- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of First-Trimester and Full-term Human Placental Hofbauer Cells
(*contributed equally to this work) Published: Vol 11, Iss 11, Jun 5, 2021 DOI: 10.21769/BioProtoc.4044 Views: 4654
Reviewed by: Luis Alberto Sánchez VargasWilliam C. W. ChenAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2232 Views

Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1335 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1460 Views
Abstract
The placenta is the crucial organ that regulates the health of both mother and fetus during pregnancy. The human placenta is composed of villous tree-like structures that embed into the maternal decidua. Within the stroma of the villi resides a population of fetally-derived macrophages, the Hofbauer cells (HBC). HBC are the only fetal immune cells found within the placenta in the steady-state and are thought to play a crucial role in placental function. From the 10th week of gestation, maternal blood flow into the intervillous space begins, resulting in the placental villi becoming bathed in maternal blood. To study HBC it is necessary to develop techniques that allow for their specific isolation and distinction from maternal blood monocytes and decidual macrophages. Here, we describe a protocol that explains step-by-step the strategy we have developed that allows the specific isolation of HBC.
Keywords: PlacentaBackground
The human placenta is a highly specialized organ that is crucial for both maternal and fetal health during pregnancy. It is made up of highly branched villous tree-like structures that are bathed in maternal blood from approximately the 10th week of gestation (Jauniaux et al., 2000). Anchoring villi attach the placenta to the endometrium, which is transformed during placentation into the decidua. A population of fetal tissue-resident macrophages, the Hofbauer cells (HBC), are abundant within the stroma of placental villi. Early studies determined that these cells are of fetal origin based on Y-chromosome detection in pregnancies with male fetuses (Kim et al., 2008). HBC are the only immune cell population on the fetal side of the placental barrier in the steady-state (Thomas et al., 2021). Accordingly, they are thought to have important roles in the regulation of placental growth and homeostasis and to act as immune sentinels, protecting the fetus from infection in utero. However, compared to other adult tissue-resident macrophages, HBC remain poorly understood. A range of protocols has been used to isolate HBC, as excellently reviewed elsewhere (Tang et al., 2011). However, a standard technique has not been established across the field. Many of the protocols used are suboptimal for the isolation of tissue macrophages. For example, unnecessarily long digestion times (sometimes up to 3 h) and the failure to use DNase I greatly affect HBC viability. Furthermore, HBC are typically isolated as total CD14+ cells from placental tissue for phenotypic and functional analysis. However, we have previously shown that maternal cells can comprise up to 30% of CD14+ cells isolated from first-trimester placental digests (Thomas et al., 2021). By adding common HLA allotype antibodies to our flow cytometry panel, we can distinguish maternal from fetal cells and ensure the isolation of a pure population of HBC. Therefore, in all future research of HBC using in vitro assays, additional steps that are described here should be performed to ensure that true fetally-derived HBC are studied with optimal viability.
In this article, we provide step-by-step protocols to isolate HBC from first-trimester and full-term human placental tissue with high purity. Procedures B and C describe how to obtain single-cell suspensions from first-trimester and full-term placental tissue, respectively. Procedure D describes how to obtain HBC from placental digests with high purity by fluorescence-activated cell sorting (FACS). Following these protocols allows investigating HBC properties with downstream in vitro assays to understand their functional properties further. A scheme of the overall procedure of HBC isolation from first-trimester and full-term placenta is shown in Figure 1.
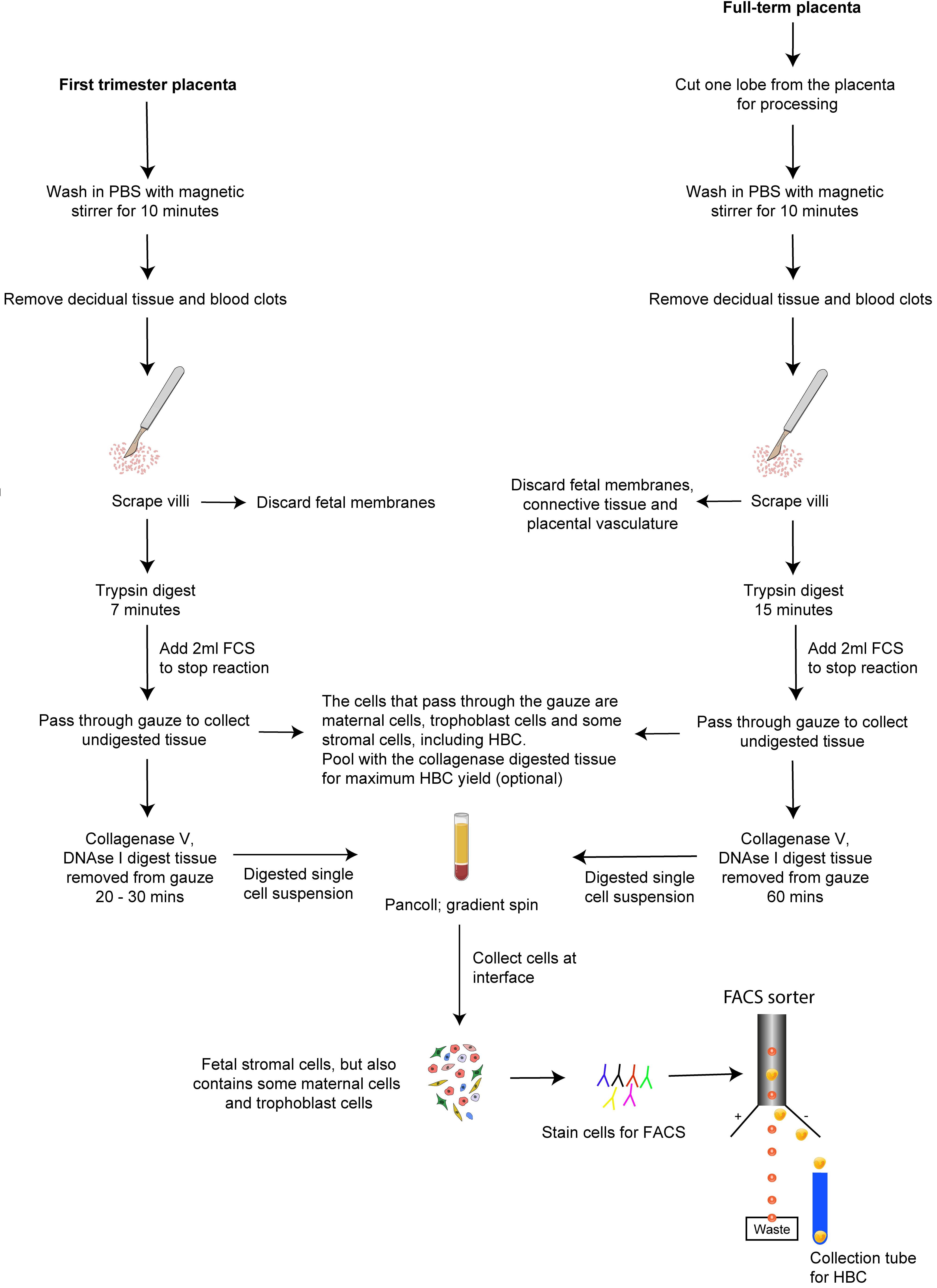
Figure 1. Isolation of HBC from human placenta. A schematic outlining the procedure for isolating HBC from first-trimester and full-term placenta.
Materials and Reagents
Disposable scalpels, No. 22 (Swann Morton, catalog number: 508)
Pasteur pipette (ELKay, catalog number: 127-P503-STR)
Muslin gauze (Winware food grade, catalog number: E948)
Cell strainers 30 µm (Miltenyi Biotec, catalog number: 130-041-407)
Cell strainers 70 µm (BD Falcon, catalog number: 352350)
Cell strainers 100 µm (BD Falcon, catalog number: 352360)
Cell strainers 400 µm (Pluriselect, catalog number: 43-50400-50)
Petri dishes 140 mm (Sterilin, catalog number: SC269)
Falcon 50 ml round bottom polypropylene test tubes (Corning, catalog number: 352070)
Polypropylene round-bottom FACS tubes 5 ml (Falcon, catalog number: 352063)
Dissection forceps (Scientific Laboratory Supplies, catalog number: SR04010)
Surgical straight scissors (SAMCO, catalog number: E101/01)
250-ml glass Duran bottles (Scientific Laboratory Supplies, catalog number: F151164 )
Funnel (Scientific Laboratory Supplies, catalog number: FUN1260)
Collagenase type V (Sigma-Aldrich, catalog number: C9263)
DAPI (4′,6-diamidino-2-phenylindole) (Sigma-Aldrich, catalog number: D8417)
DNase I (Roche, catalog number: 10104159001)
Ethylenediaminetetraacetic acid, EDTA (Sigma-Aldrich, catalog number: E9884)
Fetal calf serum (heat inactivated) (Biosera, catalog number: FB -1001)
Flow cytometry antibodies (see Table 1)
Table 1. Flow cytometric antibodies
Antibody Company Dilution Antibody CD3 FITC, clone: UCHT1 Biolegend, catalog number: 300406 1:100 Antibody CD14 PE-Dazzle 594, clone: HCD14 Biolegend, catalog number: 325634 1:100 Antibody CD19 FITC, clone: SJ25C1 Biolegend, catalog number: 363008 1:100 Antibody CD20 FITC, clone: 2H7 Biolegend, catalog number: 302304 1:100 Antibody CD45 PerCp-Cy5.5, clone: 2D1 Biolegend, catalog number: 368503 1:40 Antibody CD66b FITC, clone: G10F5 Biolegend, catalog number: 305103 1:100 Antibody CD335 FITC, clone: 9E2 Biolegend, catalog number: 331921 1:100 Antibody FOLR2 APC, clone: 94b/FOLR2 Biolegend, catalog number: 391705 1:1,000 Antibody HLA-A2 APC-Cy7, clone: BB7.2 Biolegend, catalog number: 343310 1:50 Antibody HLA-A3 BV650, clone: GAP.A3 BD Biosciences, catalog number: 747774 1:100 Antibody HLA-B7 PE, clone: BB7.1 Biolegend, catalog number: 372404 1:100 Pancoll (density 1.077 mg/ml) (Pan Biotech, catalog number: P04-601000)
Penicillin/streptomycin solution (Sigma-Aldrich, catalog number: P0781)
RPMI-1640 Medium (Sigma-Aldrich, catalog number: R7388)
Trypsin-250 (Pan Biotech, catalog number: P10-025100P)
Glucose (Sigma-Aldrich, catalog number: D9434)
NaCl (MP Biomedicals, catalog number: 151944)
KCl (MP Biomedicals, catalog number: 194738)
Disodium hydrogen orthophosphate (Fisher Scientific, catalog number: S/4520/53)
Potassium dihydrogen orthophosphate (Fisher Scientific, catalog number: P/4800/53)
Phosphate-buffered saline (PBS) 1× concentration
Human serum (Sigma-Aldrich, catalog number: H4522)
Mouse serum (Sigma-Aldrich, catalog number: M5905)
Rat serum (Sigma-Aldrich, catalog number: R9759)
Brilliant stain buffer (BD Horizon, catalog number: 563794)
Trypsin/EDTA, 0.2% solution (see Recipes)
The RF10 medium (see Recipes)
The FACS buffer (see Recipes)
Blocking buffer (see Recipes)
Equipment
BD FACSAriaTM III Cell Sorter (or similar), 4-laser cell sorter for containment level 2 samples fitted with a 100 µm nozzle (BD Biosciences, model: FACSAriaTM III)
Temperature controlled centrifuge with lids for buckets (Eppendorf Centrifuge 5810R, catalog number: F151380)
Hot plate magnetic stirrer (DLAB, catalog number: DL2255-380H)
Waterbath (VWR, catalog number: 462-0557)
Haemocytometer (VWR, catalog number: BR718605)
Light microscope (Leica, catalog number: 11964479)
Software
BD FACSDivaTM software
A collection of tools for flow cytometer and application setup, data acquisition, and analysis. It runs on the Microsoft® Windows 10 64-bit operating system.
FlowJoTM (Becton, Dickinson, and Company; 2019) software
A package for analyzing flow cytometry data. Files produced by modern flow cytometers are written in the Flow Cytometry Standard format with a .fcs file extension. FlowJo will import and analyze cytometry data regardless of which flow cytometer is used to collect the data. It is available for both Mac and PC.
Procedure
Biological samples
It is essential that all work using human tissue receives prior approval from the appropriate institutional and/or national review boards; donors must always provide informed consent. For this work, tissue samples were obtained with written informed consent from all participants under ethical approval from the East of England–Cambridge Central Research Ethics Committee (17/EE/0151).
All work with unscreened human placental tissue is regarded as a potential hazard for group 3 blood-borne pathogens and should be carried out in containment level 2 facilities.
Obtaining a single-cell suspension from first-trimester placental tissue
HBC are fetal macrophages found within the stroma of the villi, which make up the placenta. A series of enzymatic digestions ensures the breakdown of the tissue to obtain a single-cell suspension with high HBC yield and viability (see Note 2). This single-cell suspension will also contain other cell types that make up the placenta, including trophoblast cells and fibroblasts, as well as placenta-associated maternal monocytes/macrophages (PAMM) (Thomas et al., 2021). A purified population of HBC can be obtained from this digest by FACS, a method described in further detail in Procedure D.
Prepare reagents and equipment
The same materials and equipment are required for Procedures B. Media, reagents, and tools must be sterile (see Note 1).
Prepare the Trypsin/EDTA solution (see Recipes)
Prepare the RF10 medium (see Recipes)
Prepare the collagenase V stock solution (10 mg/ml) by dissolving it in RF10. Distribute the solution to 2.5 ml aliquots and store at -20 °C for up to 6 months.
Prepare the DNase I stock solution (10 mg/ml) by dissolving it in sterile water. Distribute the solution to 100 µl aliquots at -20 °C for up to 6 months.
Prepare PBS with DNase I (20 µg/ml) by diluting an aliquot of DNase I (from Procedure B1d) in 50 ml of 1× PBS. Keep sterile at 4 °C and make fresh each time (see Note 3).
Before beginning placental tissue processing, warm Trypsin/EDTA (0.2% solution), collagenase V, 1× PBS, and Pancoll to 37 °C. Set a heated shaker to 37 °C, with 500 rotations per min (RPM), and set a magnetic stirrer hot plate to 37 °C.
Processing first-trimester placental tissue to obtain a single-cell suspension
Process the placenta immediately upon receipt.
Rinse the placenta in 1× PBS gently with a magnetic stirrer for 10 min at room temperature (RT).
Transfer placenta to a Petri dish.
Remove any blood clots with forceps.
Hold down one end of the placental unit and gently scrape the villi from the chorionic membrane with a scalpel (see Figure 2A).
Discard the fetal membrane.
Using a Pasteur pipette (with the tip cut off), transfer scraped villi into 75 ml of pre-warmed 0.2% trypsin-EDTA in a sterile 250-ml glass Duran bottle. Place on the hot plate magnetic stirrer heated to 37 °C and stir gently for 7 min.
Add 2 ml of FCS to stop the trypsinisation of the tissue.
Filter the trypsined tissue through a sterile muslin gauze in a funnel.
Rinse with PBS at RT.
Keep both the digested tissue that ran through the gauze and the remaining tissue on the gauze.
Retrieve the undigested tissue from the gauze using a scalpel and place it in a 50 ml Falcon tube. Add 2.5 ml of collagenase V and 100 µl of DNase I and add PBS to a final volume of 25 ml (final concentration of 1 mg/ml Collagenase V and 40 µg/ml DNase I).
Note: The addition of DNase I is crucial; macrophage yield and viability are greatly reduced in the absence of DNase I.
Digest for 20-30 min at 37 °C on the heated shaker at 500 RPM.
Filter the resultant cell suspension through the sterile muslin gauze in a funnel. Discard any undigested tissue and centrifuge the filtrate for 5 min at 120 × g, RT, to pellet the cells. Discard the supernatant.
Pool the cells from steps i and n and wash in PBS with DNase I (see Note 3).
Note: The cells from Step B2i will mainly contain maternal and trophoblast cells. We have found that HBC are sometimes also found in this digest, and pooling cells from Steps B2i and B2n will help to maximize HBC yield. However, it not essential to pool the cells from Steps B2i and B2n, as few HBC are found in the cellular suspension from Step B2i.
Gently resuspend the cell pellet in 20 ml of PBS with DNase I.
Filter the cell suspension through a 400 µm strainer.
Layer the cell suspension onto 10 ml of Pancoll in a 50 ml Falcon tube.
Centrifuge for 20 min at 600 × g, RT, acceleration 0, break 1.
Collect the cells at the interface using a Pasteur pipette.
Wash with PBS with DNase I and centrifuge for 5 min at 120 × g, RT. Discard the supernatant.
Repeat the previous step.
Resuspend cells in PBS with DNase I and pass through a 70 µm strainer, count the cells with a haemocytometer, and place them on ice (see Notes 2 and 5).
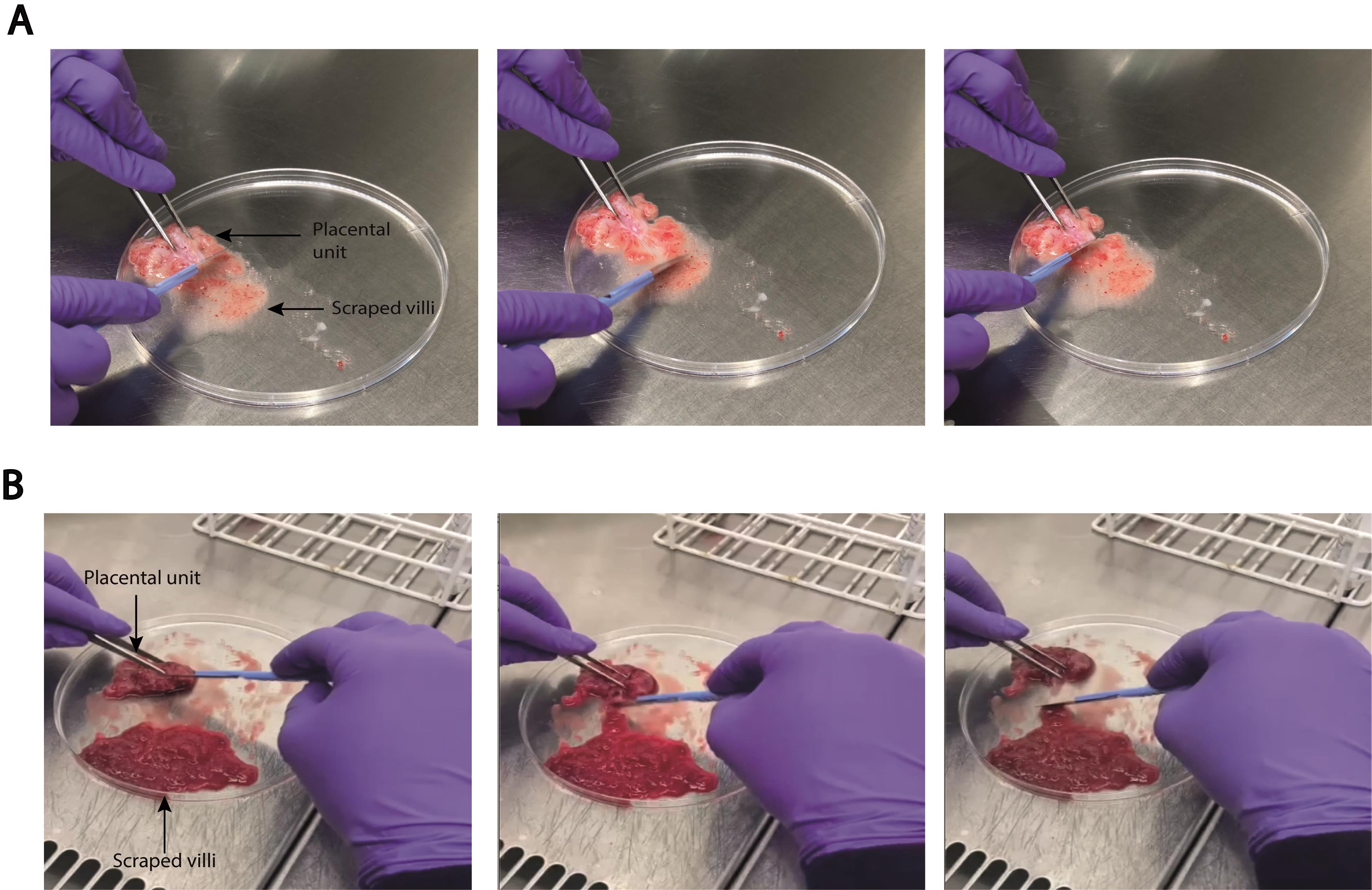
Figure 2. Scraping of placental villi. Images showing scraping of placental villi from (A) first-trimester and (B) full-term placenta.
Obtaining a single-cell suspension from full-term placental tissue
Human placenta collected from full-term pregnancies differs dramatically from those in the first trimester. As pregnancy progresses, placental villi continue sprouting, and the vascular network matures to increase the surface area and vascularisation of the organ and meet the needs of the growing fetus. Full-term placental villi have a more compact fibrous stroma with muscular arteries, which are absent in the first trimester (Castellucci and Kaufmann, 1982). For this reason, the digestion of full-term placental tissue for the isolation of HBC differs from that described in Procedure B for first-trimester placenta and is outlined in detail below.
Prepare reagents and equipment
Prepare reagents and equipment as described in Step B1.
Process full-term placental tissue to obtain a single-cell suspension
Process the placenta immediately upon receipt.
Cut out a lobe from the whole placenta using surgical scissors.
Note: For consistency between donors, always isolate the lobe from the same region.
Rinse the placenta in PBS gently with a magnetic stirrer for 10 min at RT.
Transfer to a Petri dish.
Remove any blood clots with forceps.
Remove and discard a thin layer (~1 mm) of tissue from the basal side (side which faces the mother) using surgical scissors.
Note: This step ensures the removal of any decidual tissue from the placenta.
Hold down one end of the placental unit and gently scrape the villi from the chorionic membrane with a scalpel (see Figure 2B).
Discard the fetal membrane and any thick connective tissue fibers and visible blood vessels.
Using a Pasteur pipette (with the tip cut off), transfer scraped villi into 75 ml of pre-warmed 0.2% trypsin-EDTA in a sterile glass Duran bottle (250 ml). Place on the hot plate magnetic stirrer heated to 37 °C and stir gently for 15 min (see Note 4).
Add 2 ml of FCS to stop the trypsinisation of the tissue.
Filter the trypsined tissue through a sterile muslin gauze in a funnel.
Rinse the gauze with PBS at RT.
Keep both the digested tissue that ran through the gauze and the remaining tissue on the gauze.
Retrieve the undigested tissue from the gauze and place in a 50 ml Falcon tube. Add 2.5 ml of collagenase V and 200 µl of DNase I and add PBS to a final volume of 25 ml (final concentration of 1 mg/ml collagenase V and 80 µg/ml DNase I).
Note: The addition of DNase is crucial; macrophage yield and viability are greatly reduced in the absence of DNase I.
Digest for 60 min at 37 °C on the heated shaker at 500 RPM (see Note 4).
Filter the digested tissue through a sterile muslin gauze in a funnel. Discard any undigested tissue and centrifuge the filtrate for 5 min at 120 × g, RT, to pellet the cells. Discard the supernatant.
Repeat Steps B2o-B2w.
Isolating Hofbauer cells from first-trimester and full-term placental digests
In this protocol, single-cell digests obtained from Procedures B and C are stained with fluorescent-conjugated antibodies for the isolation of HBC by FACS. The protocol includes an optimized panel and gating strategy that allows isolating a highly purified population of HBC without contamination of maternal monocytes/macrophages that are found in placental digests.
Prepare reagents and equipment (see Note 1)
Prepare the FACS buffer (see Recipes). Keep sterile and store at 4 °C for up to 1 month.
Prepare blocking buffer (see Recipes). Filter through a 0.2 µm sterile filter and store at 4 °C for up to 1 month.
Note: All serum is heat inactivated by placing in a water bath at 56 °C for 30 min.
Prepare 0.9 mM DAPI in sterile water. Store at -20 °C indefinitely.
Prepare RF10 as described in Step B1b.
Isolate HBC by FACS
Pass cells through a 30 µm filter into a polypropylene FACS tube.
Centrifuge for 5 min at 120 × g, 4 °C. Discard supernatant.
Resuspend cells in 150 µl of human blocking buffer per 10 × 106 cells.
Make antibody cocktail in 50 µl of Brilliant stain buffer per tube (follow dilutions in Table 1, for a final volume of 200 µl). The same antibody cocktail is used for first-trimester and full-term placental cells.
Add the antibody cocktail to the cells and mix by gently vortexing.
Incubate on ice for 20 min.
Add 2 ml of cold FACS buffer and centrifuge for 5 min at 120 × g, 4 °C. Discard supernatant.
Repeat previous step.
Resuspend the cells in 1 ml of cold FACS buffer.
Stain cells with DAPI (1:10,000) for 10 min prior to starting the sorting.
See representative images of the gating strategy used to identify HBC within first-trimester and full-term placental digests in Figure 3 (see Note 6).
Cells are sorted at 4 °C into appropriate collection tubes and buffers.
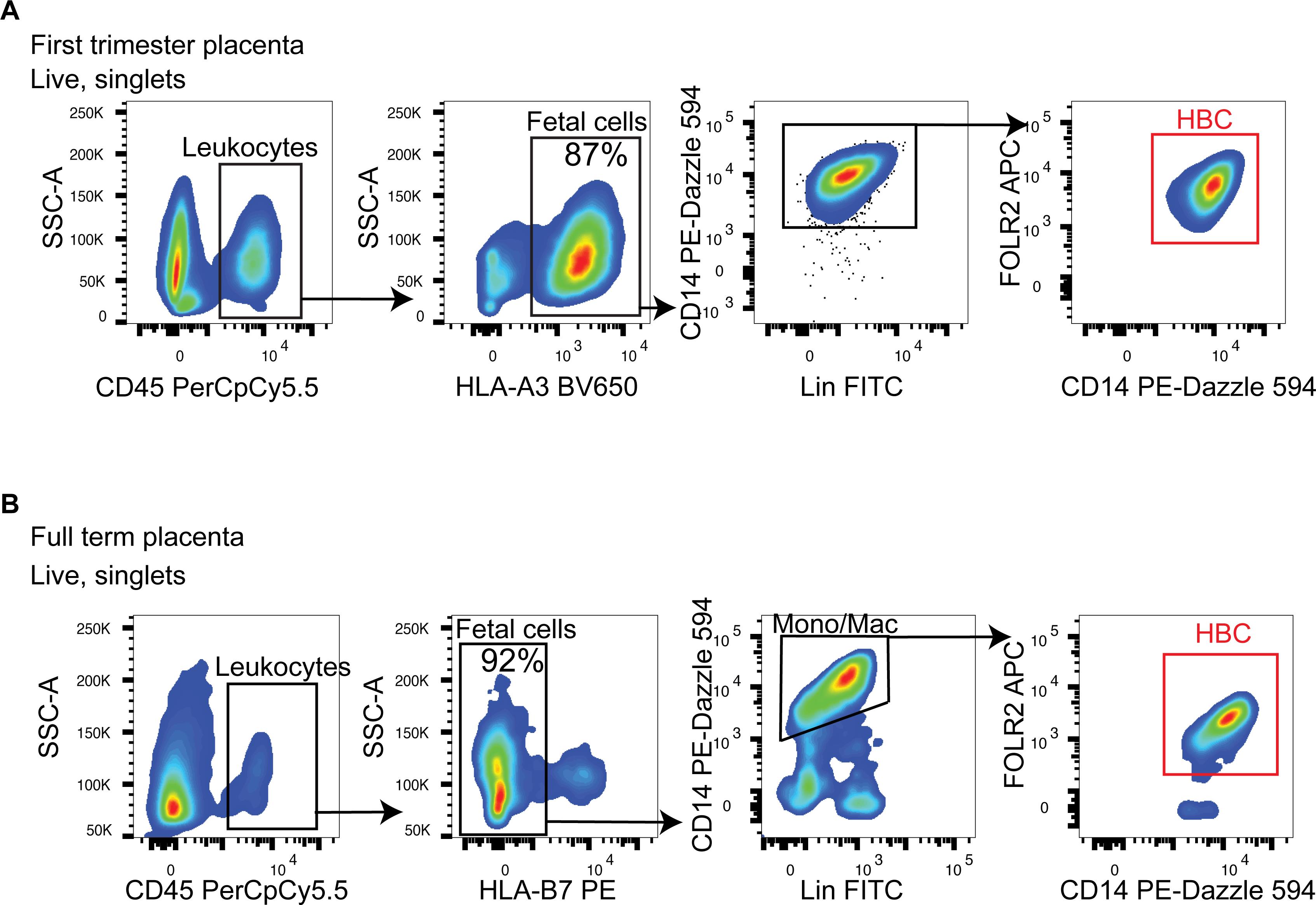
Figure 3. Fluorescent activated cell sorting gating strategy to isolate HBC. (A, B) Representative images of the gating strategy used to distinguish fetal from maternal cells using antibodies to common HLA allotypes and identity CD45+CD14+FOLR2+ HBC (red gate) within placental digests using BD FACSDiva and Flowjo software. (A) First-trimester placental samples; fetal cells are HLA-A3+, and maternal cells are HLA-A3-. (B) Full-term placental cells; fetal cells are HLA-B7-, and maternal cells are HLA-B7+. Lineage markers (Lin) include CD3, CD19, CD20, CD66b, and CD335 in FITC.
Data analysis
Isolation of viable cells from placental tissues
Our protocol for generating single-cell suspensions from placental digests yields cells with ~90% viability from first-trimester placental tissue and ~80% viability from full-term placental tissue. After enriching viable HBC by cell sorting, we obtain HBC with ~95% viability as determined by cell counting with a hematocytometer and microscope (see Note 5).
Flow cytometric gating strategy to distinguish HBC from maternal macrophages
To separate contaminating maternal CD14+ monocytes and macrophages from fetal CD14+ HBC, we developed a flow cytometric gating strategy that can be easily adapted to the configuration of different cell sorters (Table 1). We demonstrate that anti-HLA typing antibodies can be used to distinguish the more abundant fetal CD14+ HBC from the less abundant maternal CD14+ monocytes and macrophages from both first-trimester and full-term placental digests (Figure 3) (see Note 6).
Notes
There are several parameters critical to the success of the protocols described here.
It is essential to use sterile reagents and equipment throughout the protocols as HBC are responsive to bacterial products, which can alter the properties of the cells (Thomas et al., 2021). Once a single-cell suspension has been obtained, it is essential to keep it at 4 °C to avoid macrophage loss through adherence to plastic and minimize cell activation.
The addition of DNase I during the digestion and subsequent PBS washing steps is crucial to ensure high HBC viability.
The longer digest with both trypsin and collagenase V for full-term placental tissue is necessary to ensure complete digestion of the tissue and successful isolation of HBC. However, over-digestion and prolonged exposure to digestion enzymes reduce HBC viability.
The total cell counts from first-trimester and full-term placental tissue digests vary between samples. Generally, a total of 20 × 106-50 × 106 cells can be expected from placental tissue that has been digested successfully. The yield of HBC sorted by FACS also varies between samples but is ~1-3% of all cells.
To ensure the isolation of HBC by FACS with high purity, we include three HLA allotyping antibodies in our flow cytometric panel. Because HBC are fetal cells containing genetic material from the mother and father, one can expect an HLA allotype mismatch between the fetus and the mother in some cases. In these cases, we can ensure that sorted HBC are of fetal origin without the contamination of maternal cells, which are often present in placental digests (Figure 3). The specificity of the anti-HLA antibodies used in our staining panel has been validated by quantitative PCR (qPCR) on DNA from blood samples of HLA-typed donors. However, a shortcoming of the protocol described here is that if there is no HLA allotype mismatch between the fetus and mother for HLA-B7, HLA-A3, or HLA-A2, the protocol will fail to separate maternal from fetal cells. In this scenario, it is necessary to include additional markers in the flow cytometric panel that are expressed specifically by HBC. For example, we have previously shown that first trimester HBC do not express HLA-DR, whereas maternal macrophages do. Therefore, the addition of HLA-DR to the flow cytometric panel will allow the separation of first-trimester HBC from maternal macrophages in the absence of an HLA allotype mismatch (Thomas et al., 2021). HLA-DR cannot be used to distinguish maternal cells from full-term placental HBC because HBC express HLA-DR by full-term (Sutton et al., 1983).
Recipes
Trypsin/EDTA [(0.2% solution); 0.2% (w/v) Trypsin - 250/0.02% (w/v) EDTA/PBS solution)]
0.3 g Glucose
12 g NaCl
0.3 g KCl
1.725 g disodium hydrogen orthophosphate
0.3 g potassium dihydrogen orthophosphate
2 g trypsin
0.2 g EDTA
Dissolve chemicals in 1 L of water
Sterile-filter the solution using a 0.22 µm filter unit
Distribute the stock solution to 75 ml aliquots and store at -20 °C for up to 6 months
The RF10 medium
Roswell Park Memorial Institute (RPMI) 1640 Medium
10% heat-inactivated fetal calf serum
1× penicillin/streptomycin solution
100 units of penicillin, and 0.1 mg/ml streptomycin
The FACS buffer
2% heat-inactivated fetal calf serum and 2 mM EDTA in 1× PBS. Keep sterile and store at 4 °C for up to 1 month.
Blocking buffer
5% human serum, 1% rat serum, 1% mouse serum, 5% FCS, and 2 mM EDTA in 1× PBS
Acknowledgments
We thank the following for assistance: 1. The Flow Cytometry Core Facility at the Department of Pathology; 2. Lucy Gardner, Imogen Duncan, and Ritu Rani for their help collecting and processing placental samples; 3. Donors who participated in this study and the hospital staff; 4. Professor Stephen Charnock-Jones and Dr. Irving Aye Department of Obstetrics & Gynaecology, University of Cambridge, for help collecting full-term placental samples. This work was supported by the Wellcome Trust, Royal Society, Centre for Trophoblast Research, and Department of Pathology, University of Cambridge, UK. N.McG, is funded by a Wellcome Sir Henry Dale and Royal Society Fellowship (grant number 204464/Z/16/Z). J.T is funded by a Wellcome Trust PhD Studentship (grant number 215226/Z/19/Z).
This protocol was developed and adapted based on the previous work of others (Tang et al., 2011) and our own findings (Thomas et al., 2021).
Competing interests
The authors have no conflicts of interest to declare.
References
- Castellucci, M. and Kaufmann, P. (1982). A three-dimensional study of the normal human placental villous core: II. Stromal architecture. Placenta 3(3): 269-285.
- Jauniaux, E., Watson, A. L., Hempstock, J., Bao, Y. P., Skepper, J. N. and Burton, G. J. (2000). Onset of maternal arterial blood flow and placental oxidative stress. A possible factor in human early pregnancy failure. Am J Pathol 157(6): 2111-2122.
- Kim, J. S., Romero, R., Kim, M. R., Kim, Y. M., Friel, L., Espinoza, J. and Kim, C. J. (2008). Involvement of Hofbauer cells and maternal T cells in villitis of unknown aetiology.Histopathology 52(4): 457-464.
- Sutton, L., Mason, D. Y. and Redman, C. W. (1983). HLA-DR positive cells in the human placenta. Immunology 49(1): 103-112.
- Tang, Z., Abrahams, V. M., Mor, G. and Guller, S. (2011). Placental Hofbauer cells and complications of pregnancy. Ann N Y Acad Sci 1221: 103-108.
- Thomas, J. R., Appios, A., Zhao, X., Dutkiewicz, R., Donde, M., Lee, C. Y. C., Naidu, P., Lee, C., Cerveira, J., Liu, B., Ginhoux, F., Burton, G., Hamilton, R. S., Moffett, A., Sharkey, A. and McGovern, N. (2021). Phenotypic and functional characterization of first-trimester human placental macrophages, Hofbauer cells. J Exp Med 218(1).
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Appios, A., Thomas, J. R. and McGovern, N. (2021). Isolation of First-Trimester and Full-term Human Placental Hofbauer Cells. Bio-protocol 11(11): e4044. DOI: 10.21769/BioProtoc.4044.
Category
Cell Biology > Cell isolation and culture > Cell isolation
Developmental Biology > Cell growth and fate > Proliferation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








