in their reduced states (i.e., cysteine residues are present as thiols). Cytosolic enzymes maintain these precursors in their reduced state to achieve efficient translocation into mitochondria ( Durigon et al., 2012 ; Banci et al., 2013 ; Habich et al., 2019a ). To generate reduced radiolabeled precursors, perform the following steps in a hypoxic glove box and cabinet with 0.5% residual O2. Sulfur-35 [35S] is a low energy beta emitter. Refer to the general safety precautions when working with radioactivity.
Plan the synthesis of radioactively labeled precursors well. In general, synthesis should take place at least on the day before mitochondria isolation and protein import.
To synthesize 50 µl of the [35S]-labeled precursor lysate, mix 40 µl of TNT® Quick Coupled Transcription/Translation Mix and 2 µl of EasyTagTM EXPRESS 35S Protein Labeling Mix with 1 µg of DNA template. Fill the mixture up to 50 µl with nuclease-free double-distilled water.
Incubate for 90 min at 30°C in the hypoxic glove box cabinet under low O2 concentration.
To stop the reaction, add 1 µl of 200 mM L-methionine.
Incubate for 5 min at 37°C.
Add 1 µl 50 mM of EDTA, which initiates the dissociation of ribosomes.
Store the lysates at -80°C. See Note 9.
To test the efficiency of the quick transcription/translation reaction before the mitochondrial import process, resuspend 0.5 µl of the radiolabeled lysate in 20 µl of 1× sample buffer containing 50 mM DTT.
Boil all samples for 5 min at 95°C.
Analyze the samples by SDS-PAGE.
Transfer the proteins onto a nitrocellulose blotting membrane and dry it.
Expose the nitrocellulose blotting membrane to an autoradiography film/screen.
After 16 h, develop the autoradiography film using an imaging system (e.g., Typhoon). The protein signal should be well visible at the correct molecular weight. If the signal is very strong after 16 h, then dilute the lysate accordingly. If the signal is weak, the synthesis might not have worked properly, or the protein of interest contains too few methionine residues. See Note 10.
In vitro import of [35S]-labeled precursors into isolated mitochondria
In the import experiment, the translocation efficiency of mitochondrial precursors across the outer mitochondrial membrane is monitored. Non-imported pre-proteins are accessible to degradation by treatment with the serine protease, proteinase K, whereas translocated precursors are protected inside the organelle. Different variations of the protocol test the impact of the membrane potential or specific components of the import machinery on precursor import (see Note 11). For example, the impact of the membrane potential can be tested by using the proton gradient uncoupler CCCP. The import of proteins to the correct intramitochondrial localization can be tested by mitochondrial fractionation after the import.
Isolated mitochondria from HEK293 cells are very delicate. Be careful when pipetting mitochondria and perform the import reaction on the same day of the mitochondrial isolation.
Plan the layout of the experiment for determining the import kinetics, e.g., how many time points are required to assess mitochondrial import. Good time points for an initial experiment are, for example, 1, 10, and 30 min. Additionally, controls are required. The “input” control allows calculating the import efficiency and comparing biological replicates. The “Triton X-100” control allows assessing whether unfolded/folded precursors are susceptible to protease treatment. This is important as resistance to degradation will prevent distinguishing between imported and non-imported protein (= degraded by protease). The “no membrane potential” control is important to test whether the import of the specific precursor of interest depends on the membrane potential. All these samples and controls add up to the total number of import reactions necessary.
Distribute 20 µg of mitochondria per import in reaction tube (import can be performed independently for each time point, i.e., one reaction tube per time point or using a master mix from which aliquots are drawn at each time point). Centrifuge at 8,000 × g for 5 min at 4°C using a centrifuge (Eppendorf 5417R). Remove supernatant. The pellet contains mitochondria. For the “normal” import kinetics assessment, mix carefully 20 µg pelleted crude mitochondria with 4 µl [35S]-labeled precursor lysate (potentially diluted after the lysate test, see Step B13) using a cut 10 µl pipette tip. Avoid vortexing since this breaks the mitochondrial membranes. For each time point, prepare an individual reaction tube on ice. See Note 12.
For the “no membrane potential” control: mix carefully 20 µg of pelleted crude mitochondria with 1 μl of 20 mM CCCP. Allow 5 minutes incubation time. Add 4 µl of [35S]-labeled precursor lysate.
The import reaction starts as soon as mitochondria and lysate are mixed. During the import reaction, place the samples at 30°C while shaking at 600 rpm in a ThermoMixer.
Stop the reaction at desired time points by placing the samples on ice. Directly centrifuge 5 min at 8,000 × g. Remove supernatant. The pellet contains mitochondria with imported protein.
For proteinase K treatment, resuspend mitochondria in 400 µl of 1× M buffer containing 20 µg of ml-1 proteinase K.
Incubate for 20 min on ice.
For the “Triton X-100” control, resuspend mitochondria in step 6 in 400 µl of 1× M buffer containing 20 µg ml-1 of proteinase K and 0.5% Triton X-100.
Stop the digest by adding 2 µl of 200 mM PMSF (1 mM final).
Centrifuge at 10,000 × g for 5 min at 4°C using a centrifuge (Eppendorf 5417R). Remove supernatant. The pellet contains mitochondria with imported protein.
Add 400 µl of buffer M containing 1 mM PMSF and spin at 10,000 × g for 5 min at 4°C using a centrifuge (Eppendorf 5417R). Remove supernatant. The pellet contains mitochondria with imported protein. Centrifuge at 10,000 × g for 1 min at 4°C using a centrifuge (Eppendorf 5417R). Remove residual supernatant. The pellet contains mitochondria with imported protein.
Resuspend the pellet in 40 µl of 1× sample buffer containing 50 mM DTT. See Note 13.
Boil all samples for 5 min at 95°C.
Analyze the samples by SDS-PAGE.
Transfer the proteins onto a nitrocellulose blotting membrane and dry it.
Expose the nitrocellulose blotting membrane to an autoradiography film/screen.
After one day up to several days, develop the autoradiography film/screen using an imaging system (e.g., Typhoon). An example of such an experiment is shown in Figure 2.
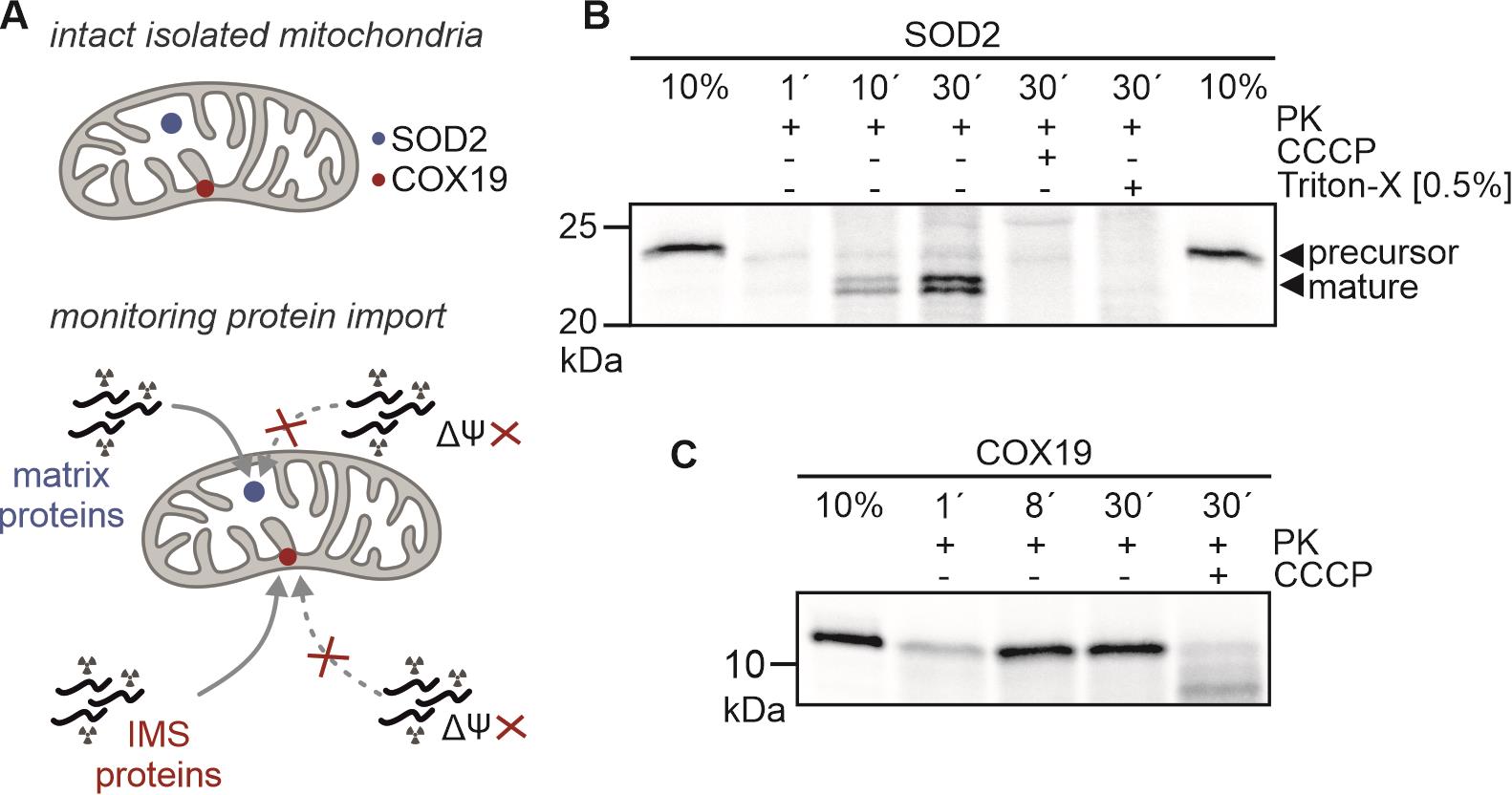
Figure 2. In organello import assay of a mitochondrial matrix protein SOD2 (containing a mitochondrial targeting signal, MTS) and an IMS protein, Cox19 (no MTS). A. SOD2 and COX19 are localized in matrix and IMS, respectively. The mitochondrial membrane potential is important for the import of matrix proteins and also for IMS proteins without MTS in human cells. B. Import kinetic of radiolabelled SOD2 into isolated HEK293 mitochondria. The SOD2 precursor is processed upon reaching the matrix (mature form). Depletion of the membrane potential (CCCP) prevents import. Mitochondria-localized SOD2 is not resistant to proteinase K (PK) treatment as its signal is lost upon Triton X-100 treatment. Imported SOD2 was visualized by autoradiography (18 h exposure). C. Import of Cox19 into HEK293 mitochondria. COX19 does not contain an MTS. Thus, imported and non-imported COX19 migrate at the same height. COX19 import is dependent on the membrane potential (lower signal in CCCP control).
Data analysis
The protein signals were quantified using ImageQuant TL 8.1 (GE Healthcare Life Science) and plotted using Microsoft Excel. Experiments are usually repeated three times and presented as averages with standard deviation.
Notes
Cells with impaired import machinery (e.g., obtained through CRISPR-Cas9-mediated deletion of import machinery components) might have difficulties growing in normal medium. In this case, uridine (50 µg/ml final, Sigma-Aldrich, #U3003), sodium pyruvate (Sigma-Aldrich, #S8636), and non-essential amino acids (Sigma-Aldrich, #M7145) can be added.
From 3 × 150 mm plates of confluent HEK293 cells, a total of 0.5-1.0 mg crude mitochondria can be obtained. Different tissue culture cells can give quite different yields. For example, for HeLa cells, a total of 0.2-0.3 mg crude mitochondria can be obtained.
The TNT® Quick Coupled Transcription/Translation System can also be ordered to accommodate a T7 promotor depending on your starting plasmid.
The EasyTagTM EXPRESS 35S Protein Labeling Mix, [35S]-, 7mCi, 1175 Ci/mmol from Perkin Elmer works best for the in organello import assay.
Keep Proteinase K in single-use aliquots and avoid multiple freeze-thaw cycles as this abolishes the enzymatic activity. Aliquots can be stored at -20°C for several months.
The volume of the 1× M buffer used for suspending cells before potter homogenization depends on the cell amount used. For 3 × 150 mm plates, use 5 ml of 1× M buffer. For more cells, multiply this amount accordingly. For fewer cells, still use 5 ml of 1× M buffer.
Incubation at this step leads to cell swelling, which is beneficial for cell lysis during potter homogenization. While swelling, the potter homogenizer can be assembled by screwing the PTFE plunger to the electric stirrer. Set the electric stirrer to 1,000 rpm. Before you start to homogenize, test if the plunger is tightly assembled and does not show any horizontal movement. Move the cylinder containing the cell suspension only when the plunger is rotating to avoid vacuum and air bubbles. Also, avoid creating a vacuum with the up-stroke to keep the mitochondria intact during homogenization. This is done by performing the up-stroke movement very slowly.
From this point on, the 1× M buffer does not contain protease inhibitor cocktail to avoid inhibition of proteinase K in the import assay.
Note that the half-life of radioactive decay of 35S is 87 days. Radioactive lysates should be used as fresh as possible to obtain maximum signal intensities. Moreover, a longer lysate storage time will lead to the oxidation of cysteine residues, which might prevent import.
If the (mature) protein does not contain enough methionine residues (and less critical, enough cysteines), additional methionine residues might be added to increase signal strength (e.g., addition of 4 methionine residues at the C-terminus). Alternatively, precursors might also be translated non-radioactively (or expressed and purified from E. coli and unfolded) and then detected by other methods, including through immunoblot against an additional tag.
Variation of the described import reaction might be implemented to analyze import pathways in mechanistic detail.
Analyzing the submitochondrial localization of the imported protein: Perform a submitochondrial fractionation after import, either by digitonin titration or hypoosmotic swelling combined with PK treatment, to remove the outer membrane and IMS proteins.
Interaction of precursor with import machinery components: Perform immunoprecipitation experiments after import, possibly using crosslinking approaches to stabilize the interactions.
Analysis of redox processes during import: Perform imports in the presence of different amounts of membrane-permeable (DTT) or impermeable (TCEP, glutathione) reducing agents. A dependency on these agents indicates the occurrence of redox processes during import (e.g., because of oxidative folding of the precursor).
Analysis of import pathways: Numerous cell lines lacking components of the import machinery have been generated ( Chiusolo et al., 2017 ; Habich et al., 2019a ; Richter et al., 2019 ); performing import experiments into mitochondria isolated from these cells allows mapping the specific import pathway in mechanistic detail.
In the import reaction with freshly isolated mitochondria, the addition of energy-regenerating components (like ADP, malate, NADH, etc.) appears unnecessary. Still, for some precursor proteins, this might be considered to further boost import efficiency.
For SDS-PAGE, 20 µl of the sample is sufficient for a good detection with a short exposure time.
Recipes
Dulbecco’s modified Eagle medium (DMEM) medium-complete
Add 10% fetal calf serum and 1% penicillin/streptomycin to a fresh bottle of DMEM high glucose. Store at 4°C and prewarm to 37°C before use. See Note 1.
Dulbecco's Phosphate Buffered Saline (PBS)
Dissolve one bottle PBS powder in double-distilled water according to the manufacturer’s description
Sterilize by autoclaving
Store at 4°C and prewarm to 37°C before use
Trypsin-EDTA solution
Dilute 10× Trypsin-EDTA solution 1:10 with PBS
Store at 4°C and prewarm to 37°C before use
Aliquots can be kept frozen at -20°C
1× buffer M
220 mM mannitol
70 mM Sucrose
5 mM HEPES-KOH, pH 7.4
1 mM EGTA-KOH, pH 7.4
Prepare freshly and store at 4°C before use
1× sample buffer
2% SDS
60 mM Tris(hydroxymethyl)aminomethane (Tris-HCl) pH 6.8
10% glycerol
0.005% bromophenol blue
50 mM dithiothreitol (DTT)
20 µg ml-1 peqGOLD proteinase K
PeqGOLD proteinase K in SEH buffer. See Note 5.
SEH buffer
250 mM Sucrose
1 mM EDTA
20 mM HEPES/KOH pH 7.4
20 µg/import mitochondria
Isolated mitochondria must be used immediately for import experiments. After isolation, mitochondria concentration is assessed with the Bradford assay. The concentration of mitochondria for each import reaction is calculated from this and directly pipetted (20 µg/import). Mitochondria can be diluted in buffer M if required.
Acknowledgments
The Deutsche Forschungsgemeinschaft (DFG) funds the research in the Laboratory of JR (RI2150/2-2 – project number 251546152, RI2150/5-1 – project number 435235019, CRC1218 / TP B02 – project number 269925409, and RTG2550/1 – project number 411422114). The protocol was used in the following original research papers: Saita et al. (2018), MacVicar et al. (2019), and Murschall et al. (2020).
Competing interests
The authors declare that they have no competing interests.
References
- Banci, L., Barbieri, L., Luchinat, E. and Secci, E. (2013). Visualization of redox-controlled protein fold in living cells. Chem Biol 20(6): 747-752.
- Chacinska, A., Koehler, C. M., Milenkovic, D., Lithgow, T. and Pfanner, N. (2009). Importing mitochondrial proteins: machineries and mechanisms. Cell 138(4): 628-644.
- Chiusolo, V., Jacquemin, G., Yonca Bassoy, E., Vinet, L., Liguori, L., Walch, M., Kozjak-Pavlovic, V. and Martinvalet, D. (2017). Granzyme B enters the mitochondria in a Sam50-, Tim22- and mtHsp70-dependent manner to induce apoptosis.Cell Death Differ 24(4): 747-758.
- Durigon, R., Wang, Q., Ceh Pavia, E., Grant, C. M. and Lu, H. (2012). Cytosolic thioredoxin system facilitates the import of mitochondrial small Tim proteins.EMBO Rep 13(10): 916-922.
- Endo, T. and Tamura, Y. (2018). Shuttle mission in the mitochondrial intermembrane space. EMBO J 37(4).
- Habich, M., Salscheider, S. L., Murschall, L. M., Hoehne, M. N., Fischer, M., Schorn, F., Petrungaro, C., Ali, M., Erdogan, A. J., Abou-Eid, S., Kashkar, H., Dengjel, J. and Riemer, J. (2019a). Vectorial Import via a Metastable Disulfide-Linked Complex Allows for a Quality Control Step and Import by the Mitochondrial Disulfide Relay. Cell Rep 26(3): 759-774 e755.
- Habich, M., Salscheider, S. L. and Riemer, J. (2019b). Cysteine residues in mitochondrial intermembrane space proteins: more than just import. Br J Pharmacol 176(4): 514-531.
- Hansen, K. G. and Herrmann, J. M. J. T. p. j. (2019). Transport of proteins into mitochondria. 38(3): 330-342.
- Hartl, F. U., Pfanner, N., Nicholson, D. W. and Neupert, W. (1989). Mitochondrial protein import. Biochim Biophys Acta 988(1): 1-45.
- MacPherson, L. and Tokatlidis, K. (2017). Protein trafficking in the mitochondrial intermembrane space: mechanisms and links to human disease.Biochem J 474(15): 2533-2545.
- MacVicar, T., Ohba, Y., Nolte, H., Mayer, F. C., Tatsuta, T., Sprenger, H. G., Lindner, B., Zhao, Y., Li, J., Bruns, C., Kruger, M., Habich, M., Riemer, J., Schwarzer, R., Pasparakis, M., Henschke, S., Bruning, J. C., Zamboni, N. and Langer, T. (2019). Lipid signalling drives proteolytic rewiring of mitochondria by YME1L. Nature 575(7782): 361-365.
- Mokranjac, D. and Neupert, W. (2007). Protein import into isolated mitochondria. Mitochondria, Springer: 277-286.
- Murschall, L. M., Gerhards, A., MacVicar, T., Peker, E., Hasberg, L., Wawra, S., Langer, T. and Riemer, J. (2020). The C-terminal region of the oxidoreductase MIA40 stabilizes its cytosolic precursor during mitochondrial import. BMC Biol 18(1): 96.
- Pfanner, N., Warscheid, B. and Wiedemann, N. J. (2019). Mitochondrial protein organization: from biogenesis to networks and function. Nat Rev Mol Cell Biol 20(5): 267.
- Richter, F., Dennerlein, S., Nikolov, M., Jans, D. C., Naumenko, N., Aich, A., MacVicar, T., Linden, A., Jakobs, S., Urlaub, H., Langer, T. and Rehling, P. (2019). ROMO1 is a constituent of the human presequence translocase required for YME1L protease import. J Cell Biol 218(2): 598-614.
- Saita, S., Tatsuta, T., Lampe, P. A., Konig, T., Ohba, Y. and Langer, T. (2018). PARL partitions the lipid transfer protein STARD7 between the cytosol and mitochondria. EMBO J 37(4).
- Schmidt, O., Pfanner, N. and Meisinger, C. (2010). Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol 11(9): 655-667.
- Spinelli, J. B. and Haigis, M. C. (2018). The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol 20(7): 745-754.
- Tang, B. L. and Tang (2015). Membrane Trafficking: Second Edition. Springer.
- Vafai, S. B. and Mootha, V. K. (2012). Mitochondrial disorders as windows into an ancient organelle. Nature 491(7424): 374-383.
- Weckbecker, D. and Herrmann, J. M. (2013). Methods to study the biogenesis of membrane proteins in yeast mitochondria. Methods Mol Biol 1033: 307-322.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Murschall, L. M., Peker, E., MacVicar, T., Langer, T. and Riemer, J. (2021). Protein Import Assay into Mitochondria Isolated from Human Cells. Bio-protocol 11(12): e4057. DOI: 10.21769/BioProtoc.4057.
Category
Biochemistry > Protein > Posttranslational modification
Cell Biology > Organelle isolation > Mitochondria
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








