- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Real-time Base Excision Repair Assay to Measure the Activity of the 8-oxoguanine DNA Glycosylase 1 in Isolated Mitochondria of Human Skin Fibroblasts
(*contributed equally to this work) Published: Vol 11, Iss 6, Mar 20, 2021 DOI: 10.21769/BioProtoc.3954 Views: 4445
Reviewed by: Michael T VelingAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protocol for Quantifying γH2AX Foci in Irradiated Cells Using Immunofluorescence and Fiji Software
Lu Deng [...] Lingying Wu
Aug 20, 2025 2422 Views

Colocalizing Telomeres With PML or γH2AX Foci by IF-FISH in Mouse Brain Neurons
Anna Konopka
Nov 5, 2025 1506 Views

A Quantitative DNA Fiber Assay to Monitor Replication Fork Progression, Protection, and Restart
Debanjali Bhattacharya and Ganesh Nagaraju
Feb 5, 2026 148 Views
Abstract
7,8-dihydro-8-oxoguanine (8-oxoG) is one of the most common and mutagenic oxidative DNA damages induced by reactive oxygen species (ROS). Since ROS is mainly produced in the inner membranes of the mitochondria, these organelles and especially the mitochondrial DNA (mtDNA) contained therein are particularly affected by this damage. Insufficient elimination of 8-oxoG can lead to mutations and thus to severe mitochondrial dysfunctions. To eliminate 8-oxoG, the human body uses the enzyme 8-oxoguanine DNA glycosylase 1 (OGG1), which is the main antagonist to oxidative damage to DNA. However, previous work suggests that the activity of the human OGG1 (hOGG1) decreases with age, leading to an age-related accumulation of 8-oxoG. A better understanding of the exact mechanisms of hOGG1 could lead to the discovery of new targets and thus be of great importance for the development of preventive therapies. Because of this, we developed a real-time base excision repair assay with a specially designed double-stranded reporter oligonucleotides to measure the activity of hOGG1 in lysates of isolated mitochondria. This system presented here differs from the classical assays, in which an endpoint determination is performed via a denaturing acrylamide gel, by the possibility to measure the hOGG1 activity in real-time. In addition, to determine the activity of each enzymatic step (N-glycosylase and AP-lyase activity) of this bifunctional enzyme, a melting curve analysis can also be performed. After isolation of mitochondria from human fibroblasts using various centrifugation steps, they are lysed and then incubated with specially designed reporter oligonucleotides. The subsequent measurement of hOGG1 activity is performed in a conventional real-time PCR system.
Keywords: DNA repairBackground
The human body represents a permanent case of damage. Each of the approximately 1013 body cells suffers tens of thousands to about 100,000 damages to its DNA per day (Janion, 2001; Jackson and Bartek, 2009). 7,8-dihydro-8-oxoguanine (8-oxoG) is one of the most common oxidative DNA damages induced by reactive oxygen species (ROS). By oxidizing the base guanine to 7,8-dihydro-8-oxoguanine, 8-oxoG, in contrast to guanine, is now able to hybridize not only with cytosine but also with adenine. This can lead to a complete base exchange after the second replication and thus trigger serious mutations. The replacement of a guanine/cytosine base pair by thymine/adenine is one of the most frequent mutations in human cancer. This so-called transversion also occurs frequently in the tumor suppressor gene p53 (Hirano, 2008). For this reason, and based on the fact that between 1,000 and 2,000 8-oxoG lesions per day occur in a healthy cell, while in tumor cells the number of 8-oxoG lesions increases up to 1,000,000, 8-oxoG is considered to be one of the major causes of cancer (Ravanat et al., 2000; Nakabeppu, 2014). Furthermore, 8-oxoG is associated with aging processes and degenerative diseases (such as Alzheimer's disease and Parkinson's disease) (Sampath et al., 2012; Nakabeppu, 2014) and thus shows great pathophysiological relevance.
To counteract these processes, the human body uses the enzyme 8-oxoguanine DNA glycosylase 1 (OGG1), which is the main antagonist to oxidative damage to DNA. Before replication, it removes 8-oxoG by means of base excision repair (BER), replaces it with a guanine base and thus prevents the development of mutations (Radicella et al., 1997; Hamann et al., 2009). However, previous studies indicate that the activities of glycosylases and thus also the activity of the human OGG1 (hOGG1) decrease with increasing age (Loft and Poulsen, 1996; Gorbunova et al., 2007). This has the effect on an organism that sooner or later, as a result of the reduced hOGG1 activity and the resulting decrease in the ability to perform BER processes, an age-related accumulation of 8-oxoG occurs. A better understanding of the exact mechanisms of hOGG1 could lead to the discovery of new targets and thus be of great importance for the development of preventive therapies.
For the measurement of the hOGG1 activity we developed an assay that exploits the principle of enzymatic removal of 8-oxoG from DNA as well as the cut into its sugar-phosphate backbone by hOGG1 as a part of the BER. The principle of the enzymatic removal of 8-oxoG from DNA as well as the incision in its sugar-phosphate backbone by hOGG1 during a BER is used. Lysates from isolated mitochondria are incubated with specially designed reporter oligonucleotides for this assay. Each of the two double-stranded constructs consists of 33 nucleotides and identical sequence. In addition, both carry a 6-carboxyfluorescein (6-FAM) molecule at each end of the reporter strand and a black hole quencher (BHQ) at each end of the complementary strand (Figure 1). Thus, in each double-stranded construct, the 6-FAM molecule (fluorescent dye) is opposite of BHQ which suppresses the fluorescence of the dye. The DNA glycosylase present in the lysates from the mitochondria recognizes the damage to the reporter oligonucleotide (CN-8-oxoG) and first cuts out the oxidized base, followed by a cut in the phosphate backbone. The incised DNA strand then dissociates from the counter strand at the incubation temperature of 37 °C and a fluorescence signal can be measured. The strength of the signal correlates with the repair activity or AP lyase activity of the hOGG1 enzyme. The use of an undamaged reporter oligonucleotide (CN-CTRL) serves as a control to exclude unspecific digestion of the oligonucleotides (Figure 2A). To determine the resulting oligonucleotide fragments, a melting curve analysis is performed at a temperature of 95 °C to 20 °C after completion of the real-time activity measurement. For this purpose, the first derivation of the trend of the fluorescence signal as a function of temperature is recorded (-(d/dT)). The resulting minima represent the melting temperatures of the different fragments (Figure 2B).
Materials and Reagents
2-Mercaptoethanol (Carl Roth, catalog number: 4227.1 )
Benzamidine (Carl Roth, catalog number: CN38.1 )
BHQ (single-stranded) ([BHQ1]GGTATTATTATTATTGCGTTATTATTATTATGG[BHQ1] 100 μM) (Sigma-Aldrich)
Bovine serum albumin (BSA) (New England BioLabs®, catalog number: B9000S )
Cell culture flasks T25, T75, T175 (Sarstedt, catalog numbers: 83.3910.302 , 83.3911.302 , 833912.302 )
CHAPS (Carl Roth, catalog number: 1479.1 )
CN-8-oxoG (single-stranded) ([6FAM]CCATAATAATAATAAC[8-oxo-dG]CAATAATAATAATA
CC[6FAM] 100 µM) (Sigma-Aldrich)
CN-CTRL (single-stranded) ([6FAM]CCATAATAATAATAACGCAATAATAATAATACC[6FAM] 100 µM) (Sigma-Aldrich)
cOmpleteTM Mini Protease Inhibitor Cocktail (Roche Diagnostics, catalog number: 04693124001 )
DMSO (Carl Roth, catalog number: A994.2 )
Dulbecco’s Modified Eagle’s Medium–high glucose (Sigma-Aldrich, catalog number: D7777-10L )
EGTA (Carl Roth, catalog number: 3054.1 )
Fetal bovine serum (FBS) (Gibco®, catalog number: 26140079 )
Gentamycin (10 mg/ml) (Gibco®, catalog number: 11500506 )
Glycerol (Carl Roth, catalog number: 6967.1 )
Injection cannula (G 20 × 1½”) (B. Braun, catalog number: 4657519 )
KCl (Carl Roth, catalog number: 6781.3 )
KH2PO4 (Carl Roth, catalog number: 3904.2 )
LightCycler® 480 Multiwell Plate 96 (white) (Roche Diagnostics, catalog number: 04729692001 )
LightCycler® 480 Sealing Foil (Roche Diagnostics, catalog number: 04729757001 )
MgCl2 (Carl Roth, catalog number: KK36.1 )
Na2HPO4·12H2O (Carl Roth, catalog number: N350.1 )
NaCl (Carl Roth, catalog number: 3957.3 )
NEBufferTM2 (New England BioLabs®, catalog number: B7002S )
Pipette filter tips: 2.5 µl, 10 µl, 20 µl, 200 µl, 1,000 µl (Sarstedt, catalog numbers: 70.1130.217 , 70.1130.215 , 70.1114.215 , 70.760.216 , 70.762.216 )
QuinoMit®–carrier control (MSE Pharmazeutika)
QuinoMit® Q10-Fluid–ubiquinol (MSE Pharmazeutika)
Reaction Tubes: 0.2 ml, 1.5 ml (Sarstedt, catalog numbers: 72.737.002 , 72.706.201 )
Recombinant Human OGG1 protein (100 µg) (Abcam, catalog number: ab98249 )
Syringe 2 ml (Injekt® 2 ml) (B. Braun, catalog number: 4606027V )
Tris-HCl (Carl Roth, catalog number: 9090.3 )
Tri-sodium citrate dihydrate (≥99%) (Carl Roth, catalog number: 3580.4 )
Trypsin-EDTA (0.5%) (Gibco®, catalog number: 10779413 )
1× PBS (see Recipes)
CHAPS buffer (see Recipes)
Carrier control (10 mM) (see Recipes)
Ubiquinol formulation (10 mM) (see Recipes)
DMEM for Fibroblasts (see Recipes)
Reaction buffer (see Recipes)
2 M tri-sodium citrate dihydrate (see Recipes)
Equipment
Centrifuge Eppendorf 5427 R (Eppendorf, catalog number: 5409000210 )
Centrifuge Eppendorf 5804 R (Eppendorf, catalog number: 5805000010 )
Freezer (Liebherr Comfort (Liebherr, catalog number: not available)
Incubator at 37 °C with 5% CO2, 90% humidity (HERA Cell 240, catalog number: 2510-413-01 )
LightCycler® 480 (Roche Diagnostics, catalog number: 05015278001 )
PCR-Cooler (Eppendorf, catalog number: 3881000031 )
pH Meter FiveEasyTM F20 (Mettler Toledo, catalog number: 30266626 )
Pipettes, Eppendorf Research® Plus: 0.1 µl 10 µl, 20 µl, 200 µl, 1,000 µl (Eppendorf, catalog numbers: 3123000012 , 3123000020 , 3123000039 , 3123000055 , 3123000063 )
Sonoplus Ultraschall-Homogenisator HD 3100 (Bandelin Electronic)
Thermocycler MKR13 HLC (DITABIS, catalog number: MKR 13 )
Ultrapure water (ddH2O) preparation unit, Purelab flex 4 (Veolia Water Technologies)
Software
GraphPad Prism, Version 8.2.1 (GraphPad Software®)
LightCycler® 480 Software (instrument software), Version 1.5.0.39 (Roche Diagnostics)
Procedure
Cell culture
First cultivate fibroblasts at 37 °C in the presence of 5% CO2 and 90% humidity in an appropriate cell specific medium with or without test substances.
After an appropriate incubation period, remove the cells with Trypsin-EDTA and transfer 3.0 × 106 cells into a new 1.5 ml reaction tube.
Centrifuge the cells for 5 min at 1,200 × g and 4 °C and resuspend the resulting pellet in 1 ml 1× PBS (washing step).
Isolation and lysis of mitochondria
Take 3.0 × 106 fibroblasts in 1× PBS and centrifuge it for 5 min at 1,200 × g and 4 °C.
Discard the supernatant and resuspend the resulting pellet in 200 µl CHAPS lysis buffer (pH 7.4) supplemented with 3 μl of 2 M tri-sodium citrate dihydrate and incubate on ice for 45 min.
Homogenize the sample with a syringe and a cannula (diameter = 0.9 mm) on ice by pipetting up and down ten times.
To completely separate the mitochondria from the remaining cell structures, centrifuge the samples again for 10 min at 1,200 × g and 4 °C.
Transfer the supernatants with the mitochondria into a new 1.5 ml reaction tube (pre-cooled on ice).
Lyse the isolated mitochondria in a further lysis step on ice with pulsed ultrasound at 20 watts for 1 min (0.5 s pulse on, 1.0 s pulse off).
After a further centrifugation step for 45 min at 21,000 × g and 4 °C, the supernatants can be stored on ice until further use within a few hours. Keep supernatants at -20 °C for overnight or longer storage.
Preparation of the reporter oligonucleotides
Mix 5 µl of each of the single-stranded reporter oligonucleotides CN-8-oxoG ([6FAM]CCATAATAATAATAAC[8-oxo-dG]CAATAATAATAATACC[6FAM]) (100 µM) and CN-CTRL ([6FAM]CCATAATAATAATAACGCAATAATAATAATACC[6FAM]) (100 µM) each in a separate 0.2 ml reaction tube with 10 μl of the BHQ-1-tagged complementary strands ([BHQ1]GGTATTATTATTATTGCGTTATTATTA TTATGG[BHQ1]) (100 μM) (all Sigma Aldrich GmbH) and 35 μl NEBufferTM2. A schematic representation of the reporter oligonucleotides can be seen in Figure 1.
Incubate the oligo mixtures for 15 min at 95 °C in the thermocycler, followed by cooling down of the finished constructs in the freezer at -20 °C (annealing during cooling phase–annealing temperature = 50 °C). The subsequent storage of the finished constructs take place at -20 °C.
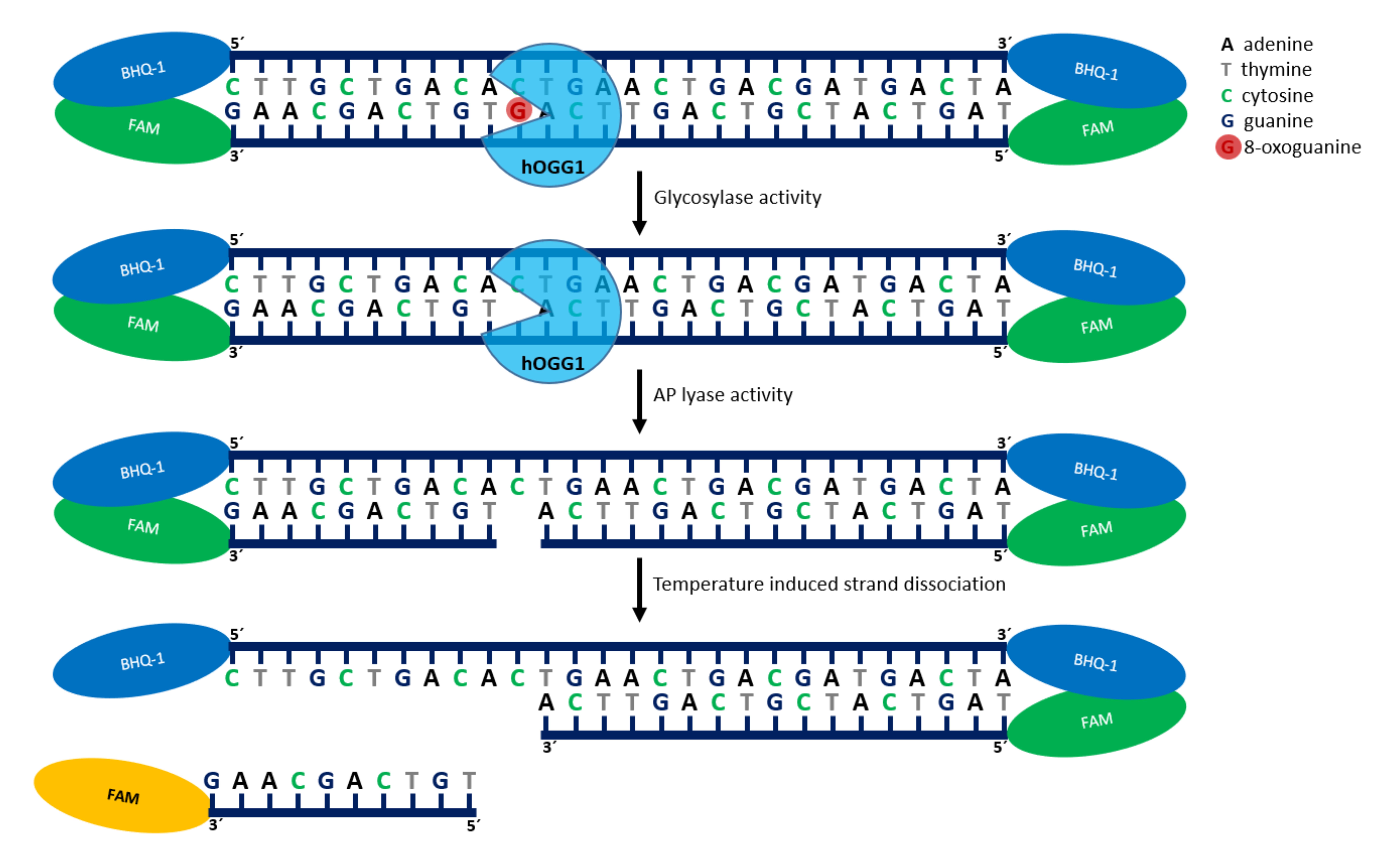
Figure 1. Basic principle of the assay. A double-stranded nucleotide strand carrying an 8-oxoG base (shown in red) is used to detect the activity of hOGG1. The strand with the damage carries at both ends a 6-FAM molecule as a fluorescence reporter and on the opposing quencher are added at both ends. In the uncut reporter oligonucleotide both strands are hybridized with each other whereby the fluorescence signal of the 6-FAM molecules is intercepted by the quenchers. If the strand with the damage is cut, the two nucleotide strands dissociate from each other and the fluorescence signal can be measured.
Base excision repair assay
Pipette the samples and controls into a white 96-well plate as shown in Table 1, homogenizing the samples by pipetting up and down (two wells are required per approach – one for CN-8-oxoG and one for CN-CTRL).
Table 1. Components of the different reaction approaches
Positive control Negative control Sample Reaction buffer 94.3 µl 94.3 µl 75.3 µl Tri-sodium citrate dihydrate [2M] 3.7 µl 3.7 µl 3.7 µl Reporter constructs [CN-8-oxoG 0.1 µM/
CN-CTRL 0.2 µM]1 µl (CN-8-oxoG)/
(CN-CTRL)1 µl (CN-8-oxoG)/
(CN-CTRL)1 µl (CN-8-oxoG)/
(CN-CTRL)Recomb. OGG1 1 µl -- -- ddH2O -- 1 µl -- Sample -- -- 20 µl Seal the plate with a sealing foil and centrifuge the plate for 5 min at 720 × g and 4 °C.
For a real-time detection of the base excision activity, perform the measurement of the fluorescence signals in a Light Cycler 480 (Roche Diagnostics) with a total of 80 cycles of 1 min each at 37 °C. Excitation of the 6-FAM molecule occurs at 465 nm and detection of the fluorescence signals in the wavelength range of 483-533 nm (Figure 2A).
To determine the resulting oligonucleotide fragments, perform a melting curve analysis at a temperature of 95 °C to 20 °C after completion of the real-time activity measurement (Figure 2B).
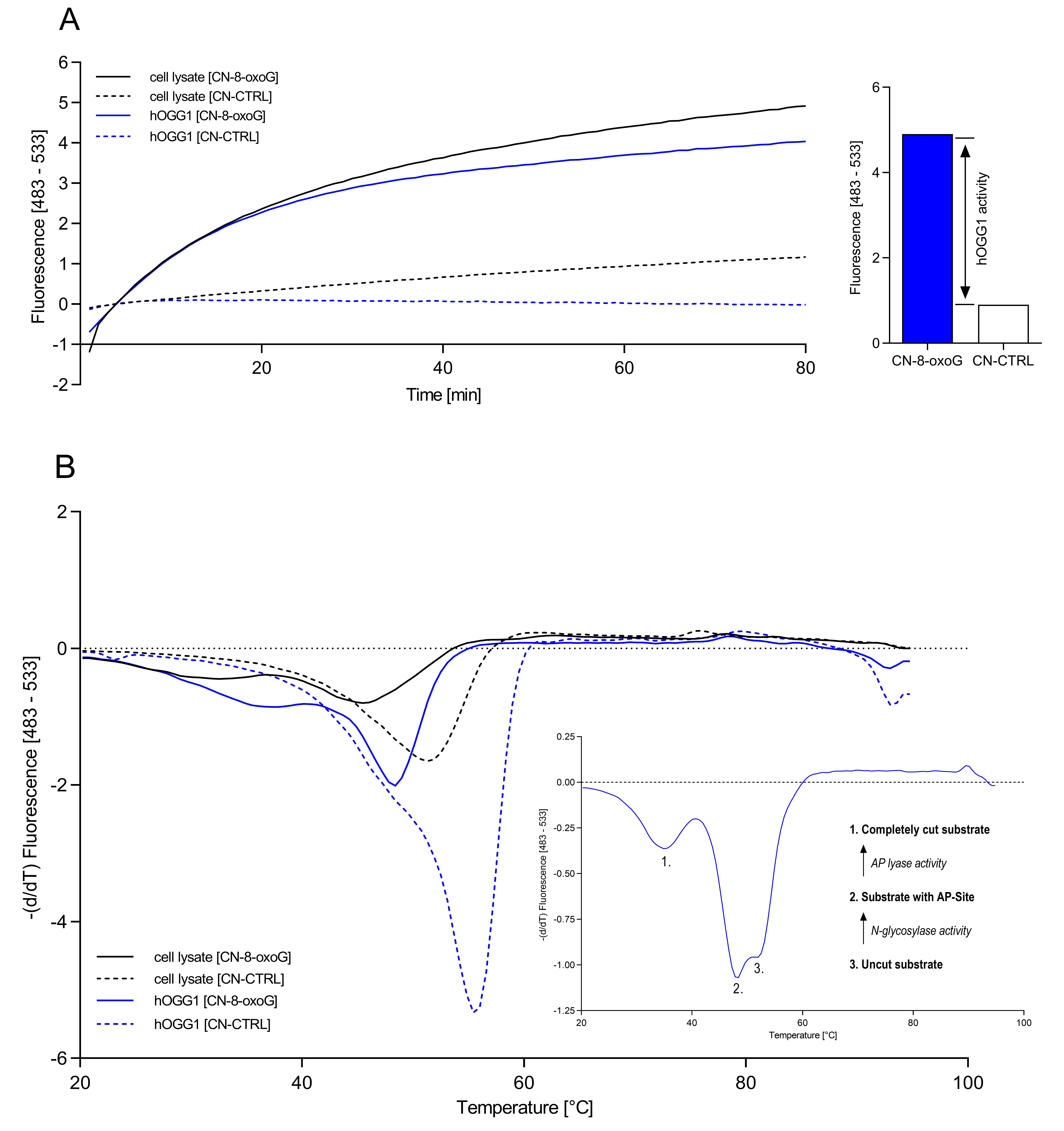
Figure 2. Schematic representation of a determination of the hOGG1 activity by fluorescence and melting curve analysis. A. Fluorescence curves of the reporter oligonucleotides CN-8-oxoG (with damage) and CN-CTRL (without damage) after digestion with 1.6 units of commercially purchased hOGG1 as well as hOGG1 obtained from cell lysates over a period of 80 cycles of 1 min each at 37 °C. B. Melting curves of the reporter oligonucleotides CN-8-oxoG and CN-CTRL after digestion with 1.6 units of commercially purchased hOGG1 and hOGG1 obtained from cell lysates.
Data analysis
The base excision repair activity of hOGG1 can be determined by means of the end point (maximum fluorescence) of the fluorescence curve of the defective reporter oligonucleotide (CN-8-oxoG) minus the nonspecific digestion (CN-CTRL) (Figure 2A).
A fragment analysis can be carried out after completion of the real-time activity measurement using the function of the recorded melting curves. For this, the first negative derivative of the trend of the fluorescence signal is recorded at a temperature change from 95 °C to 20 °C (-(d/dT)). The resulting minima represent the melting temperatures of the different fragments. The peak at position 1 represents a complete cut substrate of hOGG1 activity. In this substrate, both the modified base (8-oxoG) was excised from the DNA and the sugar-phosphate backbone of the DNA was cut. The peak labeled at position 2 represents substrate with an AP site in which the N-glycosylase function of the enzyme was already active but the AP lyase was not. At position 3 there is uncut substrate, in which neither the glycosylase nor the ligase function became active. Depending on where the larger peak is located, statements can be made about the activity of the respective enzymatic steps of hOGG1 (Figure 2B).
Data can be analyzed using GraphPad Prism, Version 8.2.1 (GraphPad Software®). Values are presented as mean ± SEM, or individual values. Three replicates per approach are recommended.
Representative data
Measurement of base excision repair activity of hOGG1 in lysates of isolated mitochondria (Figure 3) according to the proceeding steps. In this case however, to test the effect of CoQ10 on the enzymatic activity of hOGG1, an ubiquinol formulation (QuinoMit® Q10-Fluid–ubiquinol, MSE Pharmazeutika) and a carrier control (CC) (QuinoMit®–carrier control, MSE Pharmazeutika) were additionally added to the different approaches directly before the measurement. This was done because it is suspected that hOGG1, like other redox-dependent transcription factors are actively regulated by the redox-active properties of antioxidants. The carrier control was administrated exactly as described for QuinoMit® Q10-Fluid. QuinoMit® Q10-Fluid and carrier control are emulsions of phospholipid nanoparticles (30-90 nm). Both were always freshly weighed and diluted with ultrapure water. The exact composition of these test compounds can be found in Table 2 below.
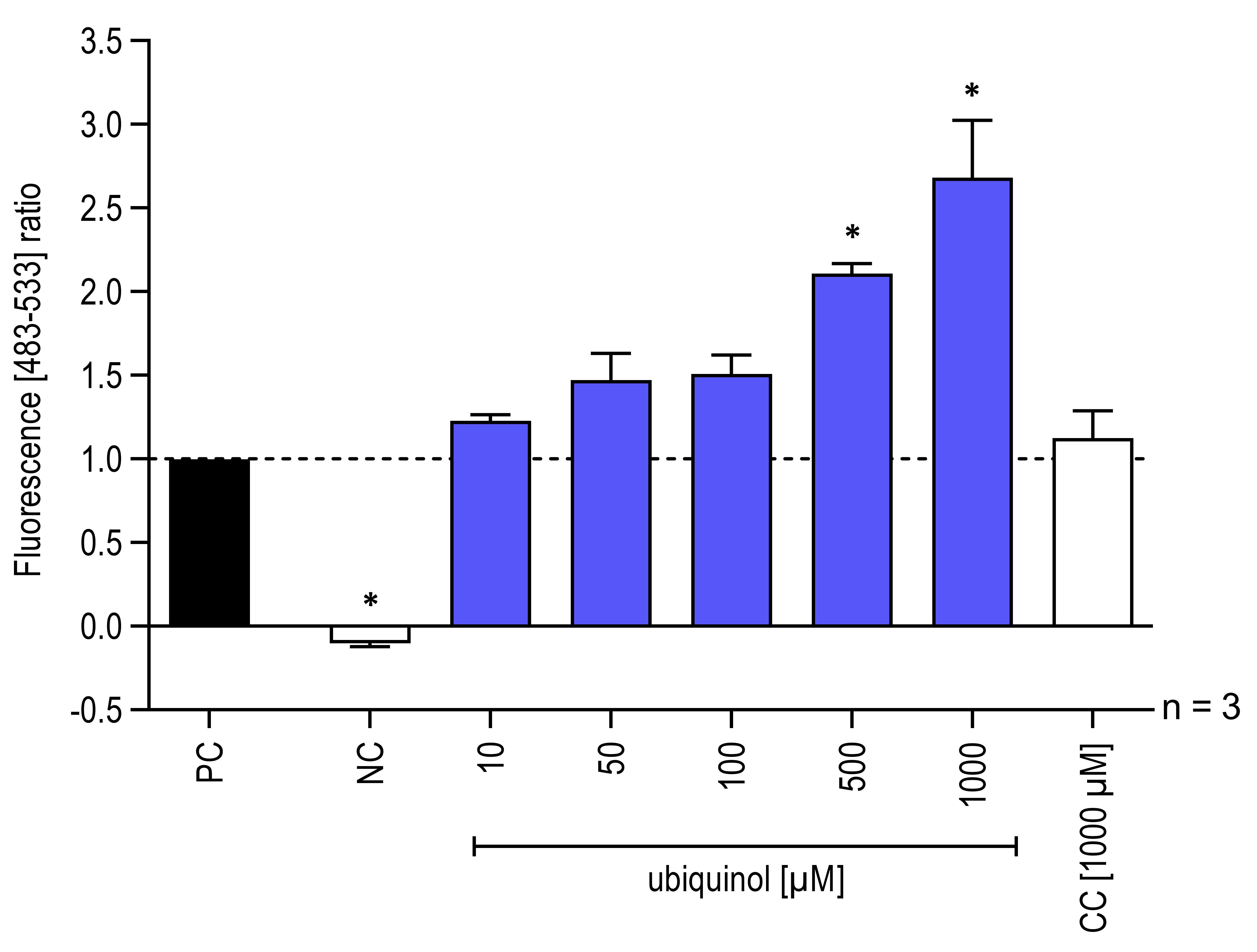
Figure 3. Influence of ubiquinol on hOGG1 enzymatic activity isolated from mitochondria. Addition of different concentrations (10-1,000 μM) of ubiquinol and carrier control (CC) to reporter oligonucleotides and hOGG1 from mitochondrial lysates, isolated from 3.0 × 106 human fibroblasts. As a negative control (NC) one approach with test solution and reporter oligonucleotides but without hOGG1 were included. The fluorescence signals were recorded with a LightCycler® 480 in a wavelength range between 483 and 533 nm. The fluorescence signals of the digested reporter oligonucleotides based on the hOGG1 activity are shown (n = 3, mean ± SEM, one-way ANOVA with Bonferroni’s multiple comparison test, *P < 0.05). Values normalized to the positive control (PC = all assay components without test solution). PC correspond to 1.0. Data obtained from Schniertshauer et al. (2020).
Table 2. Exact composition of QuinoMit® Q10-Fluid and carrier control
| CoQ10 formulations and carrier control | ||
| QuinoMit® Q10-Fluid ubiquinol (red.) | carrier control | |
| Glycerin | + | + |
| Ubiquinol | + | - |
| Deionized water | + | + |
| Soy lecithin | + | + |
| Niacinamide | + | + |
Notes
Measurement of base excision repair activity in lysates of isolated mitochondria we described here was highly reproducible.
Tri-sodium citrate is required to prevent unspecific digestion of the reporter oligonucleotides by DNases contained in the cell lysates. The optimal concentration of tri-sodium citrate dihydrate for inhibition of DNase was established for fibroblasts and could be adapted to the use of other cell lines.
The assay presented here addresses the activity of the repair enzyme hOGG1. An adaptation to other repair enzymes of BER should be possible and depends on the reporter oligonucleotides used.
Reporter oligonucleotides used here are not commercially available, but were designed by Sigma Aldrich and generated on behalf of a customer.
This method was developed on primary fibroblasts but according to initial findings it also works for lysates of PBMCs. Further cell lines must first be established.
The measurement as described here was performed in a light cycler, but can be adapted to any other RT-PCR system.
Measurement of fluorescence signals in the wavelength range between 483 and 533 is based on the fluorescent markers used (6-FAM; Ex 465 nm/Em 510 nm).
Preparation of the approaches in the 96-well plate should be carried out on cold packs (PCR-Cooler) to avoid premature enzyme activity as far as possible.
The increase in fluorescence in the real-time image is caused–as described above–by the incised fragments. These fragments have a melting temperature of about 36 °C for the reporter oligonucleotides, while the uncut oligonucleotide has a melting temperature of about 52 °C.
Human fibroblasts were isolated from skin biopsies which were received from the Kreiskrankenhaus Sigmaringen, general surgery unit, Germany; from Aesthetic Perfection Lake Constance, plastic surgery unit, Germany; or from the Chirurgische Gemeinschaftspraxis Dr. Fuhrer, H. Nonnenmacher, Dr. Astfalk und Dr. Fauser, Reutlingen, Germany. All experiments were conducted in accordance with the Declaration of Helsinki and approved by the Ethics Commission of the State Medical Association of Baden-Württemberg, Germany (187-03). Patients were informed in advance and gave their written consent to the use of their samples. Cells were isolated as described by Burger et al. (2010).
Recipes
1× PBS
8 g NaCl
0.20 g KCl
2.88 g Na2HPO4·12H2O
1.24 g KH2PO4
Adjust to 1 L ddH2O, pH 7.4
2 M tri-sodium citrate dihydrate
5.882 g tri-sodium citrate dihydrate
Adjust to 10 ml ddH2O
Carrier control (10 mM)
0.904 g QuinoMit®–carrier control
Adjust to 5 ml ddH2O, pH 7.4
CHAPS buffer
1.25 ml Glycerol (80%)
1 ml CHAPS (5%)
100 µl Tris-HCl (1 M)
100 µl EGTA (0.1 M)
10 µl MgCl2 (1 M)
5 µl Benzamidine (0.2 M)
3.52 µl 2-Mercaptoethanol (14.19 M)
Adjust to 10 ml ddH2O, pH 7.4
Add one tablet cOmpleteTM Mini Protease Inhibitor Cocktail
DMEM for Fibroblasts
500 ml DMEM
50 ml FBS
2.5ml Gentamycin (50 µg/ml)
Reaction buffer
320 µl DMSO
160 µl NEBufferTM2
16 µl BSA (20 µg/µl)
752 µl ddH2O
Ubiquinol formulation (10 mM)
0.904 g QuinoMit® Q10-Fluid
Adjust to 5 ml ddH2O, pH 7.4
Acknowledgments
The authors thank Dr. Franz Enzmann and Dr. Alexander Bürkle for scientific advice. This study was supported by the BMBFFHprofUnt2012 “MitoFunk” (03FH022PX2) and by the Baden-Württemberg Ministry of Science, Research and Art.
Competing interests
Jörg Bergemann has consulting contracts with MSE Pharmazeutika GmbH, Bad Homburg, Germany, and Beiersdorf AG, Hamburg, Germany.
References
- Burger, K., Matt, K., Kieser, N., Gebhard, D. and Bergemann, J. (2010). A modified fluorimetric host cell reactivation assay to determine the repair capacity of primary keratinocytes, melanocytes and fibroblasts. BMC Biotechnol 10: 46.
- Gorbunova, V., Seluanov, A., Mao, Z. and Hine, C. (2007). Changes in DNA repair during aging. Nucleic Acids Res 35(22): 7466-7474.
- Hamann, I., Schwerdtle, T. and Hartwig, A. (2009). Establishment of a non-radioactive cleavage assay to assess the DNA repair capacity towards oxidatively damaged DNA in subcellular and cellular systems and the impact of copper. Mutat Res 669(1-2): 122-130.
- Hirano, T. (2008). Repair system of 7, 8-dihydro-8-oxoguanine as a defense line against carcinogenesis.J Radiat Res 49(4): 329-340.
- Jackson, S. P. and Bartek, J. (2009). The DNA-damage response in human biology and disease. Nature 461(7267): 1071-1078.
- Janion, C. (2001). Some provocative thoughts on damage and repair of DNA. J Biomed Biotechnol 1(2): 50-51.
- Loft, S. and Poulsen, H. E. (1996). Cancer risk and oxidative DNA damage in man. J Mol Med (Berl) 74(6): 297-312.
- Nakabeppu, Y. (2014). Cellular levels of 8-oxoguanine in either DNA or the nucleotide pool play pivotal roles in carcinogenesis and survival of cancer cells. Int J Mol Sci 15(7): 12543-12557.
- Radicella, J. P., Dherin, C., Desmaze, C., Fox, M. S. and Boiteux, S. (1997). Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae.Proc Natl Acad Sci U S A 94(15): 8010-8015.
- Ravanat, J. L., Di Mascio, P., Martinez, G. R., Medeiros, M. H. and Cadet, J. (2000). Singlet oxygen induces oxidation of cellular DNA. J Biol Chem 275(51): 40601-40604.
- Sampath, H., Vartanian, V., Rollins, M. R., Sakumi, K., Nakabeppu, Y. and Lloyd, R. S. (2012). 8-Oxoguanine DNA glycosylase (OGG1) deficiency increases susceptibility to obesity and metabolic dysfunction.PLoS One 7(12): e51697.
- Schniertshauer, D., Gebhard, D., van Beek, H., Noth, V., Schon, J. and Bergemann, J. (2020). The activity of the DNA repair enzyme hOGG1 can be directly modulated by ubiquinol.DNA Repair (Amst) 87: 102784.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Schniertshauer, D., Gebhard, D. and Bergemann, J. (2021). Real-time Base Excision Repair Assay to Measure the Activity of the 8-oxoguanine DNA Glycosylase 1 in Isolated Mitochondria of Human Skin Fibroblasts. Bio-protocol 11(6): e3954. DOI: 10.21769/BioProtoc.3954.
Category
Molecular Biology > DNA > DNA damage and repair
Cell Biology > Cell-based analysis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









