- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro and In vivo CD8+ T Cell Suppression Assays
(*contributed equally to this work) Published: Vol 11, Iss 10, May 20, 2021 DOI: 10.21769/BioProtoc.4020 Views: 7086
Reviewed by: Luis Alberto Sánchez VargasMarieta RusevaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protocol for Screening Host-Targeting Antivirals (HTAs) Using Human PBMCs and pDCs
Zhao Xuan Low [...] Pouya Hassandarvish
Mar 5, 2025 3061 Views

Isolation and Ex Vivo Testing of CD8+ T-Cell Division and Activation Using Mouse Splenocytes
Melissa Dolan [...] John M.L. Ebos
Aug 20, 2025 3859 Views

Dual Phospho-CyTOF Workflows for Comparative JAK/STAT Signaling Analysis in Human Cryopreserved PBMCs and Whole Blood
Ilyssa E. Ramos [...] James M. Cherry
Nov 20, 2025 2336 Views
Abstract
CD8+CD28− T suppressor cells (Ts) have been documented to promote immune tolerance by suppressing effector T cell responses to alloantigens following transplantation. The suppressive function of T cells has been defined as the inhibitory effect of Ts on the proliferation rate of effector T cells. 3H-thymidine is a classical immunological technique for assaying T cell proliferation but this approach has drawbacks such as the inconvenience of working with radioactive materials. Labeling T cells with CFSE allows relatively easy tracking of generations of proliferated cells. In this report, we utilized antigen presenting cells (APCs) and T cells matched for human leukocyte antigen (HLA) class I or class II to study CD8+CD28- T cell suppression generated in vitro by this novel approach of combining allogeneic APCs and γc cytokines. The expanded CD8+CD28- T cells were isolated (purity 95%) and evaluated for their suppressive capacity in mixed lymphocyte reactions using CD4+ T cells as responders. Here, we present our adapted protocol for assaying the Ts allospecific suppression of CFSE-labeled responder T cells.
Keywords: CD8+CD28- T cellsBackground
T regulatory cells (Tregs) with dedicated suppressor function play a crucial role in the homeostatic control of immunity in the periphery. Regulatory CD8+ T cells have also been demonstrated to play an important role in neonatal tolerance and autoimmune diseases (Tang et al., 2005). There are two broad categories of immune regulation by Tregs: non-specific and antigen-specific. Non-specific immunosuppression potentially causes general immunosuppression and produces undesirable side effects, such as infectious diseases. These Tregs include CD8+CD25+, CD8+CD122+, CD45RClow, and IL-2/GM-CSF-induced CD8+ Tregs. On the contrary, antigen-specific Tregs are primed during the immune response to foreign or self-antigens and subsequently specifically downregulate that immune response. These Tregs include CD8+CD28-, CD8+CD75s+, plasmacytoid dendritic cell (DC2)-induced CD8+, CD8+CD45RChigh Tc1, and TCR peptide-specific CD8αα Tregs. CD8+CD28− Tregs have been recently documented to play an important role in alloimmunity. In our previous studies, we have expanded large numbers of human CD8+CD28− Tregs in a relatively short period of time by stimulating CD8+ T cells with APCs following supplementation with the triple common gamma chain cytokines IL-2, IL-7, and IL-15 in vitro; however, the detailed characteristics of the expanded CD8+CD28− Tregs were unclear. Moreover, the principal function of this population when transferred in vivo was yet to be examined. Measurement of suppression has been achieved through the co-culture of Tregs and T effector cells. Methods include the detection of cell proliferation, cytokine production, and activation markers (CD25 or CD134) (Long et al., 2017). CFSE-based co-culture has become the gold standard for proliferation assays and has been used successfully to assess the function of Tregs. Here, based on CFSE co-culture assays, we show that the in vitro-expanded CD8+CD28− Tregs maintain allospecific suppressive capacity both in vitro (Figure 1) and in vivo.
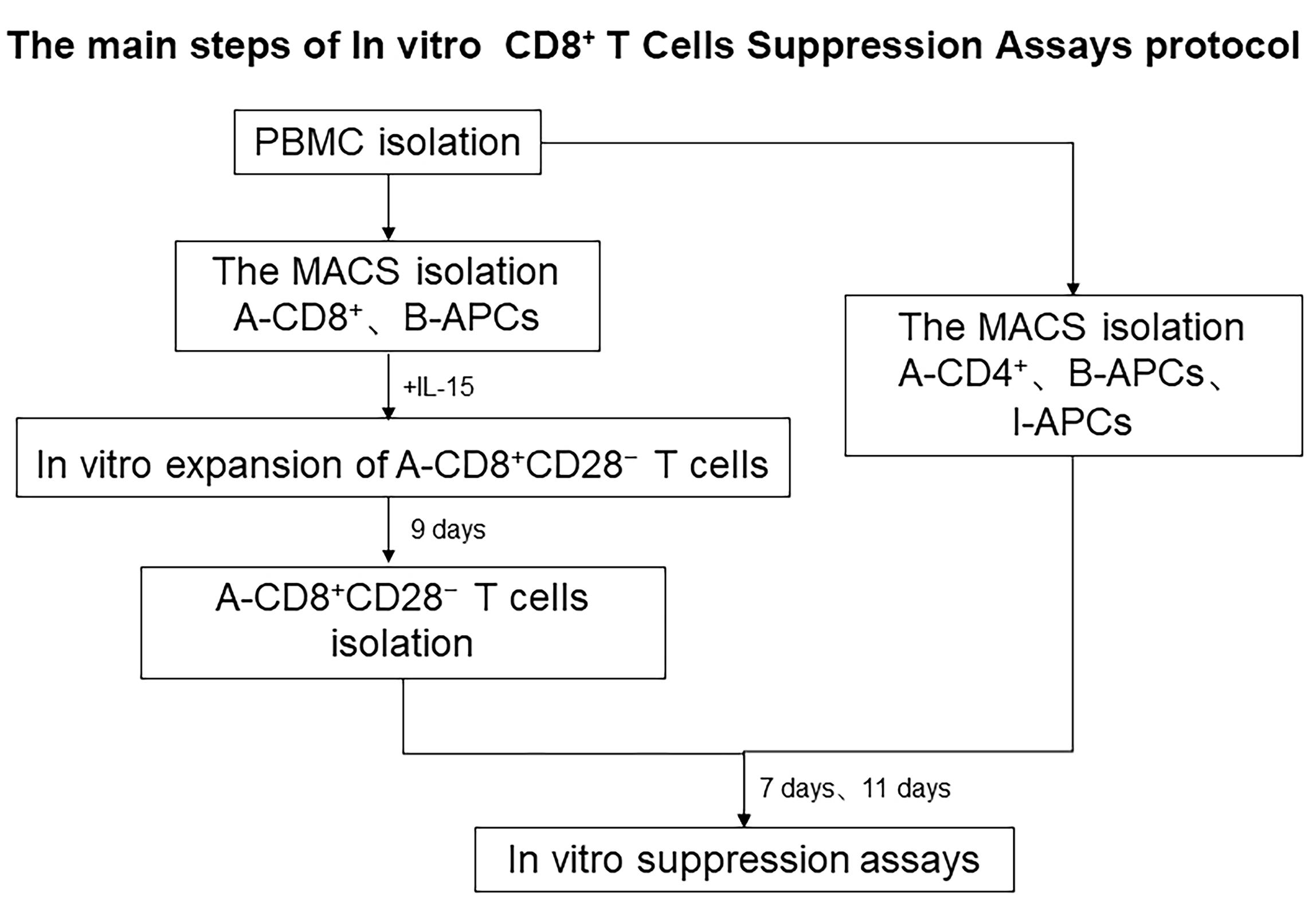
Figure 1. The main steps of the in vitro CD8+ T cell suppression assay protocol
Materials and Reagents
Heparin sodium anticoagulation tubes, 5 ml (JiangSu, YuLi)
Centrifuge tubes, 50 ml (Corning, catalog number: 430829)
Centrifuge tubes, 15 ml (Corning, catalog number: 430791)
24-well round-bottomed plates (Corning, catalog number: 3524)
96-well round-bottomed plates (Corning, catalog number: 3799)
MidiMACS separator (Miltenyi, catalog number: 130-042-302)
LS column (Miltenyi, catalog number: 130-042-401)
Disposable syringe with a needle (ShuangGe, China)
70-μm cell strainer (Biologix, catalog number: 15-1070)
NOG mice (Beijing Vital River Laboratory Animal Technology Co Charles River Laboratories)
Ficoll-Hypaque solution (Haoyang Biologiacal, TBD sciences, catalog number: LTS1077)
RPMI 1640, 500 ml (ThermoFisher, Gibco, catalog number: C11875500BT)
FBS Qualified Australia Origin (ThermoFisher, Gibco, catalog number: 10099141 C)
Bovine serum albumin, BSA (SIJIA, catalog number: N0008-1)
Phosphate-buffered saline, PBS (ThermoFisher, Gibco, catalog number: C10010500BT)
CD8 MicroBeads, human (Miltenyi, catalog number: 130-045-201)
CD28 MicroBeads, human (Miltenyi, catalog number: 130-093-247)
CD2 MicroBeads, human (Miltenyi, catalog number: 130-091-114)
CD4 MicroBeads, human (Miltenyi, catalog number: 130-045-101)
IL-2 (PeproTech, catalog number: AF-200-02-50)
IL-7 (PeproTech, catalog number: AF-200-07-50)
IL-15 (PeproTech, catalog number: AF-200-15-50)
10× RBC lysis buffer (ThermoFisher, Invitrogen, catalog number: 00-4300-54)
Trypan Blue solution 0.4%, liquid (MERCK,Sigma-Aldrich, catalog number: T8154-100ML)
7-AAD viability stain solution (ThermoFisher, Invitrogen, catalog number: 00-6993-50)
CFDA, SE (ThermoFisher, Invitrogen, catalog number: C1157)
Flow cytometry antibodies
AlexaFluorTM 700 mouse anti-human CD3 monoclonal antibody, OKT3 (ThermoFisher, eBioscience, catalog number: 56-0037-42)
efluor 450 mouse anti-human CD8a monoclonal antibody, SK1 (ThermoFisher, eBioscience, catalog number: 48-0088-41)
APC-mouse anti-human CD28 monoclonal antibody, CD28.2 (ThermoFisher, eBioscience, catalog number: 17-0289-42)
APC-mouse anti-human CD4 monoclonal antibody, OKT4 (ThermoFisher,eBioscience, catalog number: 17-0048-42)
FITC mouse anti-human CD2 monoclonal antibody, RPA-2.10 (ThermoFisher, eBioscience, catalog number: 11-0029-42)
PE-mouse anti-human CD45 monoclonal antibody, HI30 (ThermoFisher,eBioscience, catalog number: 12-0459-42)
Fixable viability stain (FVS) 620 100 μg (BD Pharmingen, catalog number: 564996)
NaCl
KCl
Na2HPO4
KH2PO4
EDTA
1× PBS (pH 7.4) (see Recipes)
D-PBS (pH 7.4) (see Recipes)
1% BSA-PBS (see Recipes)
0.5% BSA-PBS (see Recipes)
Equipment
Centrifuge (Eppendorf, model: 5810R)
Electronic balance (Sartorius, model: BP61)
Microelectronic balance (OHAUS, model: AX124ZH)
Hemocytometer
Constant temperature water box
Incubator (Thermo, model: Thermo3111)
FACS LSRFortessa (BD)
Finnpipette (Eppendorf)
Magnetic stirrer (BG-stirrelDB)
Optical microscope (CONIC, XDS-1B)
Clean bench
Software
Flowjo vX.0.7
SPSS 20.0
GraphPad Prism 5.01
Procedure
Peripheral blood mononuclear cell (PBMC) isolation
Isolate PBMCs from samples acquired in heparinized tubes from healthy volunteers. PBMCs are isolated by density gradient centrifugation using Ficoll-Hypaque solution.
Dilute the blood sample 1:2 with the same volume of RPMI 1640.
Add a volume of Ficoll-Hypaque solution equal to that of the blood sample in a 50-ml centrifuge tube.
Add the diluted blood sample carefully and slowly to the surface of the separation fluid. Centrifuge at 800 × g for 20-30 min at room temperature.
Note: The blood sample volume determines the centrifugal conditions; read the separation solution instructions.
Carefully absorb the second annular opalescent lymphocyte layer into another centrifuge tube, add 10-20 ml RPMI 1640, mix well, and centrifuge the cell suspension at 580 × g for 10 min at room temperature.
Aspirate the supernatant completely. Wash the cell pellet by adding 10 ml RPMI 1640 and centrifuging at 290 × g for 10 min at room temperature.
Repeat Step A5.
MACS separation
CD8+ cells (A-CD8+) and CD4+ cells (A-CD4+) are isolated from PBMCs of individual A by positive selection using the MACS system. APCs are obtained by depletion of CD2+ cells from PBMCs of individual B (B-APCs) or individual I (I-APCs). Donors were selected according to their HLA-A, -B, and -DR compatibility or incompatibility based on the specific requirements of individual experiments. Several groups of individuals designated as A, B, and I, which are fully HLA-A, -B, and -DR mismatched were screened out from 130 volunteers and used for independent experiments.
Note: Keep the cells and the buffer cold. Centrifuge at 4°C.
Add 10 ml 0.5% BSA-PBS buffer to resuspend PBMCs (from Step A6) and determine the cell number.
Centrifuge the cell suspension at 300 × g for 10 min. Aspirate the supernatant completely.
Resuspend the cell pellet in 80 μl 0.5% BSA-PBS buffer per 107 cells.
Add 20 μl appropriate Microbeads per 107 cells.
Mix well and incubate for 20 min in the refrigerator (2-8°C).
Wash the cells by adding 1-2 ml 0.5% BSA-PBS buffer per 107 cells and centrifuge at 300 × g for 10 min. Aspirate the supernatant completely.
Resuspend up to 108 cells in 500 μl 0.5% BSA-PBS buffer.
Note: For higher cell numbers, scale up the buffer volume accordingly.
Place the LS column in the magnetic field of a MidiMACS separator.
Note: Choose an appropriate MACS column and MACS separator according to the numbers of total and positive cells. For details, see the Microbead instructions.
Prepare the column by rinsing with 3 ml 0.5% BSA-PBS buffer.
Apply the cell suspension to the LS column.
Collect the unlabeled cells that pass through and wash the column with 3 ml 0.5% BSA-PBS buffer. Collect the total effluent; this is the unlabeled cell fraction (such as CD2- cells).
Wash the column by adding 3 ml 0.5% BSA-PBS buffer twice. Only add fresh 0.5% BSA-PBS buffer when the column reservoir is empty.
Remove the column from the separator and place on a suitable collection tube.
Pipette 5 ml buffer onto the column. Immediately flush out the magnetically labeled cells by firmly pushing the plunger into the column (such as CD8+ cells, CD4+ cells).
Perform flow cytometry analysis to evaluate the purity of the sorted cell suspensions; the purity should be ≥97%.
In vitro expansion of CD8+CD28− T cells
A-CD8+ cells are isolated by positive selection using the MACS system according to Procedure B. B-APCs (CD2- cells) from HLA-A, -B, -DR mismatched individual B are isolated by depletion of CD2+ cells using CD2 microbeads.
Centrifuge the A-CD8+ cells and B-APCs obtained above at 300 × g for 10 min. Aspirate the supernatant completely.
Resuspend in 5 ml RPMI1640 and determine the cell number.
Centrifuge the A-CD8+ cells and B-APCs at 300 × g for 10 min. Aspirate the supernatant completely.
Resuspend the cells in cell culture medium (RPMI 1640 supplemented with 15% FBS) to achieve A-CD8+ at 2 × 106 cells/ml and B-APCs at 1 × 106 cells/ml.
Seed the A-CD8+ cells (2 × 106/well) and B-APCs (1 × 106/well) onto 24-well flat-bottomed plates at a ratio of 2:1 in a total volume of 2 ml, in the presence of IL-2 at 20 U/ml, IL-7 at 50 ng/ml, and IL-15 at 50 ng/ml, in an incubator at 37°C and a humidified 5.5% CO2 atmosphere.
Change semi-culture medium (1 ml) after 3 days. After 5 days and 7 days of coculture, mix well and divide one well into two.
On day 9, harvest the cells into 50-ml centrifuge tubes. Add 10 ml 0.5% BSA-PBS buffer to wash the cells and determine the cell number.
Centrifuge the cell suspension at 300 × g for 10 min at 4°C. Aspirate the supernatant completely.
Resuspend the cell pellet in 40 μl 0.5% BSA-PBS buffer per 107 cells.
Add 10 μl CD28-PE per 107 cells.
Mix well and incubate for 10 min in the refrigerator (2–8°C).
Wash the cells by adding 1-2 ml 0.5% BSA-PBS buffer per 107 cells and centrifuge at 300 × g for 10 min at 4°C. Aspirate the supernatant completely.
Resuspend the cell pellet in 80 μl 0.5% BSA-PBS buffer per 107 cells.
Add 20 μl anti-PE microbeads per 107 cells.
Mix well and incubate for an additional 15 min in the refrigerator (2-8°C).
Wash the cells by adding 1-2 ml 0.5% BSA-PBS buffer per 107 cells and centrifuging at 300 × g for 10 min at 4°C. Aspirate the supernatant completely.
Resuspend up to 108 cells in 500 μl 0.5% BSA-PBS buffer.
Note: For higher cell numbers, scale up the buffer volume accordingly.
Place the LS column in the magnetic field of a MidiMACS separator.
Note: Choose an appropriate MACS column and MACS separator according to the numbers of total and positive cells. For details, see the microbead instructions.
Prepare the column by rinsing with 3 ml 0.5% BSA-PBS buffer.
Apply the cell suspension to the column.
Collect the unlabeled cells that pass through and wash the column with 3 ml 0.5% BSA-PBS buffer. Collect the total effluent; this is the unlabeled cell fraction (CD28- cells).
Perform flow cytometry analysis to evaluate the purity of CD28- cell suspensions; the purity should be ≥95%.
In vitro suppression assays
Responder CD4+ cells are isolated from PBMCs of individual A (A-CD4+ T cells) by positive selection using CD4 microbeads. As stimulators, APCs cells from individual B (B-APCs) or from HLA-A, -B, -DR fully mismatched indifferent individual I (I-APCs) are isolated by depletion of CD2+ cells using CD2 microbeads.
Centrifuge the A-CD4+ cell suspension (from Step B14) at 300 × g for 10 min at 4°C. Aspirate the supernatant completely.
Wash the freshly isolated CD4+ cells with 10 ml PBS to remove any proteins. Centrifuge at 400 × g for 10 min at 4°C.
Resuspend the cells in 1 ml PBS.
Add CFSE working dilution to a final concentration of 0.5 μM at room temperature.
Incubate for 7 min in the incubator (37°C); work gently and protect from light.
Add 1 ml FBS and 9 ml RPMI 1640 to terminate the reaction, then centrifuge at 300 × g for 10 min. Aspirate the supernatant completely.
Wash the cells once again with RPMI 1640. Aspirate the supernatant completely.
Resuspend in 5 ml RPMI 1640 and count the cells.
Note: After labeling with CFSE, perform flow cytometry analysis to ensure that the cells are stained.
Centrifuge the A-CD8+CD28- cells, B-APCs, and I-APCs obtained above at 300 × g for 10 min. Aspirate the supernatant completely.
Resuspend in 5 ml RPMI 1640 and count the corresponding cells.
Centrifuge the A-CD8+CD28- cells, B-APCs, I-APCs, and A-CD4+CFSE at 300 × g for 10 min. Aspirate the supernatant completely.
Resuspend the cell pellet in culture medium (RPMI 1640 supplemented with 15% FBS) to achieve A-CD4+CFSE at 1 × 106 cells/ml, B-APCs or I-APCs at 1 × 106 cells/ml, and A-CD8+CD28- cells at 2.5 × 105 cells/ml.
Seed the A-CD4+CFSE cells (5 × 104/well), B- APCs or I-APCs (5 × 104/well), and CD8+CD28- cells (2.5 × 104/well) onto 96-well round-bottomed plates in a total volume of 200 μl cell culture medium containing 15% FBS.
Note: Responder CD4+ T cells, stimulated with B-APCs or I-APCs, only served as positive controls (B-APCs were used as priming cells in vitro to expand CD8+CD28− T cells from individual A, whereas I-APCs had never had immune recognition by CD8+CD28− T cells from individual A during the in vitro expansion period).
Change semi-culture medium (100 μl) after 3 days, 5 days, and 9 days of coculture.
After 7 or 11 days of coculture, harvest, stain, and detect the CD4+ cells for CFSE dilution by flow cytometry.
In vivo suppression assays
Isolate the responder CD4+ T cells and APCs (Procedure A and B).
Mix a total of 4 × 106 human CD4+ T cells with an equal number of APCs, either B-APCs or I-APCs, and combine with 2 × 106in vitro-expanded human CD8+CD28− T cells in a total volume of 1.5 ml PBS. The responder CD4+ T cells alone plus B-APCs are used in the same fashion as a positive control.
Administer the cell mixture for each group to NOG mice by intraperitoneal injection.
Sacrifice the mice on day 11 post-injection.
Isolate the spleen and homogenize to generate a single-cell suspension.
Centrifuge the cell suspension at 500 × g for 5 min at 4°C. Aspirate the supernatant completely.
Add 3 ml 1× RBC lysis buffer to lyse the red cells, mix well, and incubate for 5 min at room temperature.
Add 2 ml PBS, centrifuge at 350 × g for 10 min at room temperature. Aspirate the supernatant completely.
Stain the cells and detect those expressing CD4+ by flow cytometry.
Note: You can also detect human CD8+ and CD4+ cells in mouse spleen tissue by immunohistochemical techniques.
Staining cells for analysis by flow cytometry
Flow cytometry analysis is performed on cells harvested from the culture under different conditions (Procedures D and E).
Place the cells to be analyzed into 12 × 75 mm polystyrene tubes.
Wash the cells in 3 ml 1% BSA-PBS buffer at 350 × g for 10 min at 4°C.
Discard the supernatant and quickly vortex to loosen the pelleted cells.
Add 2 μl antibodies (such as anti-CD4), mix well, and incubate for 30 min at 4°C in the dark.
Wash the cells in 3 ml 1% BSA-PBS buffer at 350 × g for 10 min at 4°C.
Discard the supernatant and quickly vortex to loosen the pelleted cells.
Add 3 μl 7-AAD Viability Staining Solution and incubate for 10 min at room temperature in the dark.
Resuspend the cells in an appropriate volume of 1% BSA-PBS and detect by flow cytometry.
Data analysis
Data analysis is performed using offline analysis software such as Flowjo. As shown in Figure 2, the identification of CD4+ T cells begins with the creation of gates to isolate the lymphocyte population (FSC-A; SSC-A), followed by rigorous doublet exclusion (FSC-W vs. FSC-H and SSC-W vs. SSC-A). Exclude cells that are positive for dead staining and then gate on the target population of cells.
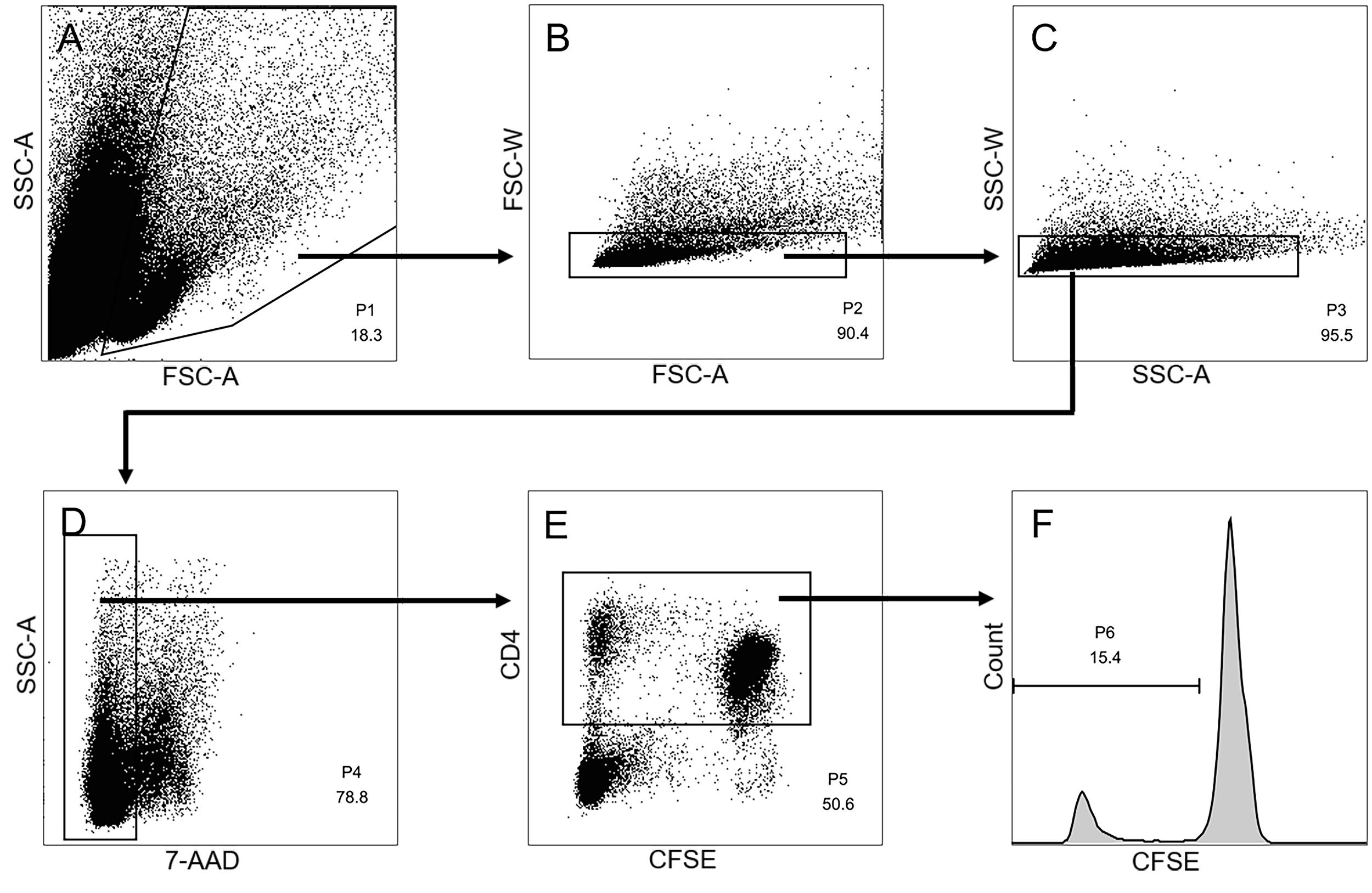
Figure 2. Gating strategy to identify CD4+ cells in suppression assays. A. Lymphocyte population identified in FSC-A vs. SSC-A plot. B-C. This is followed by doublet discrimination using FSC and SSC area and width plots. D. After staining with 7-AAD, live cells are then analyzed. E. Live CD4+ T cells are gated. F. The proliferation of CD4+ T cells was measured by CFSE dilution.The suppressive percentage is calculated as follows: % suppression = [1 – (% CD4+ T cell proliferation in the presence of CD8+CD28− T cells)/(% CD4+ T cell proliferation in the absence of CD8+CD28− T cells)] × 100.
Statistical analysis is carried out using the SPSS version 22 software. For comparison of different groups, where appropriate, an independent-samples t-test and nonparametric test are used to determine statistical significance. Graphs are created using the GraphPad Prism version 5.01 software.
Recipes
1× PBS (pH 7.4)
NaCl 8 g
KCl 0.2 g
Add ultrapure H2O to a total volume of 1 L
D-PBS (pH 7.4)
NaCl 8 g
KCl 0.2 g
Na2HPO4 1.15 g
KH2PO4 0.2 g
Add ultrapure H2O to a total volume of 1 L
1% BSA-PBS
BSA 10 g
NaCl 8 g
KCl 0.2 g
Na2HPO4 1.44 g
KH2PO4 0.24 g
Add ultrapure H2O to a total volume of 1 L
0.5% BSA-PBS
EDTA 2 mM
BSA 5 g
NaCl 8 g
KCl 0.2 g
Na2HPO4 1.44 g
KH2PO4 0.24 g
Add ultrapure H2O to a total volume of 1 L
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China [No. 81270839, 81428007] and the Natural Science Foundation of Guangdong Province [2017A030313524]. The protocols outlined here were originally described in several papers from the Yu lab (Yu et al., 2011; Feng et al., 2018; Liu et al., 2020).
Competing interests
The authors declare no competing interests, financial or non-financial.
Ethics
All experiments using human cells were carried out in accordance with the recommendations of the Ethical Review Board of Southern Medical University (Guangzhou, China); all subjects gave written informed consent in accordance with the Declaration of Helsinki. All animal experiments were approved by the Guidelines for the Care and Use of Animals established by the Animal Care and Use Committee of Southern Medical University.
References
- Feng, F., Liu, Y., Liu, G., Zhu, P., Zhu, M., Zhang, H., Lu, X., Liu, J., Luo, X. and Yu, Y. (2018). Human CD8+CD28- T suppressor cells expanded by IL-15 in vitro suppress in an allospecific and programmed cell death protein 1-dependent manner. Front Immunol 9: 1442.
- Liu, G., Yu, Y., Feng, F., Zhu, P. and Liu, Y. (2020). Human CD8+CD28- T suppressor cells expanded by common gamma chain (gammac) cytokines retain steady allospecific suppressive capacity in vivo. BMC Immunol 21(1): 23.
- Tang, X. L., Smith, T. R. and Kumar, V. (2005). Specific Control of Immunity by Regulatory CD8 T Cells. Cell Mol Immunol 2(1):11-19.
- Long, A. E., Tatum, M., Mikacenic, C. and Buckner, J. H. (2017). A novel and rapid method to quantify Treg mediated suppression of CD4 T cells.J Immunol Methods 449: 15-22.
- Yu, Y., Zitzner, J. R., Houlihan, J., Herrera, N., Xu, L., Miller, J., Mathew, J. M., Tambur, A. R. and Luo, X. (2011). Common gamma chain cytokines promote rapid in vitro expansion of allo-specific human CD8+ suppressor T cells.PLoS ONE 6(12): e28948.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Xie, L., Liu, G., Liu, Y. and Yu, Y. (2021). In vitro and In vivo CD8+ T Cell Suppression Assays. Bio-protocol 11(10): e4020. DOI: 10.21769/BioProtoc.4020.
Category
Immunology > Immune cell function > Antigen-specific response
Immunology > Immune cell function > Cytokine
Cell Biology > Cell-based analysis > Cell-to-cell interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









