- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantitation of Secretory Granule Size in Drosophila Larval Salivary Glands
Published: Vol 11, Iss 11, Jun 5, 2021 DOI: 10.21769/BioProtoc.4039 Views: 3877
Reviewed by: Gal HaimovichSteve JeanPradeep Kumar Bhaskar

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Live Imaging of Axonal Transport in the Motor Neurons of Drosophila Larvae
Tsuyoshi Inoshita [...] Yuzuru Imai
Dec 5, 2017 9852 Views

Characterization of Biological Motion Using Motion Sensing Superpixels
Felix Y. Zhou [...] Xin Lu
Sep 20, 2019 5934 Views

Live Imaging and Analysis of Meiotic Cytokinesis in Drosophila Testes
Govind Kunduri and Jairaj K. Acharya
Jan 20, 2024 3062 Views
Abstract
Maturation of secretory granules is a crucial process that ensures the bioactivity of cargo proteins undergoing regulated secretion. In Drosophila melanogaster, the larval salivary glands produce secretory granules that are up to four-fold larger in cross-sectional area after maturation. Therefore, we developed a live imaging microscopy approach to quantitate the size of secretory granules with a view to identifying genes involved in their maturation. Here, we describe the procedures of larval salivary gland dissection and sample preparation for live imaging with a fluorescence confocal microscope. Furthermore, we describe the workflow for measuring the size of secretory granules by cross-sectional surface area and statistical analysis. Our live imaging microscopy method provides a reliable read-out for the status of secretory granule maturation in Drosophila larval salivary glands.
Keywords: DrosophilaBackground
Regulated secretion is a process during which biologically active molecules such as hormones, digestive enzymes, and mucus are secreted from specialized secretory cells in a coordinated manner. Hence, regulated secretion is critical for maintaining physiological homeostasis in animals. Examples include the release of insulin after a meal, the release of mucin in response to pathogenic microorganisms, and the release of sweat during elevated body temperature. These biologically active molecules are produced by endocrine or exocrine cells and stored in long-lasting secretory organelles termed secretory granules.
Biogenesis of secretory granules begins at the trans-Golgi network, where cargoes of secretory granules aggregate and bud off as immature secretory granules (Tooze, 1991 and 1998; Borgonovo et al., 2006). Immature secretory granules then undergo a maturation process to become fully functional and competent for secretion. The maturation process includes homotypic fusion of immature secretory granules, removal of unwanted materials, and processing of cargoes (Tooze, 1991; Arvan and Castle, 1998). Failure of secretory granules to mature can lead to reduced bioactivity of their cargoes. For example, most hormones enter immature secretory granules as inactive prohormones. During maturation, the lumen of secretory granules is acidified, and prohormone convertases cleave prohormones into biologically active hormones (Moore et al., 2002). Therefore, failed granule maturation can have physiological consequences, leading to reduced activity or inefficient secretion of cargo proteins.
Although secretory granule maturation is a critical step in regulated secretion, assays for secretory granule maturation are not simple. Transmission electron microscopy is one of the standard methods to assay secretory granule maturation. Secretory granules are electron dense in electron micrographs (Nitsch and Rinne, 1981; Tatsuoka and Reese, 1989). Reduced secretory granule maturation is often correlated with a reduction in electron density or in secretory granule number (Edwards et al., 2009; Cao et al., 2013; Du et al., 2016; Emperador-Melero et al., 2018; Hummer et al., 2017; Rao et al., 2020). If antibodies are available, a decrease in hormone secretion or an increased ratio of prohormone to hormone can be measured in blood plasma or cell culture media by western blotting and densitometry or ELISA following stimulation (Cao et al., 2013; Du et al., 2016; Hummer et al., 2017). Immunofluorescence using a combination of anti-prohormone antibodies and secretory granule markers can also reveal defects in hormone processing, as indicated by an increase in intensity of the prohormone signal (Bogan et al., 2012; Cao et al., 2013).
The Drosophila larval salivary gland is a powerful genetic model for studying secretory granule biogenesis (Biyasheva et al., 2001; Burgess et al., 2011 and 2012; Torres et al., 2014; Csizmadia et al., 2018; Ma et al., 2020; Neuman et al., 2020). The larval salivary glands start producing glue protein-containing secretory granules 24 h after entry into the third instar larval stage. Immature secretory granules then mature over the next 18 h and are secreted all at once in response to a pulse of the hormone ecdysone (Biyasheva et al., 2001; Burgess et al., 2011). The secreted glue proteins adhere pupal cases onto a solid surface during metamorphosis. Size differences between immature and mature secretory granules can be 2- to 4-fold in cross-sectional surface area (5 μm2 vs. 10-25 μm2) (Ma et al., 2020). Thus, the Drosophila larval salivary gland is an excellent system to identify genes required for secretory granule maturation.
The Andres laboratory previously generated transgenic lines expressing one of the glue proteins (Sgs3) tagged with GFP or DsRed under the control of its endogenous promoter (Biyasheva et al., 2001; Costantino et al., 2008). Using these lines, secretory granules can be visualized by confocal microscopy. Our laboratory has used these transgenic lines in combination with a salivary gland-specific Gal4 driver and UAS-controlled RNAi transgenic lines to identify genes needed for secretory granule maturation. Here, we provide a detailed protocol for visualizing secretory granules via live imaging with a spinning-disc confocal microscope. Acquired data are analyzed with the imaging software Volocity 6.3 to quantitate secretory granule size. Secretory granule size distribution is then used as a readout for secretory granule maturation in the Drosophila larval salivary gland.
Materials and Reagents
Microscope slides 75 × 25 × 1 mm (VWR, catalog number: 16004-368)
Glass coverslips 18 × 18 mm No. 1.5 (VWR, catalog number: 48366-205)
Self-adhesive reinforcement labels (Avery, catalog number: 32203)
Syringe 5 ml (BD, catalog number: 309603)
Needle 18 G or lower (BD, catalog number: 305195)
Dissection needle (Fisher Scientific, catalog number: 13820024)
Petri dishes 100 × 15 mm (VWR, catalog number: 25384-088)
Petri dishes 35 × 10 mm (Corning, catalog number: 351008)
P{w+, Sgs3-GFP} (Bloomington Drosophila Stock Center, catalog number: 5884, 5885) or P{w+, Sgs3-DsRed} (Andres lab, University of Nevada, Las Vegas, USA) 3rd instar larva
High-vacuum M grease 100 g (Apiezon, Sigma, catalog number: Z273589)
Pipette tips (P200)
SYLGARDTM 184 Silicone Elastomer Kit 0.5 kg (Dow Chemical, Sigma, catalog number: 4019862)
KCl (Sigma, catalog number: P3911-500G)
NaCl (Bio Basic, catalog number: CA99501-558)
CaCl2·2H2O (Sigma, catalog number: C-5080-500G)
Tris-HCl (VWR, catalog number: 0234-1KG)
Drosophila Ringer’s solution (see Recipe 1)
Silicone dissection plate (see Recipe 2)
Equipment
Dumont #5 fine forceps (Fine Science Tools, catalog number: 11251-20)
Stereomicroscope (Leica Microsystems, model: Leica MZ6)
Spinning-disc confocal coupled with an Olympus IX81 microscope (Quorum Technologies, Puslinch, Ontario, Canada)
Software
Volocity 6.3 (Quorum Technologies, Puslinch, ON, Canada)
Adobe Creative Cloud Photoshop (Adobe, San Jose, CA, USA)
Microsoft Office 365 Excel (Microsoft, Redmond, WA, USA)
Procedure
Dissection and sample preparation
Pick 3rd instar wandering larvae with a dissection needle and transfer to a 35-mm Petri dish with Drosophila Ringer’s solution. Use fine forceps to hold the larvae and swirl in Drosophila Ringer’s solution to wash off any food. 3rd instar wandering larvae are those moving away from the food source; they can be easily located on the side of vials or bottles housing flies.
Note: It is best to pick larvae that are still moving and have food left in the gut. If molasses is included in the Drosophila food recipe, food in the gut can easily be identified by the dark brown color in the middle of the larvae. Otherwise, consumer food coloring can also be added to the Drosophila food to help identify larvae in the right stage. Larvae that have stopped moving or have no food left in the gut are ready to pupariate; therefore, their secretory granules would be secreted and would not be useful in determining secretory granule size.
Transfer individual clean larva with fine forceps to a droplet of Drosophila Ringer’s solution (50 μl) on a silicone dissection plate.
Dissect the larva under a stereomicroscope with fine forceps. Immobilize the larva by gently holding it down with forceps using the non-dominant hand. Pinch the mouth hook of the larva with the dominant hand and pull to remove the salivary glands.
Clean unwanted tissues away from the salivary glands using forceps (Figure 1A) and mount the samples as demonstrated in the illustration (Figure 1B and 1C).
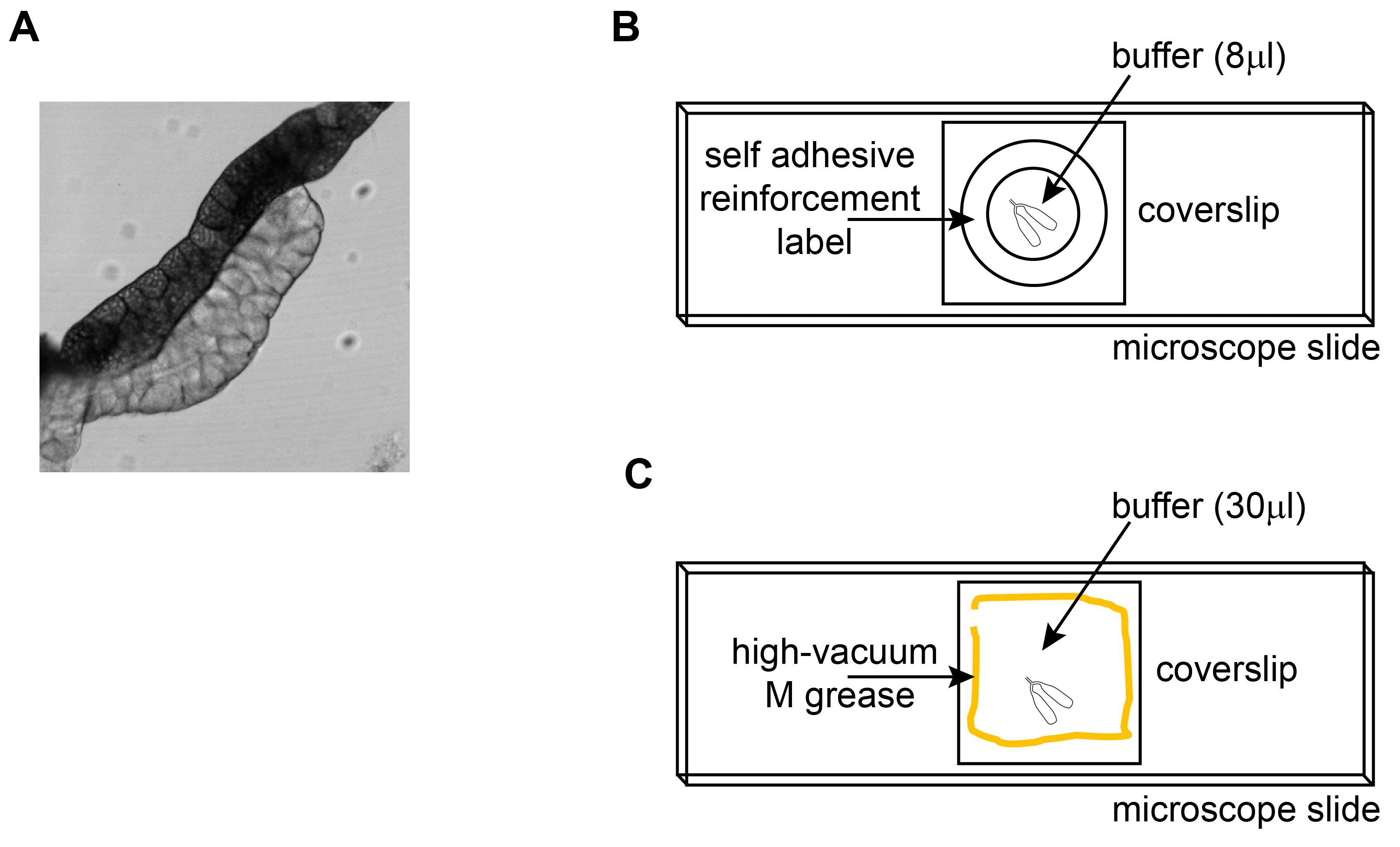
Figure 1. Mounting of Drosophila Larval Salivary Glands for Live Microscopy. A. DIC image of a larval salivary gland (translucent) with accompanying fat body (opaque) taken with 10× objective. B. Mounting of larval salivary glands on microscope slide with self-adhesive reinforcement label. C. Mounting of larval salivary glands on microscope slide with vacuum grease.Mounting the samples with self-adhesive reinforcement labels is fast and consistent (Figure 1B, step 6). However, to prevent anoxia, samples should be mounted with vacuum grease if a longer imaging time (>30 min) is desired (Figure 1C, step 7).
Place one self-adhesive reinforcement label on the microscope slide. Ensure that the label completely adheres to the slide. Pipette 8 µl Drosophila Ringer’s solution to the middle of the label. Transfer salivary glands to the solution (3-5 pairs of salivary glands per label). Place cover slip on and seal the slide with quick-drying nail polish. Samples are good for up to 30 min (following 10 min dissection time).
Dispense vacuum grease into a 5-ml syringe. Use a needle with a small gauge number. Dispense vacuum grease by drawing a square on the microscope slide using the syringe. Leave a small gap and do not complete the square. Pipette 30 µl Drosophila Ringer’s solution into the middle of the square. Transfer salivary glands to the solution (up to 8 pairs). Place cover slip on and gently push down with fine forceps until the vacuum grease seals the sample. Seal the slide with quick-drying nail polish. Samples are good for up to 50 min (following 10 min dissection time).
Imaging
Transfer slide to the stage of spinning-disc confocal microscope.
Open Volocity 6.3 in acquisition mode. Image samples with the 60× objective. Use corresponding setting to acquire images in the green or red channel for Sgs3-GFP or Sgs3-DsRed, respectively.
Capture 20 μm in Z with 0.3 µm per slice. Image at least three representative cells for each pair of glands (Figure 2).

Figure 2. Secretory Granules of Drosophila Larval Salivary Glands. A representative spinning-disc confocal image of one Z-slice of a Drosophila larval salivary gland cell expressing the secretory granule marker Sgs3-DsRed.Image at least three independent glands for each genotype.
Data analysis
Quantitation and Statistical Analysis
Launch Volocity 6.3 in analysis mode. Open the files saved from acquisition. Create a point spread function (PSF) file for deconvolution (Actions > Create New > Calculated PSF…). Enter values corresponding to the acquisition setup (Figure 3A).
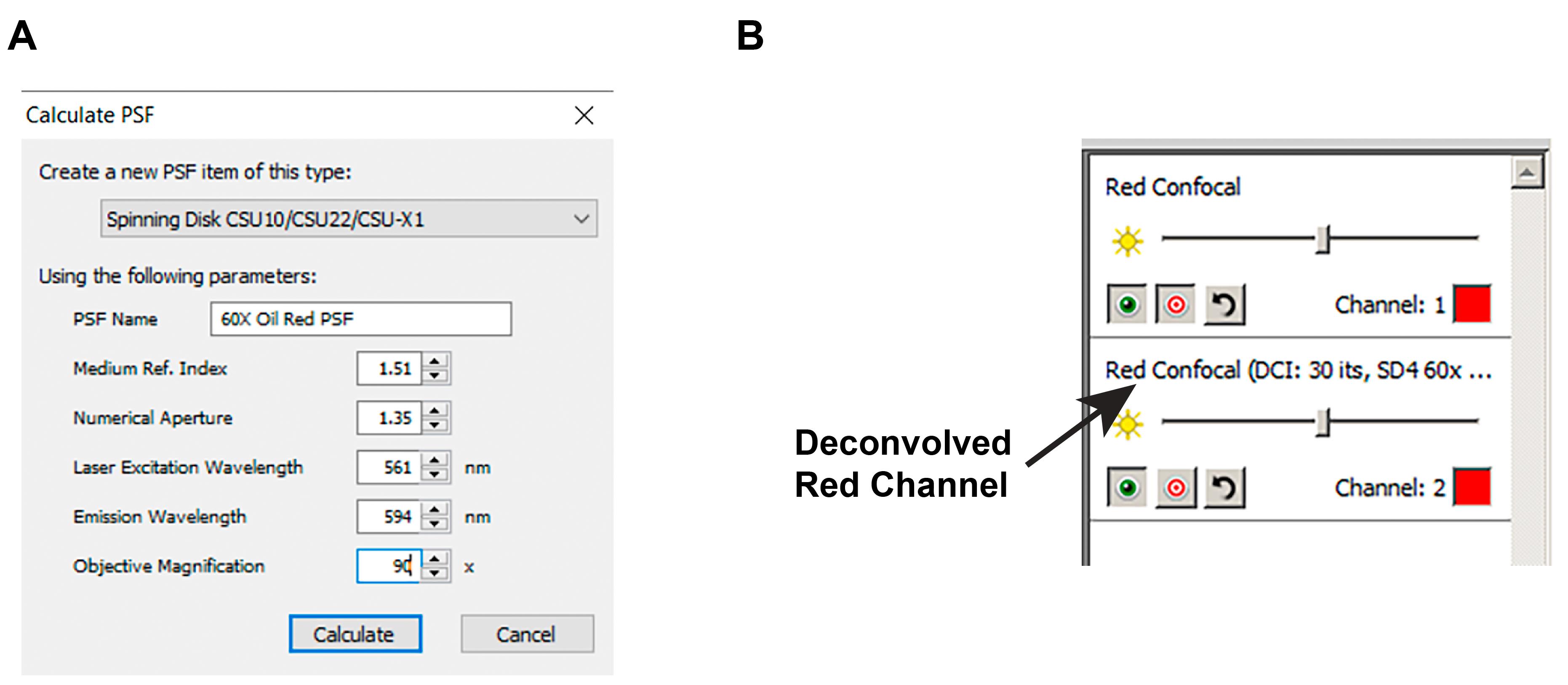
Figure 3. Deconvolution in Volocity 6.3. A. Settings for creating a new point spread function file to perform deconvolution on data from the red channel in Volocity 6.3. Values may need to be adjusted for a different setup. Here, the objective magnification is increased by 1.5-fold (60× to 90×) because the detector used in our system further magnifies the image by 1.5-fold. B. Deconvolved data will be listed as a new channel in Volocity 6.3.Run deconvolution on acquired data (Tools > Iterative Restoration…). Set the confidence limit to 99% and the iteration limit to 30. A new channel (deconvolved data) will be generated in the right panel (Figure 3B).
Split the Z-stacks into individual images (Tools > Split Volumes, select Organize the images into folders; Figure 4A). A new folder containing two subfolders will be generated in the left panel (Figure 4B). Perform analysis on deconvolved images in the second subfolder.
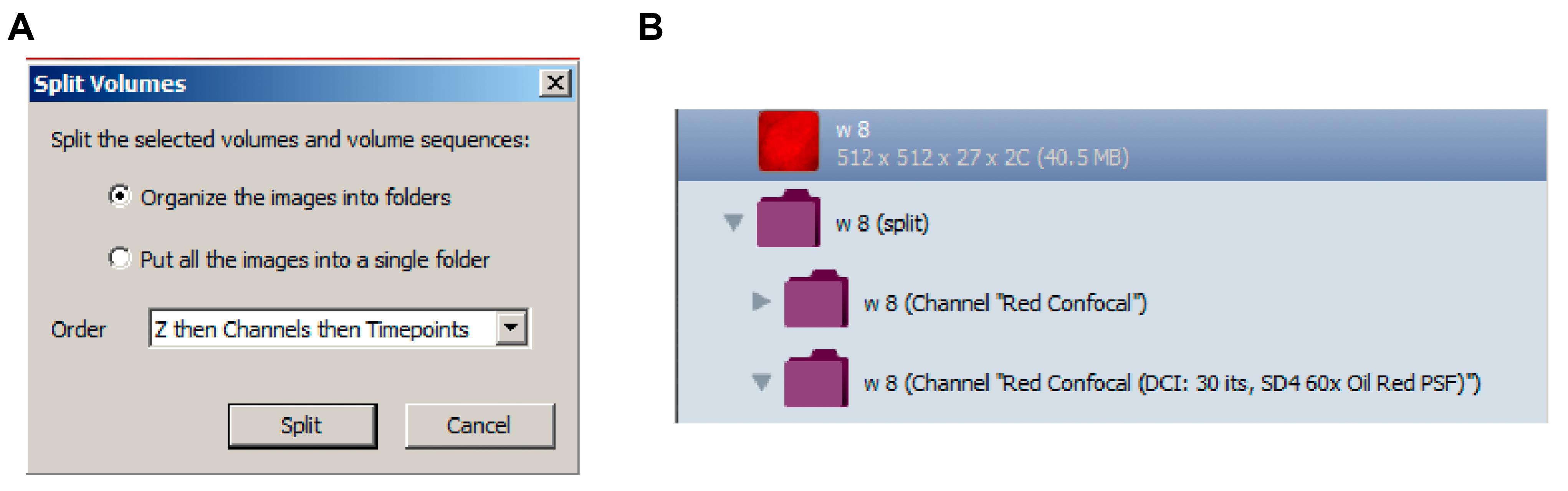
Figure 4. Split Volumes. A. Split volume function to organize Z-slices from raw data and deconvolved data into different folders. B. Folders of images created to the left panel of Volocity 6.3 after split volume function.Select the first image (z = 1) and go to the Measurements tab. Drag Find Objects and Separate Touching Objects tasks to the measurement protocol panel at the top of the screen (Figure 5). Make sure the two tasks are under the same protocol, generating only one population of measurements.
Click the gear icon beside the Find Objects function. For the Threshold, using drop down menu, select Automatic. In the Offset threshold by field, enter -10%. In the Minimum object size field, enter 4 μm2. Press OK. Under the Separate Touching Objects function, enter 12 μm2 in the Object size guide field (Figure 5). This setting works well for secretory granules in wild-type salivary glands. The values need to be adapted for genotypes with very large or very small secretory granules.
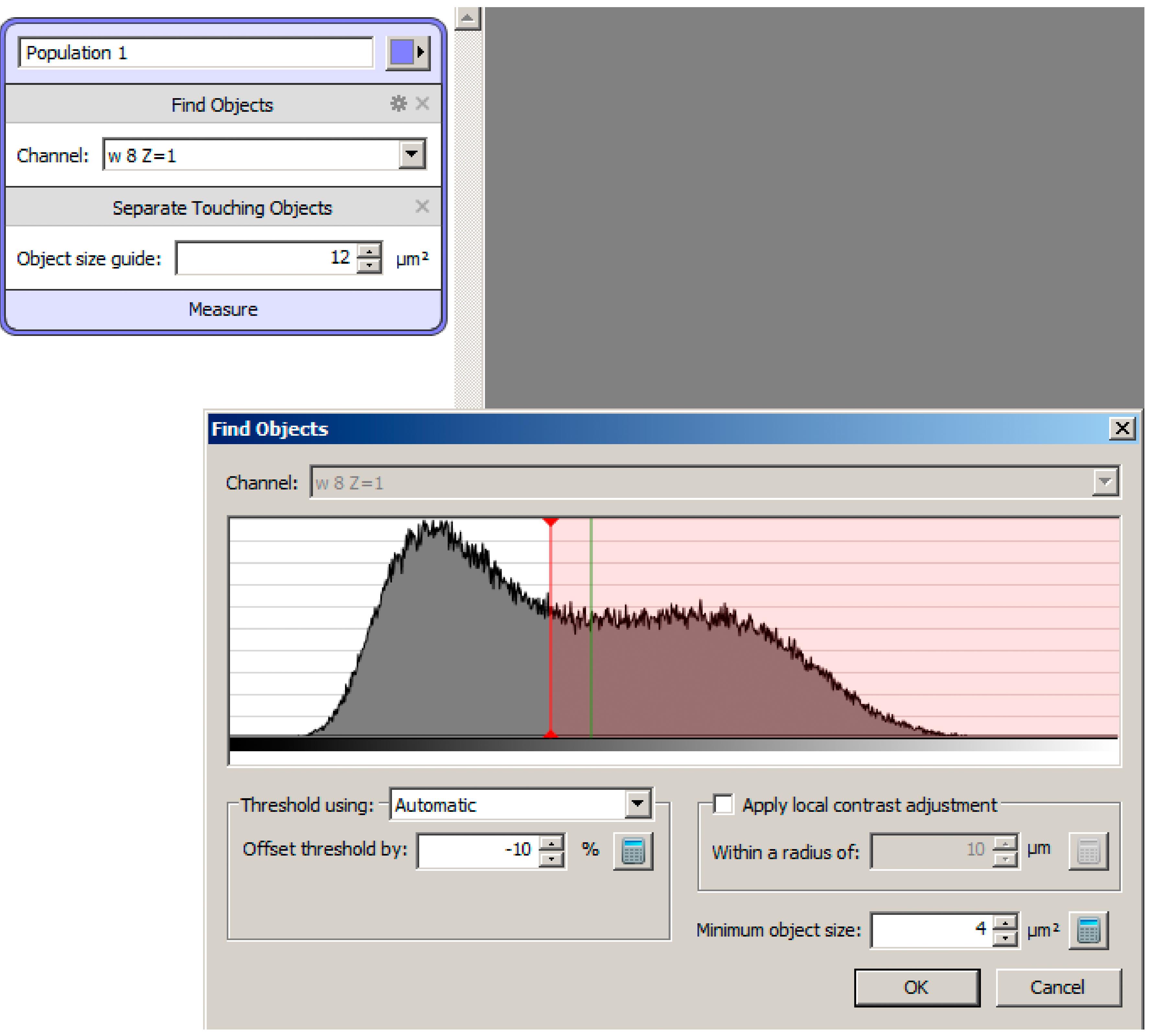
Figure 5. Functions to Measure Secretory Granules. A screen shot displaying a protocol containing Find Objects and Separate Touching Objects in Volocity 6.3 (top left). Settings for the Find Objects function by thresholding (bottom right).Click Measure under the function generated from steps 4 and 5. Check Intensity and Volume measurements and press OK. Identified objects will be outlined in the color chosen in the protocol created from steps 4 and 5 (Figure 6A). A list of objects should be populated in the bottom right panel (Figure 6B). The function can be saved (Measurements > Save Protocol…) and later restored (Measurements > Restore Protocol…).
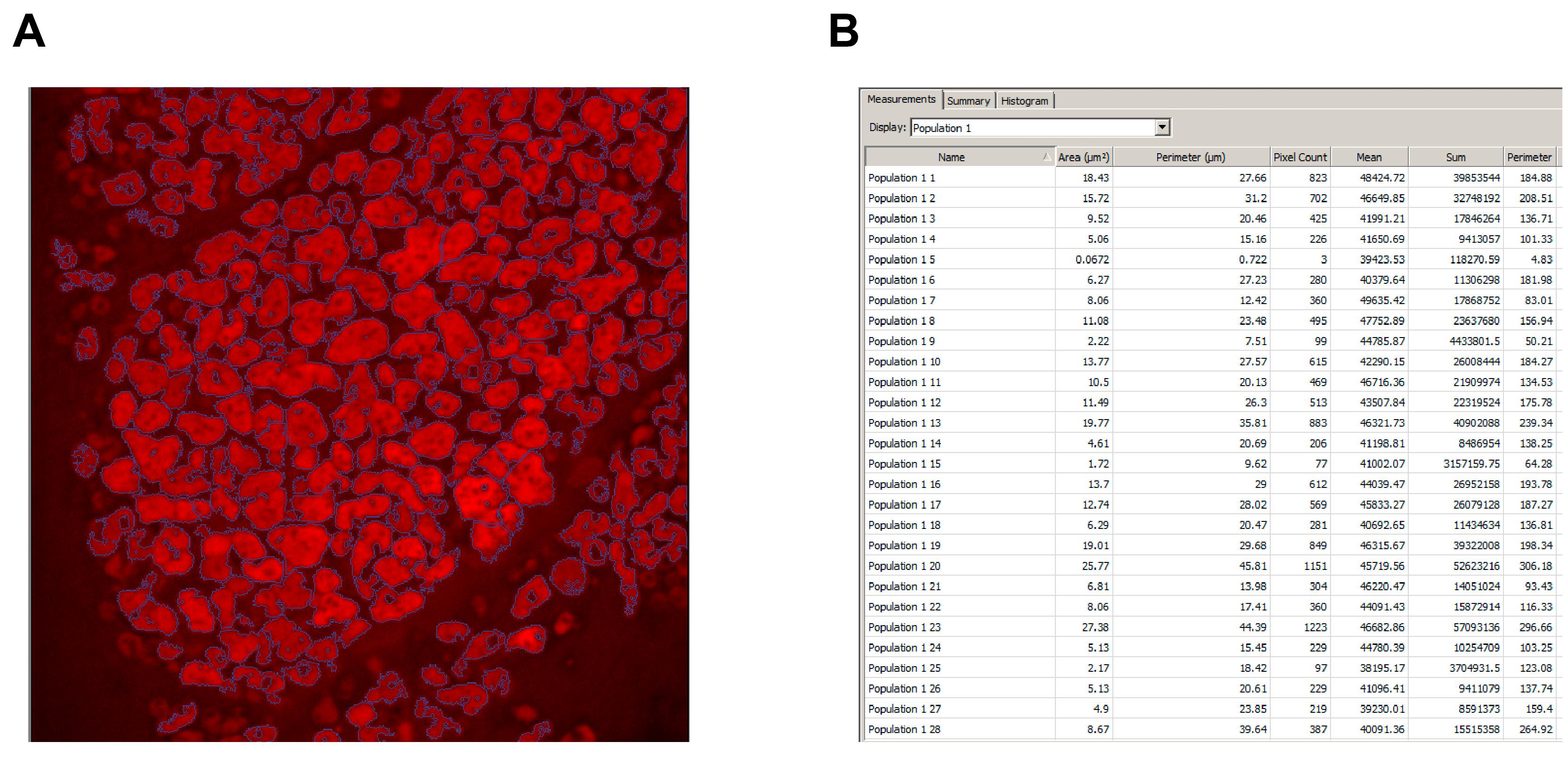
Figure 6. Measuring Secretory Granule Size. A. Secretory granules (red) outlined in purple using the protocol described in steps 4-5. B. Output of secretory granule cross-sectional area.Go to the list of objects. Double click on the Area (μm2) tab until the objects are sorted by size in descending order. Select all the data excluding the those that are less than 4 μm2. Copy the data and paste into Excel 365 for statistical analysis.
Repeat Steps 4-7 for z = 10 and z = 20 to account for variability in secretory granule cross-sectional area throughout the cell. If the measurement protocol was saved, steps 4-5 can be avoided by the Restore Protocol… function described in step 6.
Repeat Steps 4-8 for three representative cells from the same salivary gland.
Repeat Steps 4-9 for three independent salivary glands for each genotype.
Pool all the data for each genotype in Excel 365.
Box plots (Insert > Insert Statistic Chart > Box and whisker plot) can be used to visualize the distribution of secretory granule size by cross-sectional surface area (Figure 7).
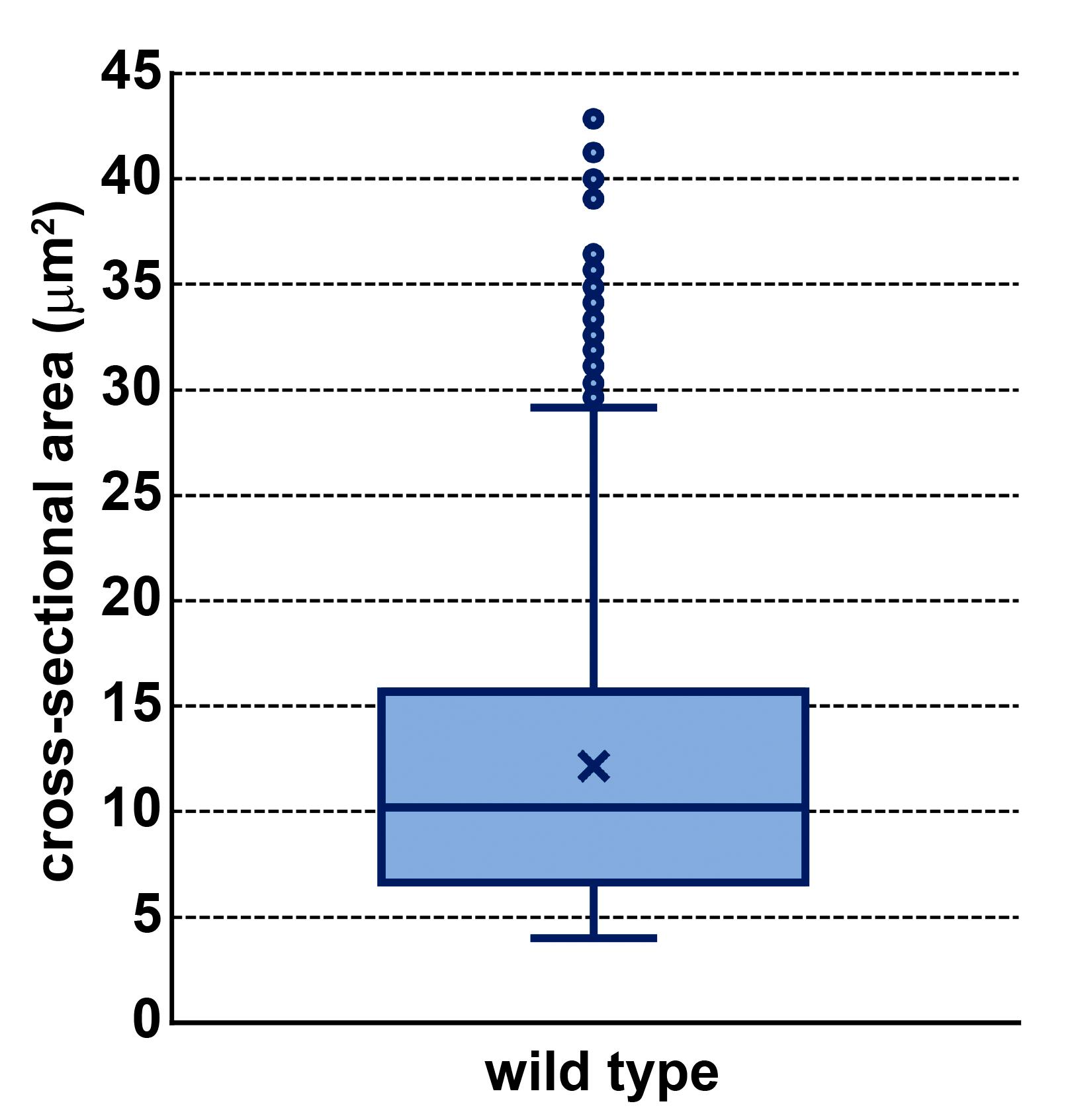
Figure 7. Box Plot Showing Secretory Granule Size Distribution. Distribution of secretory granule sizes by cross-sectional area in wild type salivary glands. Each line of the box from bottom to top represents the 25th, 50th (median), and 75th percentile of the population. X denotes the mean of the sample. Bottom and top whiskers represent minimum and maximum values determined by 1.5× the interquartile range. Dots outside of the whiskers are samples considered as statistical outliers. n = 1597 secretory granule cross-sections from 3 salivary gland cells (3 Z-slices from each cell for a total of 9 Z-slices).t-Test (Data > Data Analysis > t-Test: Two-Sample Assuming Unequal Variances) can be used to evaluate the statistical difference in secretory granule cross-sectional area between two genotypes.
Recipes
Drosophila Ringer’s solution (pH 7.2)
KCl 182 mMNaCl 46 mM
CaCl2·2H2O 3 mM
Tris-HCl 10 mM
Silicone dissection plate
Mix the base and curing agent from the SYLGARD 184 kit (10:1, weight:weight) directly in a 100-mm Petri dish with a spatula. All procedures should be conducted in a fume hood.
Allow to cure inside the fume hood for 48 h.
Acknowledgments
This protocol was adapted from our previous work (Ma et al., 2020). We acknowledge the following funding sources: SickKids Restracomp Studentship, Canadian Institutes of Health Research Strategic Training Fellowship #TGF-53877, University of Toronto Open Fellowship, and Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarships–Doctoral Program Studentship (to Cheng-I J. Ma); Canadian Institutes of Health Research Grants #PJT-162165 and #MOP-119483 and Canadian Institutes of Health Research Institute of Genetics #IG1-115714 (to Julie A. Brill).
Competing interests
There are no competing interests to declare.
References
- Arvan, P. and Castle, D. (1998). Sorting and storage during secretory granule biogenesis: looking backward and looking forward. Biochem J 332 (Pt 3): 593-610.
- Biyasheva, A., Do, T. V., Lu, Y., Vaskova, M. and Andres, A. J. (2001). Glue secretion in the Drosophila salivary gland: a model for steroid-regulated exocytosis. Dev Biol 231(1): 234-251.
- Bogan, J. S., Xu, Y. and Hao, M. (2012). Cholesterol accumulation increases insulin granule size and impairs membrane trafficking. Traffic 13(11): 1466-1480.
- Borgonovo, B., Ouwendijk, J. and Solimena, M. (2006). Biogenesis of secretory granules. Curr Opin Cell Biol 18(4): 365-370.
- Burgess, J., Jauregui, M., Tan, J., Rollins, J., Lallet, S., Leventis, P. A., Boulianne, G. L., Chang, H. C., Le Borgne, R., Kramer, H. and Brill, J. A. (2011). AP-1 and clathrin are essential for secretory granule biogenesis in Drosophila. Mol Biol Cell 22(12): 2094-2105.
- Burgess, J., Del Bel, L. M., Ma, C. I., Barylko, B., Polevoy, G., Rollins, J., Albanesi, J. P., Kramer, H. and Brill, J. A. (2012). Type II phosphatidylinositol 4-kinase regulates trafficking of secretory granule proteins in Drosophila. Development 139(16): 3040-3050.
- Cao, M., Mao, Z., Kam, C., Xiao, N., Cao, X., Shen, C., Cheng, K. K., Xu, A., Lee, K. M., Jiang, L. and Xia, J. (2013). PICK1 and ICA69 control insulin granule trafficking and their deficiencies lead to impaired glucose tolerance. PLoS Biol 11(4): e1001541.
- Costantino, B. F., Bricker, D. K., Alexandre, K., Shen, K., Merriam, J. R., Antoniewski, C., Callender, J. L., Henrich, V. C., Presente, A. and Andres, A. J. (2008). A novel ecdysone receptor mediates steroid-regulated developmental events during the mid-third instar of Drosophila. PLoS Genet 4(6): e1000102.
- Csizmadia, T., Lorincz, P., Hegedus, K., Szeplaki, S., Low, P. and Juhasz, G. (2018). Molecular mechanisms of developmentally programmed crinophagy in Drosophila. J Cell Biol 217(1): 361-374.
- Du, W., Zhou, M., Zhao, W., Cheng, D., Wang, L., Lu, J., Song, E., Feng, W., Xue, Y., Xu, P. and Xu, T. (2016). HID-1 is required for homotypic fusion of immature secretory granules during maturation. Elife 5: e18134.
- Edwards, S. L., Charlie, N. K., Richmond, J. E., Hegermann, J., Eimer, S. and Miller, K. G. (2009). Impaired dense core vesicle maturation in Caenorhabditis elegans mutants lacking Rab2. J Cell Biol 186(6): 881-895.
- Emperador-Melero, J., Huson, V., van Weering, J., Bollmann, C., Fischer von Mollard, G., Toonen, R. F. and Verhage, M. (2018). Vti1a/b regulate synaptic vesicle and dense core vesicle secretion via protein sorting at the Golgi. Nat Commun 9(1): 3421.
- Hummer, B. H., de Leeuw, N. F., Burns, C., Chen, L., Joens, M. S., Hosford, B., Fitzpatrick, J. A. J. and Asensio, C. S. (2017). HID-1 controls formation of large dense core vesicles by influencing cargo sorting and trans-Golgi network acidification. Mol Biol Cell 28(26): 3870-3880.
- Ma, C. J., Yang, Y., Kim, T., Chen, C. H., Polevoy, G., Vissa, M., Burgess, J. and Brill, J. A. (2020). An early endosome-derived retrograde trafficking pathway promotes secretory granule maturation. J Cell Biol 219 (3): e201808017.
- Moore, H. H., Andresen, J. M., Eaton, B. A., Grabe, M., Haugwitz, M., Wu, M. M. and Machen, T. E. (2002). Biosynthesis and secretion of pituitary hormones: dynamics and regulation. Arch Physiol Biochem 110(1-2): 16-25.
- Neuman, S. D., Terry, E. L., Selegue, J. E., Cavanagh, A. T. and Bashirullah, A. (2020). Mistargeting of secretory cargo in retromer-deficient cells. Dis Model Mech 14(1): dmm046417.
- Nitsch, C. and Rinne, U. (1981). Large dense-core vesicle exocytosis and membrane recycling in the mossy fibre synapses of the rabbit hippocampus during epileptiform seizures. J Neurocytol 10(2): 201-209.
- Rao, A., McBride, E. L., Zhang, G., Xu, H., Cai, T., Notkins, A. L., Aronova, M. A. and Leapman, R. D. (2020). Determination of secretory granule maturation times in pancreatic islet β-cells by serial block-face electron microscopy. J Struct Biol 212(1): 107584.
- Tatsuoka, H. and Reese, T. S. (1989). New structural features of synapses in the anteroventral cochlear nucleus prepared by direct freezing and freeze-substitution. J Comp Neurol 290(3): 343-357.
- Tooze, S. A. (1991). Biogenesis of secretory granules. Implications arising from the immature secretory granule in the regulated pathway of secretion. FEBS Lett 285(2): 220-224.
- Tooze, S. A. (1998). Biogenesis of secretory granules in the trans-Golgi network of neuroendocrine and endocrine cells. Biochim Biophys Acta 1404(1-2): 231-244.
- Torres, I. L., Rosa-Ferreira, C. and Munro, S. (2014). The Arf family G protein Arl1 is required for secretory granule biogenesis in Drosophila. J Cell Sci 127(Pt 10): 2151-2160.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ma, C. J. and Brill, J. A. (2021). Quantitation of Secretory Granule Size in Drosophila Larval Salivary Glands. Bio-protocol 11(11): e4039. DOI: 10.21769/BioProtoc.4039.
- Ma, C. J., Yang, Y., Kim, T., Chen, C. H., Polevoy, G., Vissa, M., Burgess, J. and Brill, J. A. (2020). An early endosome-derived retrograde trafficking pathway promotes secretory granule maturation.J Cell Biol 219 (3): e201808017.
Category
Developmental Biology > Morphogenesis > Organogenesis
Cell Biology > Cell imaging > Live-cell imaging
Cell Biology > Cell isolation and culture > Organ culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








