- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Simple and Robust Protocol for in vitro Differentiation of Mouse Non-pathogenic T Helper 17 Cells from CD4+ T Cells
Published: Vol 11, Iss 10, May 20, 2021 DOI: 10.21769/BioProtoc.4029 Views: 5662
Reviewed by: Guangyong PengRAMESH KUDIRAMichael EnosAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Culture of Ferret Airway Stem Cells
Ziying Yan [...] Feng Yuan
Jul 20, 2025 2411 Views

Dual Phospho-CyTOF Workflows for Comparative JAK/STAT Signaling Analysis in Human Cryopreserved PBMCs and Whole Blood
Ilyssa E. Ramos [...] James M. Cherry
Nov 20, 2025 2338 Views

Optimization of Adipogenic Differentiation Protocol for Murine and Human Cell Culture Models
Junwan Fan [...] Wenyan He
Jan 20, 2026 217 Views
Abstract
Functional and mechanistic studies of CD4+ T cell lineages rely on robust methods of in vitro T cell polarization. Here, we report an optimized protocol for in vitro differentiation of a mouse non-pathogenic T helper 17 (TH17) cell lineage. Most of the previously established protocols require irradiated splenocytes as artificial antigen presenting cells (APC) for TCR activation. The protocol described here employs plate-bound antibodies and a TH17-polarizing cytokine cocktail to activate and differentiate naïve CD4+ T (Tnai) cells, reflecting a simple and robust protocol for in vitro TH17n differentiation. Using T cells that are genetically engineered with an IL-17 reporter, this protocol may enable the rapid production of a pure population of IL17-expressing CD4+ T cells for system biology studies and high-throughput functional screening.
Keywords: TH17Background
Naive CD4+ T cells circulate through secondary lymphoid tissues after emerging from the thymus. The activation of naïve CD4+ T cells, by antigen-presenting cells that offer cognate antigens, initiates various differentiation programs that lead to the development of highly specialized T helper (TH) cell lineages (Bhaumik and Basu 2017). TH17 cells are a subset of pro-inflammatory TH cells defined by the production of interleukin 17 (IL-17) (Hartigan-O'Connor et al., 2011). To date, at least two different types of TH17 have been described. Type 1, pathogenic or disease-inducing TH17 (TH17p) cells, are induced by the combined IL-6, IL-1β, and IL-23 cytokine stimulation, and produce IL-17 together with IFN-γ, which results in tissue inflammation and autoimmunity (Cua et al., 2003; Awasthi et al., 2009; Jager et al., 2009; Lee et al., 2012; Lee et al., 2014). Type 2, non-pathogenic TH17 (TH17n) cells, are induced by TGF-β and IL-6 in vitro, and produce IL-17 but are not able to induce effective tissue inflammation or autoimmunity. TH17n cells play a protective role in mucosal tissues, promoting tissue homeostasis as well as maintaining barrier function (Bettelli et al., 2008; Ouyang et al., 2008; Korn et al., 2009; Guglani and Khader 2010; Gaffen et al., 2011). Here, we report a simple and robust protocol for generating non-pathogenic TH17 (TH17n) cells in vitro.
Materials and Reagents
48-well cell culture plates (ASi Alkali Scientific Inc., catalog number: TP9048)
CellProTM 70 μm cell strainers (ASi, Falcon®, catalog number: MT4070)
1 ml syringe (BD Tuberculin Slip Tip, catalog number: 9189520)
50 ml centrifuge tubes (ASi, catalog number: C5602)
15 ml centrifuge tubes (ASi, catalog number: C5600)
C57BL/6J mice (The Jackson Laboratory, catalog number: 000664)
C57BL/6-IL17atm1Bcgen/J mice (The Jackson Laboratory, catalog number: 018472)
RBC lysis buffer 10× (Biolegend®, catalog number: 420302)
Cell culture grade water (HyPureTM, catalog number: SH30529.02)
MojoSortTM Mouse CD4 Naïve T Cell Isolation Kit (Biolegend®, catalog number: 480040)
Coating antibodies:
Anti-mouse CD3ϵ (clone 145-2C11, Bio X Cell, catalog number: BE0001-1)
Anti-mouse CD28 (clone 37.51, Bio X Cell, catalog number: BE0015-1)
Staining antibodies:
APC anti-mouse CD4 (clone RM4-5, Biolegend, catalog number: 100516)
PE/Cy7 anti-mouse IL-17A (clone TC11-18H10.1, Biolegend, catalog number: 506922)
Cell Stimulation Cocktail Plus Protein Transport Inhibitors (Invitrogen eBioscienceTM, catalog number: 00-4975-93)
FoxP3/Transcription Factor Staining Buffer Set (Fixation Diluent, Fixation Reagent, Permeabilization kit, eBioscienceTM, catalog number: 00-5523-00)
RPMI 1640 1× with L-glutamine (Corning, catalog number: 10-040-CV)
Fetal bovine serum (FBS) (ATLANTA biologicalsTM, R&D SYSTEMS, catalog number: S12450)
L-glutamine 200 mM (Corning, catalog number: 25-005-CI)
Penicillin-streptomycin solution (PS) 100× (Corning, catalog number: 30-002-CI)
Sodium pyruvate 100 mM (Corning, catalog number: 25-000-CI)
2-mercaptoethanol (ME) 1000× (Life Technologies, Gibco®, catalog number: 21985-023)
TH17 polarization cytokines and antibodies:
Recombinant Human TGF-β1 (PeproTech, catalog number: 100-21C)
Recombinant Murine IL-6 (PeproTech, catalog number: 216-16)
Anti-mouse IFN-γ (clone XMG1.2, Bio X Cell, catalog number: BE0055)
Anti-mouse IL-2 (clone JES6-1A12, Bio X Cell, catalog number: BE0043)
Anti-mouse IL-4 (clone 11B11, Bio X Cell, catalog number: BE0045)
Phosphate-buffered saline (PBS) 1× (Corning, catalog number: 21-040-CV)
Bovine serum albumin (BSA) (Fisher BioReagentsTM, catalog number: BP1600-100)
Ethylenediamine tetraacetate acid (EDTA) 0.5 M (Fisher BioReagentsTM, catalog number: BP2482-500)
Carboxyfluorescein succinimidyl ester (CFSE) (InvitrogenTM, catalog number: C1157)
Culture medium (see Recipes)
TCR-stimulation antibody mixture (see Recipes)
RBC lysis buffer (1×) (see Recipes)
MACS buffer (see Recipes)
TH17 polarization medium (see Recipes)
Staining buffer (see Recipes)
Equipment
NovoCyte (ACEA Biosciences)
Software
FlowJo software (BD Biosciences)
Procedure
Naïve CD4+ T cell isolation and seeding.
Day -2, pre-coat 200 μl TCR-stimulating antibody mixture/well in a 48 well-plate at 4°C.
Day 0, euthanize a C57BL/6J mouse (or IL17A-IRES-GFP-KI mouse), harvest the spleen and lymph nodes (LNs), and isolate naïve CD4+ T cells using the MojoSort™ Isolation Kit according to the manufacturer’s instructions.
Note: Basic information regarding the anatomy of a laboratory mouse can be found at http://www.informatics.jax.org/cookbook/.
Count the cells and seed 2.5 × 105 cells/well in the pre-coated 48-well plate with 500 μl TH17 polarization medium at 37°C/5% CO2.
Note: To follow T cell proliferation, it is optional to stain naive CD4+ T cells with CFSE according to the manufacturer’s instructions.
FACS analysis of TH17 cell differentiation (Figure 1)
Day 3 (or anytime between day 2 and day 5; we routinely observed over 40% IL17+ cells starting at day 2 and remaining for 3 additional days until the media's nutrients are depleted), treat the cells for 4 h with Cell Stimulation Cocktail Plus Protein Transport Inhibitors according to the manufacturer’s for 4 h, and then stain the cells with the anti-mouse CD4 antibody (1:100) for 30 min at 4°C. Next, fix and permeabilize the cells using the FoxP3 Fixation/Permeabilization Kit according to the manufacturer's instructions. Finally, stain the cells with the anti-IL-17A antibody (1:100) at RT for 15 min before FACS analysis (if cultured cells are from an IL17A-IRES-GFP-KI mouse, FACS analysis can be performed directly without the procedures described above in B).
Note: Basic principles and applications of flow cytometry can be found in the literature (McKinnon, 2018).
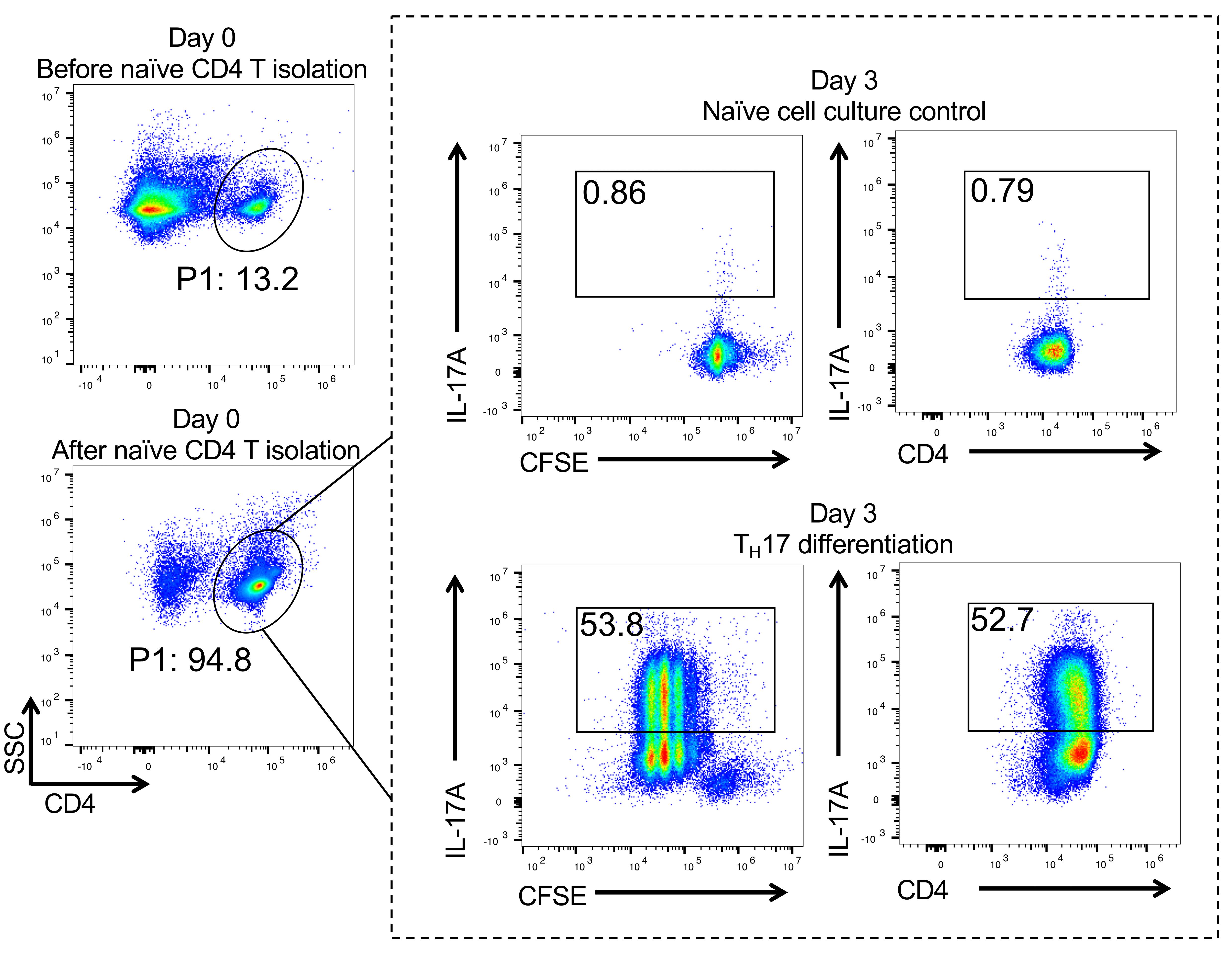
Figure 1. Naïve CD4+ T cell isolation and TH17 differentiation. Flow cytometry data showing that more than 90% naïve CD4+ T cells can be obtained after isolation on day 0 (left panel). Day 3, CFSE labeling data showing that more than 50% TH17 cells can be observed post-polarization (dotted line frame).
Data analysis
FlowJo software: Main gate P1; day 0 data, choose log SSC versus CD4 for gating CD4+ T cells. Day 3 data; within P1-CD4 gate (same gating scheme as day 0), choose log CFSE or CD4 versus IL-17A to analyze intracellular IL-17.
Recipes
Culture medium
RPMI 1640 1×
10% fetal bovine serum (FBS)
2 mM L-glutamine
1% sodium pyruvate
1% PS
1:1,000 2-ME
Store at 4°C
TCR-stimulation antibody mixture
10 μg/ml anti-mouse CD3 and 10 μg/ml anti-mouse CD28 antibodies in PBS
RBC lysis buffer (1×)
1 ml 10× buffer with 9 ml cell culture grade water
Store at 4°C
MACS buffer
500 ml PBS containing 2.5 g bovine serum albumin (BSA) and 2 ml 0.5 M EDTA
Store at 4°C, fliter using a 0.2-μm filter
TH17 polarization medium
Culture medium containing mIL-6 (50 ng/ml), hTGF-β1 (20 ng/ml), anti-mIL-2 (8 μg/ml), anti-mIL-4 (8 μg/ml), and anti-mIFN-γ (8 μg/ml)
Prepare immediately before use
Staining buffer
PBS containing 2% (w/v) BSA
Store at 4°C
Acknowledgments
The authors would like to acknowledge support by 1R21CA227926-01A1 and 1UO1CA232488-01 from the National Institutes of Health (Cancer Moonshot program), 1R01AI114581 from the National Institutes of Health, V2014-001 from the V-Foundation, and 128436-RSG-15-180-01-LIB from the American Cancer Society (to RW). With respect, the authors wish to emphasize that this protocol builds upon previous knowledge and experimental details of TH17 in vitro differentiation that has been established in the field for many years (Harrington, Hatton et al., 2005; Park, Li et al., 2005; Veldhoen; Hirota et al., 2009). This protocol was adapted with minor modification from previous study published by Wu et al. (2020).
Competing interests
The authors declare no conflicts of interest.
References
- Awasthi, A., Riol-Blanco, L., Jager, A., Korn, T., Pot, C., Galileos, G., Bettelli, E., Kuchroo, V. K. and Oukka, M. (2009). Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J Immunol 182(10): 5904-5908.
- Bettelli, E., Korn, T., Oukka, M. and Kuchroo, V. K. (2008). Induction and effector functions of TH17 cells. Nature 453(7198): 1051-1057.
- Bhaumik, S. and Basu, R. (2017). Cellular and Molecular Dynamics of Th17 Differentiation and its Developmental Plasticity in the Intestinal Immune Response. Front Immunol 8: 254.
- Cua, D. J., Sherlock, J., Chen, Y., Murphy, C. A., Joyce, B., Seymour, B., Lucian, L., To, W., Kwan, S., Churakova, T., Zurawski, S., Wiekowski, M., Lira, S. A., Gorman, D., Kastelein, R. A. and Sedgwick, J. D. (2003). Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 421(6924): 744-748.
- Gaffen, S. L., Hernandez-Santos, N. and Peterson, A. C. (2011). IL-17 signaling in host defense against Candida albicans. Immunol Res 50(2-3): 181-187.
- Guglani, L. and Khader, S. A. (2010). Th17 cytokines in mucosal immunity and inflammation. Curr Opin HIV AIDS 5(2): 120-127.
- Hartigan-O'Connor, D. J., Hirao, L. A., McCune, J. M. and Dandekar, S. (2011). Th17 cells and regulatory T cells in elite control over HIV and SIV. Curr Opin HIV AIDS 6(3): 221-227.
- Jager, A., Dardalhon, V., Sobel, R. A., Bettelli, E. and Kuchroo, V. K. (2009). Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J Immunol 183(11): 7169-7177.
- Korn, T., Bettelli, E., Oukka, M. and Kuchroo, V. K. (2009). IL-17 and Th17 Cells. Annu Rev Immunol 27: 485-517.
- Lee, Y., Awasthi, A., Yosef, N., Quintana, F. J., Xiao, S., Peters, A., Wu, C., Kleinewietfeld, M., Kunder, S., Hafler, D. A., Sobel, R. A., Regev, A. and Kuchroo, V. K. (2012). Induction and molecular signature of pathogenic TH17 cells. Nat Immunol 13(10): 991-999.
- Lee, Y., Collins, M. and Kuchroo, V. K. (2014). Unexpected targets and triggers of autoimmunity. J Clin Immunol 34 Suppl 1: S56-60.
- Ouyang, W., Kolls, J. K. and Zheng, Y. (2008). The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity 28(4): 454-467.
- Katherine M McKinnon (2018). Flow Cytometry: An Overview. Curr Protoc Immunol. 120:5.1.1-5.1.11.
- Harrington, L. E., R. D. Hatton, P. R. Mangan, H. Turner, T. L. Murphy, K. M. Murphy and C. T. Weaver (2005). Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol 6(11): 1123-1132.
- Park, H., Z. Li, X. O. Yang, S. H. Chang, R. Nurieva, Y. H. Wang, Y. Wang, L. Hood, Z. Zhu, Q. Tian and C. Dong (2005). A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol 6(11): 1133-1141.
- Veldhoen, M., K. Hirota, J. Christensen, A. O'Garra and B. Stockinger (2009). Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med 206(1): 43-49.
- Ruohan Wu*, Xuyong Chen*, Siwen Kang*, Tingting Wang, JN Rashida Gnanaprakasam, Yufeng Yao, Lingling Liu, Gaofeng Fan, Mark R. Burns, Ruoning Wang†. (2020). De novo synthesis and salvage pathway coordinately regulate polyamine homeostasis and determine T cell proliferation and function. Science Advances 6 (51): eabc4275.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Kang, S., Wu, R. and Wang, R. (2021). A Simple and Robust Protocol for in vitro Differentiation of Mouse Non-pathogenic T Helper 17 Cells from CD4+ T Cells. Bio-protocol 11(10): e4029. DOI: 10.21769/BioProtoc.4029.
- Ruohan Wu*, Xuyong Chen*, Siwen Kang*, Tingting Wang, JN Rashida Gnanaprakasam, Yufeng Yao, Lingling Liu, Gaofeng Fan, Mark R. Burns, Ruoning Wang†. (2020). De novo synthesis and salvage pathway coordinately regulate polyamine homeostasis and determine T cell proliferation and function. Science Advances 6 (51): eabc4275.
Category
Immunology > Immune cell function > Cytokine
Immunology > Immune cell staining > Flow cytometry
Cell Biology > Cell isolation and culture > Cell differentiation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









