- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Cell-free Synthesis of Correctly Folded Proteins with Multiple Disulphide Bonds: Production of Fungal Hydrophobins
Published: Vol 11, Iss 10, May 20, 2021 DOI: 10.21769/BioProtoc.4019 Views: 5893
Reviewed by: Alba BlesaRalph Thomas BoettcherMurugappan Sathappa

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2167 Views

Prokaryotic Expression and Purification of the hSox2-HMG Domain
Lijie Yang [...] Jingjun Hong
Aug 20, 2025 2367 Views

On-Column Dual-Gradient Refolding for Efficient Recovery of Insoluble Affinity-Tagged Recombinant Proteins
Anna Vlaskina [...] Maxim Patrushev
Feb 5, 2026 39 Views
Abstract
Cell-free synthesis is a powerful technique that uses the transcriptional and translational machinery extracted from cells to create proteins without the constraints of living cells. Here, we report a cell-free protein production protocol using Escherichia coli lysate (Figure 1) to successfully express a class of proteins (known as hydrophobins) with multiple intramolecular disulphide bonds which are typically difficult to express in a soluble and folded state in the reducing environments found inside a cell. In some cases, the inclusion of a recombinant disulphide isomerase DsbC further enhances the expression levels of correctly folded hydrophobins. Using this protocol, we can achieve milligram levels of protein expression per ml of reaction. While our target proteins are the fungal hydrophobins, it is likely that this protocol with some minor variations can be used to express other proteins with multiple intramolecular disulphide bonds in a natively folded state.
Graphic abstract:

Figure 1. Workflow for cell-free protein expression and single-step purification using affinity chromatography. A. E. coli S30 lysate prepared as described in Apponyi et al. (2008) can be stored for up to several years at -80°C without any loss of activity in our experience. B. The S30 lysate, plasmid DNA that encodes for the protein of interest along with an affinity tag and components required for transcription and translation are added to the reaction mix. Following a single-step protein purification, the protein of interest can be isolated for further use.
Background
The bacterial cytosol microenvironment is home to thousands of diverse reactions and is tightly regulated in a reducing redox state. This poses challenges in recombinant bacterial protein overexpression as the reducing environment can prevent the correct formation of critical disulphide bonds in proteins that require them for structure and/or function (Kadokura and Beckwith, 2010). As such, these proteins are often expressed by E. coli into insoluble inclusion bodies (Berkmen, 2012) that renders them unusable. In favorable cases, proteins from inclusion bodies can be refolded in vitro after solubilizing in strong denaturants (Qin et al., 2015). However, this is typically a lengthy and reagent consuming process and the yield of the correctly refolded protein is variable.
One alternative to conventional recombinant bacterial overexpression is cell-free synthesis, where proteins can be expressed using the bacterial transcription and protein production machinery, but without the confines of a cell (Apponyi et al., 2008; Ozawa and Loh, 2014; Gao et al., 2019; Dondapati, 2020). Cell-free protocols are particularly useful in the expression of cysteine-rich proteins, as the redox environment can be controlled by simply changing the ratios of reduced and oxidized glutathione (GSH:GSSG) and varying the concentration of reducing agents such as dithiothreitol (DTT). After optimization, the yield of protein with correctly formed disulphide bonds suitable for downstream experiments (e.g., binding studies, structural and biophysical characterisation) can be greatly increased.
Beyond proteins with disulphide bonds, cell-free protein synthesis systems can hold additional advantages over traditional recombinant protein production such as the ability to readily incorporate unnatural amino acids (Gao et al., 2019; Dondapati et al., 2020). In addition, proteins that are too toxic for a host cell can be expressed in a cell-free environment as there is no requirement to maintain cell viability and this has allowed high throughput production and screening for membrane proteins, enzymes, and other therapeutics (Khambhati et al., 2019).
Our optimized cell-free expression protocol allows direct control of the redox conditions in the reaction mixture and demonstrates a distinct advantage over recombinant expression for a class of proteins known as the fungal hydrophobins (Siddiquee et al., 2020). This class of proteins is characterized by four disulphide bonds and the correct bonding pattern is required for hydrophobins to self-assemble into amphipathic layers at hydrophilic:hydrophobic interfaces (Sunde et al., 2008; Ball et al., 2020). These unique assemblies have suggested a range of biotechnological applications from drug delivery systems to coating implants and the engineering of surfaces to increase biocompatibility (Wosten and Scholtmeijer, 2015; Berger and Sallada, 2019). While many hydrophobins can be overexpressed in E. coli to a very high yield, the protein is almost exclusively found misfolded within inclusion bodies and cannot be used for downstream applications. Therefore, a long process of in vitro refolding and multi-step purification is typically required to separate the correctly folded protein from the partially folded and misfolded species (Kwan et al., 2006; Pille et al., 2015; Pham et al., 2016). To overcome this problem, we have developed a cell-free expression system for hydrophobin production based on the published S30 E. coli lysate (Apponyi et al., 2008).
The S30 lysate is one of the most used extracts in cell free synthesis of proteins and contains the cellular transcriptional and translational machinery. A Pubmed search with “S30 cell free” as search terms gives >150 articles. While the S30 lysate can be purchased commercially (e.g., from Promega), it can be produced readily in laboratories that perform recombinant protein expression and purification at a fraction of the cost of commercially available cell-free expression kits. This cell-free reaction involves incubating the lysate and other key cofactors with a gene of interest cloned downstream of a constitutive promoter in a dialysis set up. The dialysis set up allows continuous exchange of lower molecular reagents (e.g., amino acids, rNTPs, ATP) and offers higher protein yields per unit of components that are the more expensive or time-consuming to prepare (e.g., transcription and translation machinery in the S30 extract as well as plasmid DNA).
To aid purification, we have cloned the gene into the vector pET-MCSIII that has a hexa-histidine affinity tag but lacks the lac operator (Neylon et al., 2000). The protocol presented herein can be easily amended to enable the expression of other proteins with disulphide bonds. The apparatus used in this protocol can be easily prepared using disposable plastic tubes commonly used in most scientific laboratories and can be scaled up as required. The efficiency and low cost of this set up allow multiple proteins (e.g., a protein and its mutants) to be produced in parallel cell-free reactions using one preparation of dialysis buffer. Isotopically labeled proteins can also be easily produced by substituting in labeled amino acids in the reaction mixture. In some cases, this has allowed Nuclear Magnetic Resonance (NMR) and mass spectroscopy experiments to be carried out on the protein of interest within one day of setting up the reaction and without the need for further purification.
Materials and Reagents
Eppendorf tubes 1.5 ml (Thermo Fisher Scientific, catalog number: 69715)
Eppendorf tubes 0.5 ml (Eppendorf, catalog number: 0030121023)
Greiner Bio-One 50 ml tubes
Pipette tips
S30 Lysate prepared according to Apponyi et al. (2008)
rNTP (Sigma-Aldrich, catalog numbers: A6559, C8552, G3776, U1006)
HEPES (Sigma-Aldrich, catalog number: 15630106)
Folinic acid (Sigma-Aldrich, catalog number: 47612)
cAMP (Sigma-Aldrich, catalog number: A6885)
NH4OAc (Sigma-Aldrich, catalog number: A1542)
ATP (Sigma-Aldrich, catalog number: A1852)
Creatine phosphate (Sigma-Aldrich, catalog number: 27920)
Potassium glutamate (Sigma-Aldrich, catalog number: G1501)
Creatine kinase (Sigma-Aldrich, catalog number: C9983)
Mg(OAc)2 (Sigma-Aldrich, catalog number: M5661)
tRNA (Sigma-Aldrich, catalog number: R8759)
Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D0632)
L-Glutathione reduced (GSH) (Sigma-Aldrich, catalog number: G4251)
L-Glutathione oxidised (GSSG) (Sigma-Aldrich, catalog number: G4376)
20 amino acids (Sigma-Aldrich)
10,000 MWCO SnakeSkin® dialysis membrane (Thermo Fisher Scientific, catalog number: 88243). Please note to choose a membrane cutoff that is at least three times smaller than the size of the expected protein product
NuPAGETM 4-12% Bis-Tris Protein Gel (Invitrogen, catalog number: NP0323PK2) or any SDS-PAGE gel that is suitable for the detection of the protein of interest
MES buffer (Invitrogen, catalog number: NP0002) or other compatible buffers for the running the SDS-PAGE gel
NuPAGETM LDS Sample Buffer (4×) (Thermo Fisher Scientific, catalog number: NP0007)
PBS (Sigma-Aldrich, catalog number: P4417)
Urea (Sigma-Aldrich, catalog number: U5378)
Recombinant disulphide isomerase DsbC (Note 2)
Plasmid or PCR-amplified fragment encoding for the protein of interest (Note 3)
Equipment
Standard laboratory equipment commonly available in molecular biology and protein laboratories, such as pipettes, fridges, magnetic stirrers have not been included. Equipment that have similar specifications to those listed below can also be used.
Scalpel
NanoDropTM UV-Vis Spectrophotometer (Thermo Fisher Scientific, catalog number: ND-2000)
Bunsen burner
Incubator (Eppendorf, model: Innova 44; M1282-0002)
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) apparatus (ThermoFisher Scientific, catalog number: A25977)
Software
Microsoft Excel
Procedure
Cell-free (in vitro) protein synthesis
The cell-free reaction involves incubating the S30 lysate with the DNA construct, encoding the gene of interest under a constitutive promoter, and the necessary cofactors in a dialysis setting. For more details about the DNA construct, please see Note 3.
When testing new constructs for cell-free expression, the use of reagents can be minimized by preparing small-scale cell-free reactions in a total inner volume of 50 µl and outer volume of 500 µl in the dialysis (Figure 2). Figure 2 illustrates the setup we use in our laboratory but other dialysis set ups can be used. For most proteins, a reaction volume of 50 µl should provide enough sample for running on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel to confirm the expression of the protein of interest and its relative yield across different reaction conditions.
To produce larger quantities of proteins, the inner and outer volumes can be increased accordingly. Our large-scale production typically has an inner volume of 1 ml and an outer volume of 10 ml with each 1-ml reaction producing ~0.5-2 mg of a hydrophobin protein.
To allow easy reproduction and scaling of the cell-free reactions, prepare the necessary reagents as combinations of mixtures as per Tables 1 and 2 and store at -80°C. The following protocol shows a small-scale cell-free reaction of 50 µl inner reaction volume and 500 µl outer reaction volume as described in Table 3. The concentration ranges for components that likely require optimization depending on the proteins of interest are given in Table 4. An interactive Excel spreadsheet that aids the calculation of reagents used in the reaction setup is available for download (Note 1).
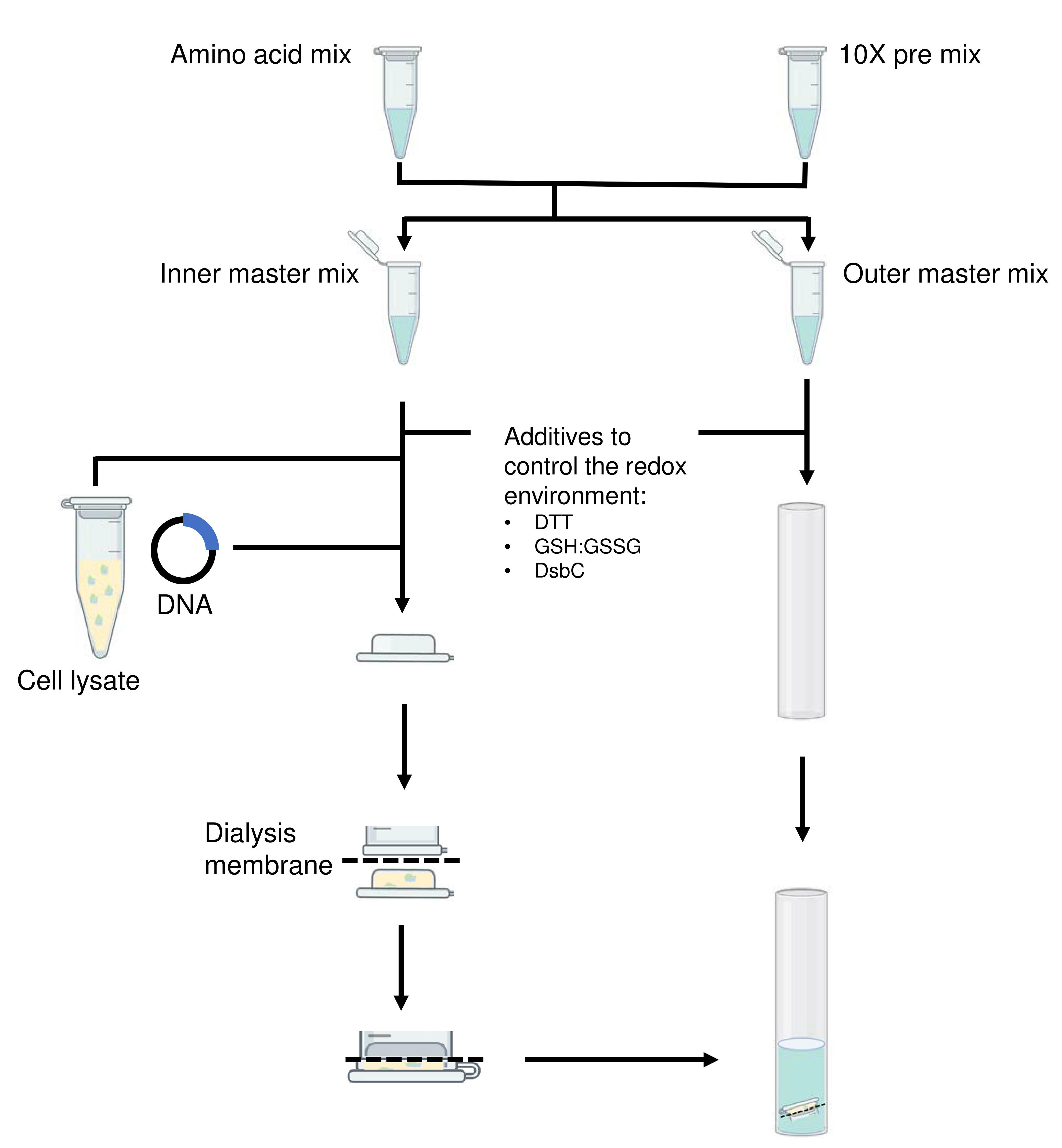
Figure 2. Flowchart illustrating the setup of a small-scale cell-free reaction. The slashed lines indicate the position of the dialysis membrane.
Preparation of reagent mixtures
Prepare 3 mixtures of the 20 amino acids into 3 separate tubes on ice according to Steps A2 to A4 below.
Add the amino acids (Ala, Arg, Gly, His, Lys, Pro, Ser, Thr, Val) and dissolve in MilliQ water to make water soluble amino acid stocks.
Add the amino acids (Asn, Asp, Cys, Glu, Gln, Leu, Met, Trp, Tyr) and dissolve in 1 M HCl to make acid soluble amino acid stocks.
Add the amino acids (Ile, Phe) and dissolve in 1 M KOH to make base soluble amino acid stocks.
Mix the 3 components listed in Table 1 to make the total amino acid mixture with each amino acid at a final concentration of 15 mM.
Table 1. Preparation of amino acids mixture for a 50-µl inner reaction
Components Stock concentration Final concentration Volume Water soluble amino acid stock 50 mM 15 mM 12 µl Acid soluble amino acid stock 50 mM 15 mM 12 µl Base soluble amino acid stock 50 mM 15 mM 12 µl MilliQ water - - Fill up to 40 µl Mix all components listed in Table 2 on ice to prepare for the 10× pre-mix.
Table 2. Preparation of 10× pre-mix for a 50 µl inner reaction
Component Stock concentration Final concentration in reaction (1×) Final concentration (10×) Volume rNTP 25 mM 0.8 mM 8 mM 19.2 µl HEPES, pH 7.58 2 M 55 mM 550 mM 16.5 µl Folinic acid 10 mM 68 µM 680 µM 4.08 µl cAMP 100 mM 0.64 mM 6.4 mM 3.84 µl NH4OAc 10 M 27.5 mM 275 mM 1.65 µl MilliQ water - - - Fill up to 60 µl The amino acid mixtures and the 10× pre-mix can be stored at -80°C for later use.
Preparation of inner and outer reaction master mixes
Thaw the amino acid mix and the 10× pre-mix on ice.
Add components of the inner reaction master mix into a new tube and outer reaction master mix into another tube (Table 3).
Table 3. Preparation of inner and outer reaction master mix for a 50 µl inner reaction
Component Stock concentration Final concentration in inner reaction Inner master mix (50 µl) Final concentration in outer reaction Outer master mix (500 µl) 10× pre-mix 10× 1× 5 µl 1× 50 µl Creatine phosphate 1 M 80 mM 4 µl 80 mM 40 µl Amino acid mixture 15 mM 1 mM 3.3 µl 1 mM 33.33 µl Potassium Glutamate 4 M 208 mM 2.6 µl 208 mM 26 µl Creatine kinase 10 mg/ml 250 µg/ml 1.25 µl - - Mg(OAc)2 1 M 15 mM 0.75 µl 19.3 mM 9.65 µl ATP 100 mM 1.2 mM 0.6 µl 1.2 mM 6 µl tRNA 17.5 mg/ml 0.175 mg/ml 0.5 µl - - Total - - 18 µl 164.98 µl Keep the mixtures on ice while setting up the reaction apparatus.
Cell-free reaction apparatus set-up
Add components of inner reaction into a tube (Table 4).
Table 4. Cell-free reaction set-up in a dialysis setting for a 50-µl inner reaction (use the provided Excel calculator and enter other volumes as required)
Component Stock concentration Final concentration in inner reaction Inner Volume Final concentration in outer reaction Outer Volume Reaction master mix - - 18 µl - 164.65 µl S30 lysate - 30% (v/v) 15 µl - - DTTa 100 mM 1.7 mM 0.85 µl 1.7 mM 8.5 µl GSHb 10 mM 1 mM 5 µl 1 mM 50 µl GSSGc 5 mM 0.1 mM 1 µl 0.1 mM 10 µl DNAd 300 ng/µl 6 ng/µl 1 µl - - DsbCe 10 mg/ml 0.3 mg/ml 1.5 µl 0.3 mg/ml 15 µl MilliQ water Fill up to 50 µl Fill up to 500 µl aDTT (0.1-1.7 mM)
bGSH (0.1-1 mM)
cGSSG (0.1-0.1 mM)
dDNA (0.3-6 ng/µl)
eDsbC (0.1-1.7 mM)
a−eCan be varied to meet the needs of the protein of interest
Add components of outer reaction into another tube (Table 4).
Adjust the outer reaction buffer to pH 7.5 before adding DTT, GSH, GSSG.
Add Disulfide bond isomerase C (DsbC) (Note 2).
Setup the apparatus for small-scale (Figure 3) or large-scale cell-free reaction.
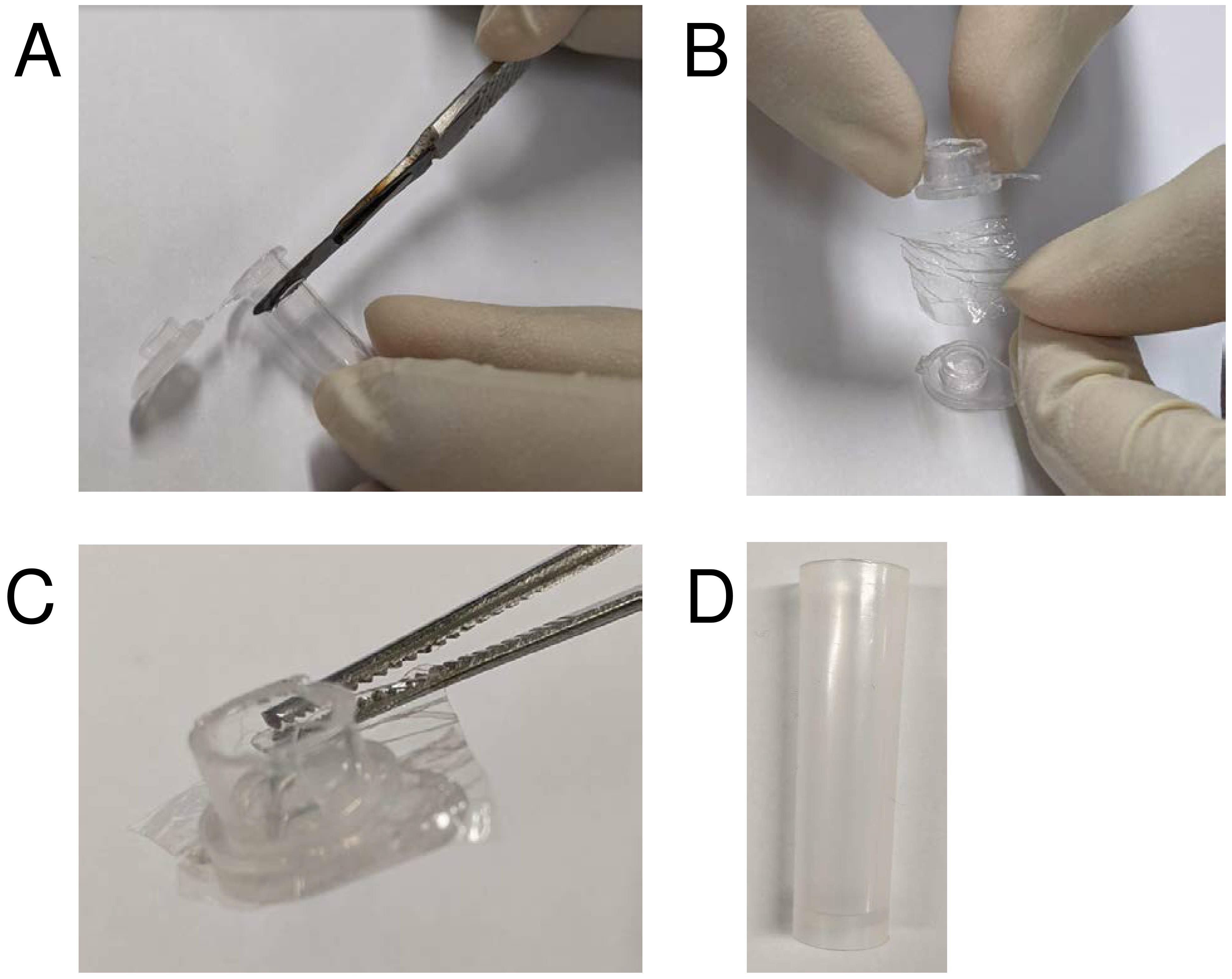
Figure 3. Preparation of a custom micro-dialysis button. A. Heat up a scalpel (e.g., using a Bunsen burner) and slice the tip of the tube and cut off the lid with scissors. B. Add inner reaction mixture to the lid, cover with a small piece of dialysis membrane. C. Seal the dialysis membrane onto the lid with the cut plastic tube, in the same orientation as per standard tube closure. Make sure the membrane is in place securely with excess dialysis membrane visible all around the lid. D. Add the outer reaction mixture to a collection tube that has a diameter slightly larger than the micro-dialysis button. Place the micro-dialysis button inside this tube.For a small-scale reaction, make a micro-dialysis button by slicing the lid of a 0.45-ml PCR tube.
Place the lid upside down on a flat surface and add 50 μl of the inner reaction mixture (or any other desirable volume that fits) to the lid.
Wet a small stretch of 10,000 MWCO SnakeSkin® dialysis membrane with water, dab off the excess water with a tissue paper and wedge the membrane between the lid and the cut tube.
Add the outer reaction mixture to a tube and place the micro-dialysis button inside with the membrane exposed to the outer reaction mixture.
Incubate the cell-free reaction for 4-20 h at 30°C and 180 rpm by taping it to a holder in a shaking incubator.
Initial analysis of protein expression
Once the reaction has been completed, remove the dialysis buttons and dab dry. Then carefully pierce the membrane with a pipette tip with pipette plunger depressed to the first stop and aspirate the inner reaction mixture for downstream analysis.
Clarify the reaction mixture by centrifugation (20,000 × g, 10 min, 4°C) and separate soluble and insoluble fractions and aliquot appropriate amounts for SDS-PAGE analysis.
For 20-µl SDS-PAGE gel samples, take 10 µl of the clarified soluble fraction and add 10 µl of 1× PBS, and take the insoluble fraction and dilute with 20 µl of 1 M Urea.
Add 1× LDS buffer from 4× LDS stock to the gel samples and heat for 10 min at 80°C.
Run SDS-PAGE analysis at 200 V for 20 min and stain according to manufacturer’s protocol.
Purification of the expressed protein
This will vary depending on the protein of interest and whether an affinity tag is present. For the hydrophobin proteins produced in our laboratory, we use Nickel-NTA Affinity Chromatography (IMAC), followed by cleavage and removal of the hexa-histidine tag when needed and then reverse-phased High Pressure Liquid Chromatography (rpHPLC). The use of isotopically labeled amino acids (e.g., with 15N) can circumvent the need for protein purification in certain applications (e.g., to check protein conformation using 1H-15N NMR spectroscopy) which can lead to a substantial time saving.
Analysis of the purified protein
The analysis of the purified protein, including measuring its yield will vary according to the specific characteristics of the protein of interest as well as the available equipment. Several techniques such as gel densitometry, Bradford assays or UV-Vis spectroscopy can be utilized to measure the yield of the purified protein. An activity assay (e.g., enzymatic activity assay) for the protein of interest should be conducted if available to assess if the produced protein is functional. With our proteins of interest, we check protein purity using SDS-PAGE, calculate the yield of the hydrophobin proteins by measuring absorbance of the purified protein solution at 280 nm using a NanoDropTM, confirm the correct folding via NMR spectroscopy, confirm the mass of the protein matches that of the calculated mass via mass spectrometry and validate its ability to assemble using Thioflavin-T assays.
Notes
An interactive cell-free reaction set-up calculator is available to download. On the left-hand side, the reaction is shown as a small-scale cell-free reaction with a 50-μl inner reaction volume. These values are used on the right-hand side within the reaction set-up calculator. Any desired inner reaction volume can be entered along with the number of reactions and the required volumes will be calculated. The desired concentrations of the redox additives (DTT, GSH:GSSG, DsbC) and the concentration of DNA can be adjusted as required (see Table 4 for suggested ranges).
The DsbC was expressed and purified as described in Matsuda et al. (2013) and dialyzed into Phosphate-buffered saline (PBS) and stored at 4°C until use. The construct we used was obtained from the Kigawa laboratory.
It is important to choose a plasmid that allows for constitutive protein expression, i.e., without the lac operator. The plasmid we used is the pET-MCSIII as this vector has been reported for the successful synthesis of a number of proteins in cell free synthesis schemes. However, we believe that any recombinant protein expression vector can be used with the protocol provided constitutive protein expression can be achieved. If an existing recombinant protein expression vector has a lac operator, we suggest removing this to increase the yield. Instead of plasmid DNA, the use of PCR-amplified DNA can also be used with this set up (Neylon et al., 2000; Wu et al., 2007).
If production of proteins with selectively labeled amino acid(s) is required, simply replace the unlabeled amino acid(s) used in Steps A2, A3 and/or A4 with the labeled amino acid(s) of choice when making up the amino acid mixture (Table 1).
Acknowledgments
This project was supported by a Discovery Project grant, DP200102463 from the Australian Research Council to A.H.K.
Competing interests
The authors declare no competing interests.
References
- Apponyi, M. A., Ozawa, K., Dixon, N. E. and Otting, G. (2008). Cell-free protein synthesis for analysis by NMR spectroscopy. Methods Mol Biol 426: 257-268.
- Ball, S. R., Pham, C. L. L., Lo, V., Morris, V. K., Kwan, A. H. and Sunde, M. (2020). Formation of Amphipathic Amyloid Monolayers from Fungal Hydrophobin Proteins. Methods Mol Biol 2073: 55-72.
- Berger, B. W. and Sallada, N. D. (2019). Hydrophobins: multifunctional biosurfactants for interface engineering. J Biol Eng 13: 10.
- Berkmen, M. (2012). Production of disulfide-bonded proteins in Escherichia coli. Protein Expr Purif 82(1): 240-251.
- Dondapati, S. K., Stech, M., Zemella, A. and Kubick, S. (2020). Cell-Free Protein Synthesis: A Promising Option for Future Drug Development. BioDrugs 34(3): 327-348.
- Gao, W., Cho, E., Liu, Y. and Lu, Y. (2019). Advances and Challenges in Cell-Free Incorporation of Unnatural Amino Acids Into Proteins. Front Pharmacol 10: 611.
- Kadokura, H. and Beckwith, J. (2010). Mechanisms of oxidative protein folding in the bacterial cell envelope. Antioxid Redox Signal 13(8): 1231-1246.
- Khambhati, K., Bhattacharjee, G., Gohil, N., Braddick, D., Kulkarni, V. and Singh, V. (2019). Exploring the Potential of Cell-Free Protein Synthesis for Extending the Abilities of Biological Systems. Front Bioeng Biotechnol 7: 248.
- Kwan, A. H., Winefield, R. D., Sunde, M., Matthews, J. M., Haverkamp, R. G., Templeton, M. D. and Mackay, J. P. (2006). Structural basis for rodlet assembly in fungal hydrophobins. Proc Natl Acad Sci U S A 103(10): 3621-3626.
- Matsuda, T., Watanabe, S. and Kigawa, T. (2013). Cell-free synthesis system suitable for disulfide-containing proteins. Biochem Biophys Res Commun 431(2): 296-301.
- Neylon, C., Brown, S. E., Kralicek, A. V., Miles, C. S., Love, C. A. and Dixon, N. E. (2000). Interaction of the Escherichia coli replication terminator protein (Tus) with DNA: a model derived from DNA-binding studies of mutant proteins by surface plasmon resonance.Biochemistry 39(39): 11989-11999.
- Ozawa, K. and Loh, C. T. (2014). Site-specific incorporation of unnatural amino acids into proteins by cell-free protein synthesis.Methods Mol Biol 1118: 189-203.
- Pham, C. L., Rey, A., Lo, V., Soules, M., Ren, Q., Meisl, G., Knowles, T. P., Kwan, A. H. and Sunde, M. (2016). Self-assembly of MPG1, a hydrophobin protein from the rice blast fungus that forms functional amyloid coatings, occurs by a surface-driven mechanism. Sci Rep 6: 25288.
- Pille, A., Kwan, A. H., Cheung, I., Hampsey, M., Aimanianda, V., Delepierre, M., Latge, J. P., Sunde, M. and Guijarro, J. I. (2015). 1H, 13C and 15N resonance assignments of the RodA hydrophobin from the opportunistic pathogen Aspergillus fumigatus. Biomol NMR Assign 9(1): 113-118.
- Qin, M., Wang, W. and Thirumalai, D. (2015). Protein folding guides disulfide bond formation. Proc Natl Acad Sci U S A 112(36): 11241-11246.
- Siddiquee, R., Choi, S. S., Lam, S. S., Wang, P., Qi, R., Otting, G., Sunde, M. and Kwan, A. H. (2020). Cell-free expression of natively folded hydrophobins. Protein Expr Purif 170: 105591.
- Sunde, M., Kwan, A. H., Templeton, M. D., Beever, R. E. and Mackay, J. P. (2008). Structural analysis of hydrophobins. Micron 39(7): 773-784.
- Wosten, H. A. and Scholtmeijer, K. (2015). Applications of hydrophobins: current state and perspectives. Appl Microbiol Biotechnol 99(4): 1587-1597.
- Wu, P. S., Ozawa, K., Lim, S. P., Vasudevan, S. G., Dixon, N. E. and Otting, G. (2007). Cell-free transcription/translation from PCR-amplified DNA for high-throughput NMR studies. Angew Chem Int Ed Engl 46(18): 3356-3358.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Siddiquee, R. and Kwan, A. H. (2021). Cell-free Synthesis of Correctly Folded Proteins with Multiple Disulphide Bonds: Production of Fungal Hydrophobins. Bio-protocol 11(10): e4019. DOI: 10.21769/BioProtoc.4019.
Category
Biochemistry > Protein > Expression
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










