- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Inkjet 3D Printing of Polymers Resistant to Fungal Attachment
(*contributed equally to this work) Published: Vol 11, Iss 9, May 5, 2021 DOI: 10.21769/BioProtoc.4016 Views: 4664
Reviewed by: Jörg ToepelMarie Marie MueheAnonymous reviewer(s)
Abstract
Inkjet 3D printing is an additive manufacturing method that allows the user to produce a small batch of customized devices for comparative study versus commercial products. Here, we describe the use of a commercial 2D ink development system (Dimatix material printing) to manufacture small batches of 3D medical or other devices using a recently characterized fungal anti-attachment material. Such printed devices may resist problems that beset commercial medical products due to colonization by the fungal pathogen Candida albicans. By sequentially introducing the cross-section bitmaps of the product’s CAD model and elevating the print head height using the auto-clicking script, we were able to create complex self-support geometries with the 2D ink development system. The use of this protocol allows researchers to produce a small batch of specimens for characterization from only a few grams of raw material. Additionally, we describe the testing of manufactured specimens for fungal anti-attachment. In comparison with most commercial AM systems, which require at least a few hundred grams of ink for printing trials, our protocol is well suited for smaller-scale production in material studies.
Keywords: Additive manufacturingBackground
Additive manufacturing (AM) or 3D printing is a convenient tool for preparing small batches of products such as biomedical or electronic devices; however, commercial AM systems normally require at least a few hundred grams of material for the smallest batch of product, which remains a challenge for research focused on materials exploration. The Dimatix material printer is a commercial ink formulation development instrument designed for 2D printing, which runs with disposable cartridges that only require 3-5 grams of ink for printing. Its printing unit also allows height adjustment up to 25 mm. With customized scripts, we were able to use this 2D printer to simulate an inkjet-based 3D printing process and prepare experimental specimens from only a few grams of raw material. This protocol has been used in our fungal anti-attachment material screening study (Vallieres et al., 2020), in which a batch of six specimens was produced using only 4 grams of ink made from the candidate monomer discovered in the materials screening. Biofilm formation by fungi such as Candida albicans is regarded as an important source of nosocomial systemic infections, with mortality rates up to 50% (Cavalheiro and Teixeira, 2018). The resistance of the printed materials to C. albicans colonization can be tested by measuring fungal biofilm formation using crystal violet, which binds to negatively charged surface molecules and polysaccharides in the extracellular matrix. The candidate anti-attachment methacrylate polymer, (R)-α-acryloyloxy-β,β-dimethyl-γ-butyrolactone) (AODMBA), was successfully printed into fungal-resistant voice prosthesis components, demonstrating the capability to use inkjet 3D printing to manufacture bespoke medical devices resistant to fungal attachment (Vallieres et al., 2020). In this paper, we detail the protocol for creating small batches of test specimens and subsequent testing against C. albicans. The preparation of inkjet-printable ink formulations and other key parameters for successful printing are described, as well as the scripts enabling the production of self-support 3D geometries. With the help of this protocol, material screening and specimen preparation can be simplified and accelerated for the discovery of printable materials for medical applications (e.g., in-dwelling devices resistant to fungal biofilms, as described here) or non-medical applications (e.g., identification of photoreactive materials for printing electronics).
Materials and Reagents
Starlab tips (Starlab, catalog numbers: S1111-1706 and S1111-6701)
90 mm Petri dishes (Fisher Scientific, catalog number: 11308283)
10 ml and 25 ml pipettes (Greiner Bio-One, catalog numbers: 607180 and 760180)
50 ml centrifuge tubes (Scientific Laboratory Supplies, catalog number: SLS8110)
2 ml reaction tubes (Greiner Bio-One, catalog number: 623201)
12-well and 96-well plates (Greiner Bio-One, catalog numbers: 665102 and 655185)
Nitrogen gas (BOC UN1066 compressed nitrogen)
5 ml syringe (BD Emerald REF 307731)
PEN film (GTS RPEN-075-0320)
Vials, 22 ml disposable scintillation vials
Ductile tape, Scotch®
Syringe filters, Millex-HA, MF-Millipore Membrane 50pk 0.45 µm
0.2 μm SartoriusTM MinistartTM syringe filter
Syringe, Braun inkjet 10 ml syringe
BD Discardit II 20 ml syringe
2,2-Dimethoxy-2-phenylacetophenone (99%, Sigma-Aldrich), stored at 4°C
(R)-α-acryloyloxy-β,β-dimethyl-γ-butyrolactone (AODMBA) (95%, Sigma, catalog number: 376361), stored at 4°C
RPMI 1640 with 20 mM HEPES and L-glutamine, without sodium bicarbonate (Sigma, catalog number: R7388)
NB: RPMI 1640 can be stored at 4°C for several months.
PBS (phosphate-buffered saline) (Fisher Scientific, catalog number: 10209252)
NB: PBS is autoclaved and stored at room temperature for several months.
Crystal violet (Sigma, catalog number: C3886)
NB: Crystal violet is made fresh each day and filter-sterilized using a BD Discardit II 20 ml syringe and a 0.2 μm SartoriusTM MinistartTM syringe filter.
Isopropanol (Fisher Scientific, Laboratory reagent grade ≥99.5%)
YPD medium (see Recipes)
Bacteriological peptone (Oxoid, catalog number: LP0037)
Yeast extract (Oxoid, catalog number: LP0021)
D-glucose anhydrous (Fisher Scientific, catalog number: G/0500/61)
Equipment
50 ml Erlenmeyer flasks (Fisher Scientific, catalog number: 15499093)
PTFE stirrer 5 mm
FujiFilm Dimatix DMP-2830 material printer
FujiFilm 10pL DMP cartridges DMC-1610/PN2100201146
Inert customized purge box PG-7-0444
UV unit: EPL UV 119-070 high-intensity unit
Stirring stage IKA RCT Basic
Fridge LEC Medical PE207C
Vacuum oven: Fistreem 31L capacity
GilsonTM PIPETMANTM Neo Pipets
Biological safety cabinet with UV unit
Heated incubator IN30 Memmert
Heated orbital shakers: New BrunswickTM Innova® 44 (for flasks) ELMI SkyLine DTS-4 Digital Thermo Shaker (for microplates)
Eppendorf centrifuge 5810
Scientific Industries SITM Vortex-GenieTM 2
Hemocytometer Weber Scientific International Ltd.
Prior Scientific Microscope PX043
Sterile tweezers
BioTek EL800 Microplate Spectrophotometer
Balance (Denver Instrument SI-234)
Software
Fujifilm DMP version 2.0.0.1
GIMP 2.8.14
AutoHotKey 1.1.32.00
Procedure
Development of an ink for inkjet-based 3D printing processes should consider not only viscosity and surface tension as per typical 2D print ink formulations (Derby, 2010) but also the reactivity or solidification speed, which is important and correlates with factors such as photoinitiator concentration and printing environment. We have described these factors in detail in our previous studies (He et al., 2016; Zhang et al., 2019). The protocol below revealed the successful candidate ink formulation developed from a (R)-α-acryloyloxy-β,β-dimethyl-γ-butyrolactone monomer printed in our recent work (Vallieres et al., 2020).
Ink preparation
Place a 5-mm PTFE stirrer into a 22-ml vial.
Place the vial on a balance and tare the weight.
Add 0.1 g 2,2-dimethoxy-2-phenylacetophenone (solid) into the vial.
Wrap the vial in aluminium film before adding liquid monomers.
Place the vial back on the balance and tare the weight.
Add 10 g (R)-α-acryloyloxy-β,β-dimethyl-γ-butyrolactone (liquid) to the vial and close the cap.
Place the vial on the stirring stage (IKA RCT Basic) and mix the contents at 800 rpm for 15 min at room temperature.
Place the vial in a shaded box to help exclude ambient light during nitrogen purging. Open the cap and purge nitrogen gas (flow rate ~10 ml/min) into the mixture for 15 min through a needle, to allow the ink to become saturated with nitrogen. This step helps to minimize the oxygen inhibition effect during printing.
Collect the ink with a 10-ml syringe in a dark room and purge through a syringe filter into a fresh vial. The filter will help to remove any solid contamination and avoid print head clogging during printing.
Cap the vial and wrap in aluminum foil to exclude ambient light that could trigger polymerization and cause ink solidification.
Store the ink for at least 12 h at 4°C before use.
Filling the ink cartridge
Carry out all the following steps in a dark room.
Carefully collect 3 ml ink using a 5-ml syringe, taking care to avoid the generation of bubbles at this stage.
Fix the flat-head needle that comes with the cartridge to the end of the syringe and carefully transfer the sampled 3 ml ink into the cartridge bag.
Wrap the cartridge in ductile tape to avoid curing triggered by ambient light during printing and assemble the print head.
Printing and post-processing
Clamp the filled print head onto the DMP 2830 printer (Figure 1).
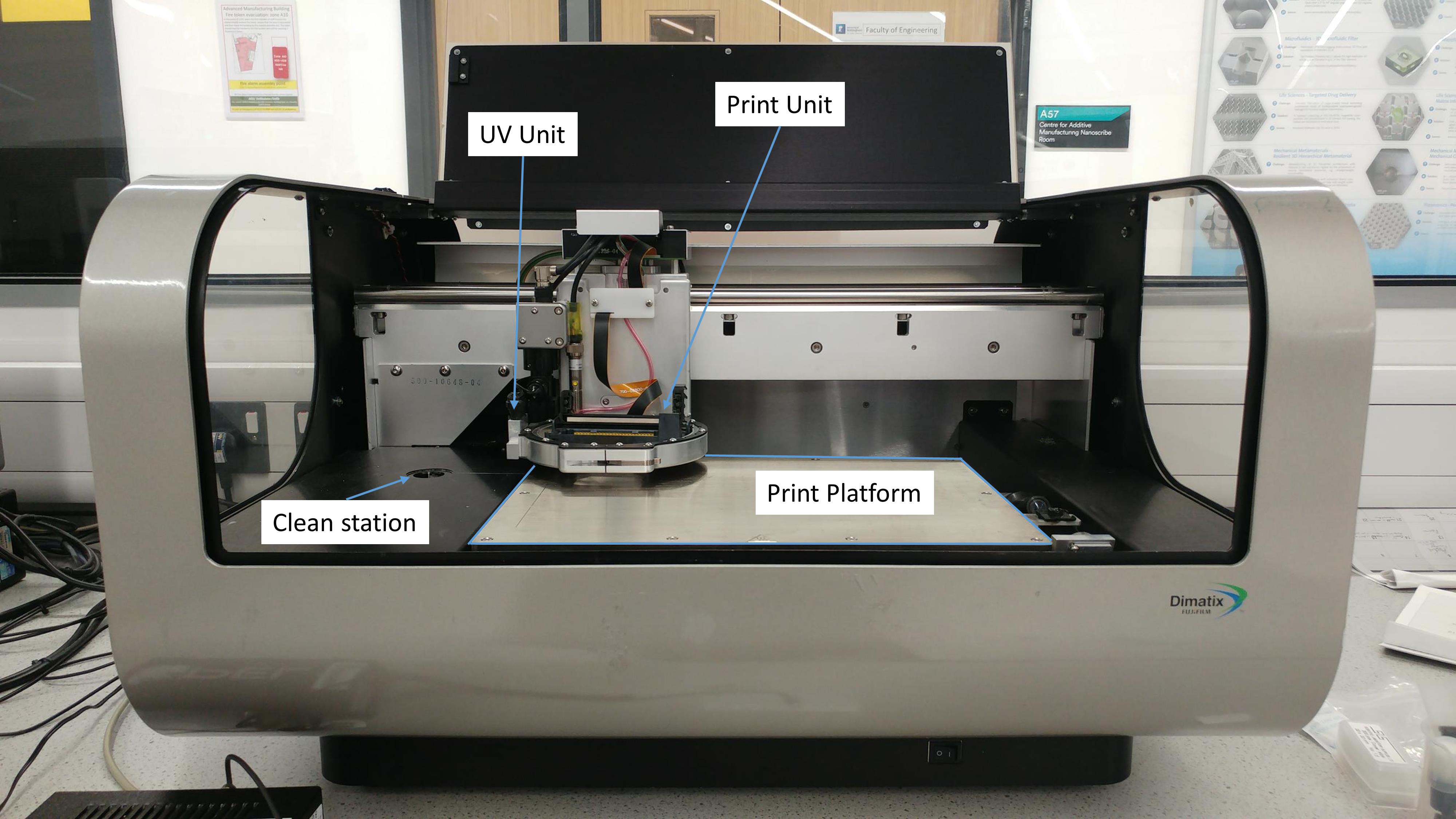
Figure 1. Interior set up of the Dimatix DMP 2830 printerAffix the ceramic cleaning pad and purge the cartridge for 2 s to remove residual air from the print head.
Cut a PEN film to A4 size and clean both sides with isopropanol.
Place the film on the printing stage and turn on the vacuum to ensure that the film is firmly held on the stage.
Install the front panel of the glovebox and purge nitrogen until the oxygen level is reduced to 0.2-0.3% (v/v).
Disable the tickle control function, monitor the drop viewer to check for droplet formation, and set the printing waveform as shown below (Table 1).
Table 1. Key parameters of the jetting and non-jetting waveforms used in (R)-α-acryloyloxy-β,β-dimethyl-γ-butyrolactone inkJetting Waveform Level (%) Slew Rate Duration (µs) Maximum Jetting Frequency (kHz) Waveform Width (µs) Section 1 0 0.65 3.584 5 11.52 Section 2 100 0.93 3.712 Section 3 13 0.6 3.392 Section 4 27 0.8 0.832 Non-Jetting Waveform Level (%) Slew Rate Duration (µs) Maximum Jetting Frequency (kHz) Waveform Width (µs) Section 1 33 1 3.712 5 11.52 Section 2 33 1 6.976 Section 3 27 1 0.832 Set the printing temperature to 48°C and the meniscus to 4.0 inches H2O. Adjust the printing voltage of each nozzle such that droplet formation is similar to that shown in Figure 2. The droplet speed needs to be between 7 m/s and 10 m/s. Twelve jets (jets 5-16) were used in our work.
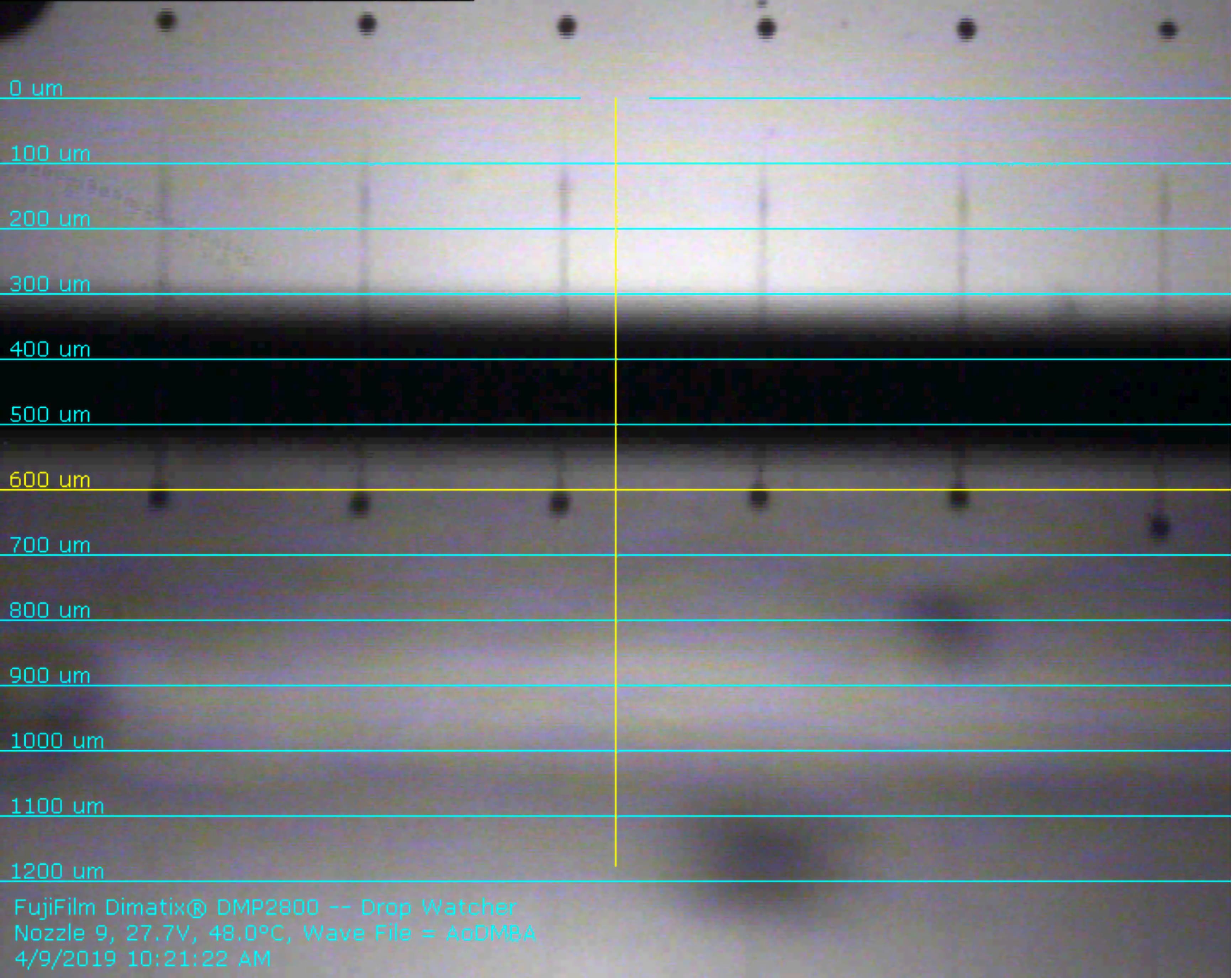
Figure 2. Exemplar droplet formation for (R)-α-acryloyloxy-β,β-dimethyl-γ-butyrolactone ink captured by the Dimatix instrument drop watcher.A full video is attached in the supplementary document.
Set the droplet spacing to 35 µm and the print head angle to 5.8 degrees.
Set the cleaning program to a 0.3 s purge and blot at the beginning of each layer, raising the print head by 12 µm after each layer is finished.
The printing pattern for each layer is a mono bitmap, which is named as number+’.bmp’ starting from 1.bmp; select 1.bmp in the printing program.
Copy the script in the supplement (Supplementary Script) into a notepad and save as an AHK file.
Run the saved script and sequentially input the ‘original height’, ‘number of layers’, and ‘height increment per layer.’ For (R)-α-acryloyloxy-β,β-dimethyl-γ-butyrolactone ink, we used 200 µm, 100 layers, and 11 µm height increment per layer.
Deposit the liquid monomer ink onto the target location using a drop-on-demand inkjet print head. The printed ink is scanned and cured via the attached UV unit as it moves with the print head (He et al., 2017).
Collect the printed samples and place in a vacuum oven (room temperature, -300 mmHg) for 24 h (Figure 3).

Figure 3. Voice prosthesis valve-flap samples collected after vacuum dryingWash the samples by sequential immersion in isopropanol and distilled water, and dry in air.
Transfer the samples to a 12-well plate and irradiate with UV-C for 20 min to sterilize the samples.
Growing the fungal cultures, preparation of inocula, and biofilm formation (use aseptic technique; a biological safety cabinet may be required depending on the fungal species)
Day 1 – Streak out strainsInoculate YPD agar medium with C. albicans cells from a stock kept at -80°C (for the stock, freeze C. albicans cells at -80°C in YPD broth with 20% glycerol) and incubate for 2 days in a static incubator at 37°C. We used the C. albicans strain CAF2-yCherry; however, the method can be applied to other C. albicans strains.
Inoculate YPD broth medium (typically 10 ml medium in a 50-ml sterile Erlenmeyer flask) with a colony of C. albicans from a 2-day-old culture on YPD agar. We recommend assaying a minimum of three independent colonies. Incubate overnight in an orbital shaker (150 rpm) at 37°C.
Harvest cells from the overnight liquid cultures by centrifugation (3,000 × g for 3 min).
Remove the supernatant and wash once in 20 ml PBS and once in 20 ml RPMI 1640 using a 25-ml pipette.
Resuspend the final cell pellets in 10 ml RPMI 1640 using a 10-ml pipette.
From the resulting cell suspensions, prepare 1:100 and/or 1:1,000 dilutions in RPMI 1640 in 2-ml reaction tubes, vortex, and count using a hemocytometer under a brightfield microscope with 40× objective.
Calculate the inoculum required to prepare 40 ml cell suspension at a final concentration of 1 × 106 ml-1 cells in RPMI 1640.
Transfer 1 ml adjusted cell suspension to each well of the 12-well plate (from Step C16).
Cover the plate with its lid, seal with tape, and incubate statically for 2 h at 37°C to allow the cells to adhere to the surface.
Use sterile tweezers to transfer the printed samples to a fresh plate and wash three times with 2 ml PBS to remove non-adhered cells.
NB: Printed samples must be transferred carefully to fresh plates using tweezers to avoid mechanically disrupting cell attachment. The transfer of samples to fresh plates (here and in Step E2) is important since free cells can also adhere to the surface of the wells, which would add to the crystal violet signal intended to reflect biofilm formation on the printed sample, impairing the analysis.
Add 1 ml fresh RPMI 1640 and incubate for 46 h at 37°C with orbital shaking at 100 rpm.
Fungal biofilm assessment (use aseptic technique and a biological safety cabinet)
Staining with crystal violet offers an approach for assessing the biomass of the biofilm. Alternatively, biofilm metabolic activity can be assayed; we detect such activity of 24 h-biofilms by measuring the reduction of XTT (tetrazolium salt, 2,3-bis[2-methyloxy-4-nitro-5-sulfophenyl]-2H-tetrzolium-5-carboxanilide). However, crystal violet is less expensive than XTT and allows for the easy visual detection of biofilms on the surface of samples (e.g., Figure 4C in Vallieres et al., 2020).
After 48 h, C. albicans should display mature biofilms. Aspirate the medium carefully so as not to touch or disrupt the biofilm.
Gently transfer the printed samples carrying biofilm to a fresh plate.
NB: As above, samples must be carefully transferred to fresh plates using tweezers to avoid mechanically disrupting the biofilm (mechanical disruption during sample transfer could lead to the biofilm detaching from the sample surface and interfering in the final assessment. Therefore, multiple specimens (n>3) are required in each test to ensure that biofilm resistance is not a result of operational error. A control group (Vallieres et al., 2020) is also needed to ensure that the biofilm formation is as expected).
Stain the biofilms with 2 ml 0.5% (w/v) crystal violet for 1 min at room temperature.
Wash three times with 2 ml PBS to remove non-adhered biofilm and excess stain.
For quantitation, add 1 ml ethanol to dissolve the crystal violet.
Transfer 100 μl reaction to a 96-well plate. Dilutions of the reaction may be needed if the concentration of crystal violet is too high. We recommend the preparation of a standard curve to estimate the level of saturation of crystal violet absorption before the assay. To do so, measure the absorbance over a range of crystal violet (in ethanol) concentrations.
Measure the absorbance at 600 nm using the BioTek EL800 microtiter plate reader.
Data analysis
From the resulting colorimetric readings after subtracting the corresponding values for the negative control (ethanol only, i.e., no crystal violet), calculate the mean (of at least three biological replicates) and SEM, and plot the data. Alternatively, results for the 3D-printed samples can be presented as a percentage of the control [in Vallières et al., 2020, the positive control was a commercial prosthesis valve flap provided by Atos Medical (raw material Silastic Q7-4735, Dow Corning)].
Notes
Crystal violet can be incompatible with some materials e.g., polyethylene glycol diacrylate (PEG575DA). Printed PEG575DA samples are completely stained with crystal violet even in the absence of C. albicans. For those materials, we recommend the assessment of fungal biofilm formation using the XTT reduction assay mentioned above.
Recipes
YPD medium
2% bacteriological peptone
1% yeast extract
2% D-glucose anhydrous
NB: Where necessary, the medium is solidified with 2% (w/v) agar (Sigma, catalog number: A7002). YPD medium is autoclaved and can subsequently be stored at room temperature for several months.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council (grant number BB/P02369X/1) and the Engineering and Physical Sciences Research Council (grant numbers EP/N006615/1 and EP/N024818/1).
Competing interests
The authors declare no competing interests.
References
- Cavalheiro, M. and Teixeira, M. C. (2018). Candida Biofilms: Threats, Challenges, and Promising Strategies. Front Med (Lausanne) 5: 28.
- Derby, B. (2010). Inkjet printing of functional and structural materials: fluid property requirements, feature stability, and resolution. Annu Rev Mater Res 40: 395-414.
- He, Y., Wildman, R. D., Tuck, C. J., Christie, S. D., and Edmondson, S. (2016). An Investigation of the Behavior of Solvent based Polycaprolactone ink for Material Jetting. Sci Rep 6: 20852.
- He, Y., Tuck, C. J., Prina, E., Kilsby, S., Christie, S. D., Edmondson, S, Hague R.J.M., Rose F.R.A.J., Wildman, R. D. (2017). A new photocrosslinkable polycaprolactone-based ink for three‐dimensional inkjet printing. J Biomedical Mater Res B App Biomater 105(6): 1645-1657.
- Vallieres, C., Hook, A. L., He, Y., Crucitti, V. C., Figueredo, G., Davies, C. R., Burroughs L., Winkler D.A., Wildman R.D., Irvine D.J., Alexander, M. R., Avery S.V., (2020). Discovery of (meth) acrylate polymers that resist colonization by fungi associated with pathogenesis and biodeterioration. Science Adv 6: eaba6574.
- Zhang, F., Saleh, E., Vaithilingam, J., Li, Y., Tuck, C. J., Hague, R. J., Wildman R.D., He, Y. (2019). Reactive material jetting of polyimide insulators for complex circuit board design. Addit Manuf 25: 477-484.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- He, Y., Vallieres, C., Alexander, M. R., Wildman, R. D. and Avery, S. V. (2021). Inkjet 3D Printing of Polymers Resistant to Fungal Attachment. Bio-protocol 11(9): e4016. DOI: 10.21769/BioProtoc.4016.
- Vallieres, C., Hook, A. L., He, Y., Crucitti, V. C., Figueredo, G., Davies, C. R., Burroughs L., Winkler D.A., Wildman R.D., Irvine D.J., Alexander, M. R., Avery S.V., (2020). Discovery of (meth) acrylate polymers that resist colonization by fungi associated with pathogenesis and biodeterioration. Science Adv 6: eaba6574.
Category
Biological Engineering > Bioprinting
Microbiology > Microbial biofilm
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










