- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ligand and Carbohydrate Engagement (LACE) Assay and Fluorescence Quantification on Murine Neural Tissue
Published: Vol 11, Iss 6, Mar 20, 2021 DOI: 10.21769/BioProtoc.3952 Views: 4674
Reviewed by: Alexandros C KokotosMarion HoggAnonymous reviewer(s)
Abstract
The interaction between cell surface heparan sulphate and diffusible ligands such as FGFs is of vital importance for downstream signaling, however, there are few techniques that can be used to investigate this binding event. The ligand and carbohydrate engagement (LACE) assay is a powerful tool which can be used to probe the molecular interaction between heparan sulphate and diffusible ligands and can detect changes in binding that may occur following genetic or pharmacological intervention. In this protocol we describe an FGF17:FGFR1 LACE assay performed on embryonic mouse brain tissue. We also describe the method we have used to quantify changes in fluorescent LACE signal in response to altered HS sulphation.
Keywords: LigandBackground
Heparan sulphate (HS) is an extracellular matrix and cell surface glycosaminoglycan molecule that is extensively modified by sulphation. HS interacts with a wide range of developmentally important signaling molecules including FGFs, Wnts, BMPs and Slits. During FGF signalling, for example, HS acts as a co-receptor, facilitating the binding of the FGF ligand to the cell surface FGFR receptor. The formation of this FGF:FGFR:HS complex is required for FGF signaling to occur (Allen et al., 2001). Differential sulphation of HS has been shown to affect the binding of FGF ligands to their FGFR cell surface receptors. In our recent paper we used a ligand and carbohydrate engagement (LACE) assay to probe the interaction between HS and FGF proteins (Clegg et al., 2019). This method involves the addition of recombinant FGF and FGFR protein to form the FGF:FGFR:HS complex on tissue sections. The recombinant FGFR can then be labelled using a fluorescently conjugated antibody allowing the visualization of the binding event (Allen et al., 2001; Allen and Rapraeger., 2003).
The LACE assay allows us to observe any change in FGF:FGFR:HS binding caused by altered HS sulphation. For example, in our recent study we were able to show reduced FGF17:FGFR1 binding in embryos lacking 2-O HS sulphation (Clegg et al., 2019). Using image analysis software, we were also able to quantify the change in fluorescent LACE signal, and in this protocol we will detail our robust quantification procedure. In our experiments we have used the LACE assay to analyze the binding of FGF17 and FGF8 to FGFR1 and FGFR3, however, this technique could be adapted to probe the binding properties and HS interaction of a wide range of different ligand and receptor pairs.
Materials and Reagents
Materials
7 ml bijou tubes (Greiner, catalog number: 189170 )
Pipette tips (Greiner, catalog number: 739288 )
1.5 ml microcentrifuge tubes (Greiner, catalog number: 616201 )
Peel-A-Way® Embedding Molds (Square-S22, Polysciences)
SterilinTM Standard 90 mm Petri Dishes (Thermo Fisher, catalog number: 101R20 )
Animals
Mice used in our original study were maintained on a CBA background. Mice used for timed matings were aged between 6 and 24 weeks.
Reagents
Sucrose (Thermo Fisher, catalog number: S25590 )
1× PBS (Thermo Fisher, catalog number: 14190094 )
Tris-HCl (Thermo Fisher, catalog number: 10812846001 )
CaCl2 (Thermo Fisher, catalog number: 10657662 )
Ethanol (Thermo Fisher, catalog number: AC615090010 )
Bovine serum albumin (Merck, catalog number: A1933 )
Paraformaldehyde (Merck, catalog number: 158127 )
OCT embedding matrix (Cellpath, catalog number: KMA-0100-00A )
Sodium borohydride (Merck, catalog number: 452882 )
Glycine (Merck, catalog number: G8898 )
Bovine serum albumin (Merck, catalog number: 05470)
Heparinase I and III blend (Millipore Sigma, catalog number: H3917 )
Recombinant Human FGFR1 beta (IIIc) Fc Chimera Protein (R&D Systems, catalog number: 661-FR-050 )
Recombinant Human/Mouse FGF-8b Protein (R&D Systems, catalog number: 423-F8 )
Recombinant Mouse FGF-17 Protein (R&D Systems, catalog number: 7400-FG )
Anti-Human IgG (Fc specific)-Cy3 antibody (Merck, catalog number: C2571 )
DAPI (Thermo Fisher, catalog number: D1306 )
Vectashield Hardset (Vector Labs, catalog number: H-1400 )
Heparinase Buffer (see Recipes)
Equipment
Pipettes
Dissecting microscope (Euromex, catalog number: DZ1100 )
Cryostat (Leica, catalog number: CM3050 S )
Incubator (37 °C)
Forceps, size 5 (Fine Science Tools, catalog number: 11251-20 )
Microscissors, 5 mm cutting edge (Fine Science Tools, catalog number: 15003-08 )
Scissors, 9 cm (Fine Science Tools, catalog number: 14060-09 )
Super-frost Plus slides (Thermo Fisher, catalog number: J1800AMNZ )
Coplin jar (Millipore Sigma, catalog number: S6016 )
Analog Rocking Platform Shaker (VWR, catalog number: 10127-872 )
Hydrophobic barrier pen (VectorLabs, catalog number: H-4000 )
Humidified chamber/slide staining tray (Heathrow Scientific, catalog number: HS15951A )
Epifluorescence or confocal microscope with image capture software (e.g., Leica AF6000 epifluorescence microscope )
Software
ImageJ/FIJI, version 1.53 (https://imagej.net/Fiji)
Procedure
Summary: This protocol is divided into two sections. In part A we describe the LACE assay beginning with the dissection of the embryonic brain, the embedding of the tissue and cryo-sectioning. We then detail the LACE assay itself including the incubation of recombinant ligand and receptor proteins with the tissue sections followed by the detection of bound receptor protein using a fluorescent conjugated antibody. In part B we describe the imaging of reacted tissue sections and the quantification of fluorescent LACE signal using FIJI software.
LACE Assay
Cull pregnant female mouse at desired embryonic stage. Using scissors, open the abdomen and remove the uterus which should be clearly visible.
Note: In our testing we have successfully performed the LACE assay using mouse brain tissue aged between E11.5 and E18.5. Other groups have successfully used embryonic heart, lung and liver tissue in addition to CHO cell lines (Allen and Rapraeger, 2003).
Place the uterus in a dish of chilled PBS. Using forceps open the uterus, carefully remove the embryos and transfer to a fresh dish of PBS.
Cut the head from the body of the embryo using scissors and transfer to fresh, chilled PBS. Use forceps to peel away the skin from the head to expose the skull, cut down the midline of the skull using microscissors and peel back the skull to expose the brain. Carefully detach the brain from the base of the head and remove using forceps (Figures 1A-1D).
Note: Take a tissue sample for genotyping if required.
Fix the brains in 4% paraformaldehyde (PFA) in PBS in 7 ml bijou tubes, overnight at 4 °C, with shaking (20 rpm).
Note: 7 ml PFA is sufficient for embryonic brains. Postnatal brains up to P3 should be fixed in 20 ml tubes, older brains should be fixed by trans-cardial perfusion.
Wash brains briefly with 1× PBS and cryoprotect tissue using 30% sucrose in PBS at 4 °C overnight, or until the tissue has sunk.
Note: PFA and PBS can be poured off without needing to change containers.
Transfer the brain to an embedding mold and cover the tissue in 50% OCT embedding matrix; 50% Sucrose/PBS. Position the tissue at the desired orientation and freeze on dry ice (Figures 1E, 1F).
Section tissue using a cryostat at a thickness of 10 µm and collect sections on Superfrost Plus slides. (Figures 1G-1I).
Note: Sections can be stored at -20 °C until needed.
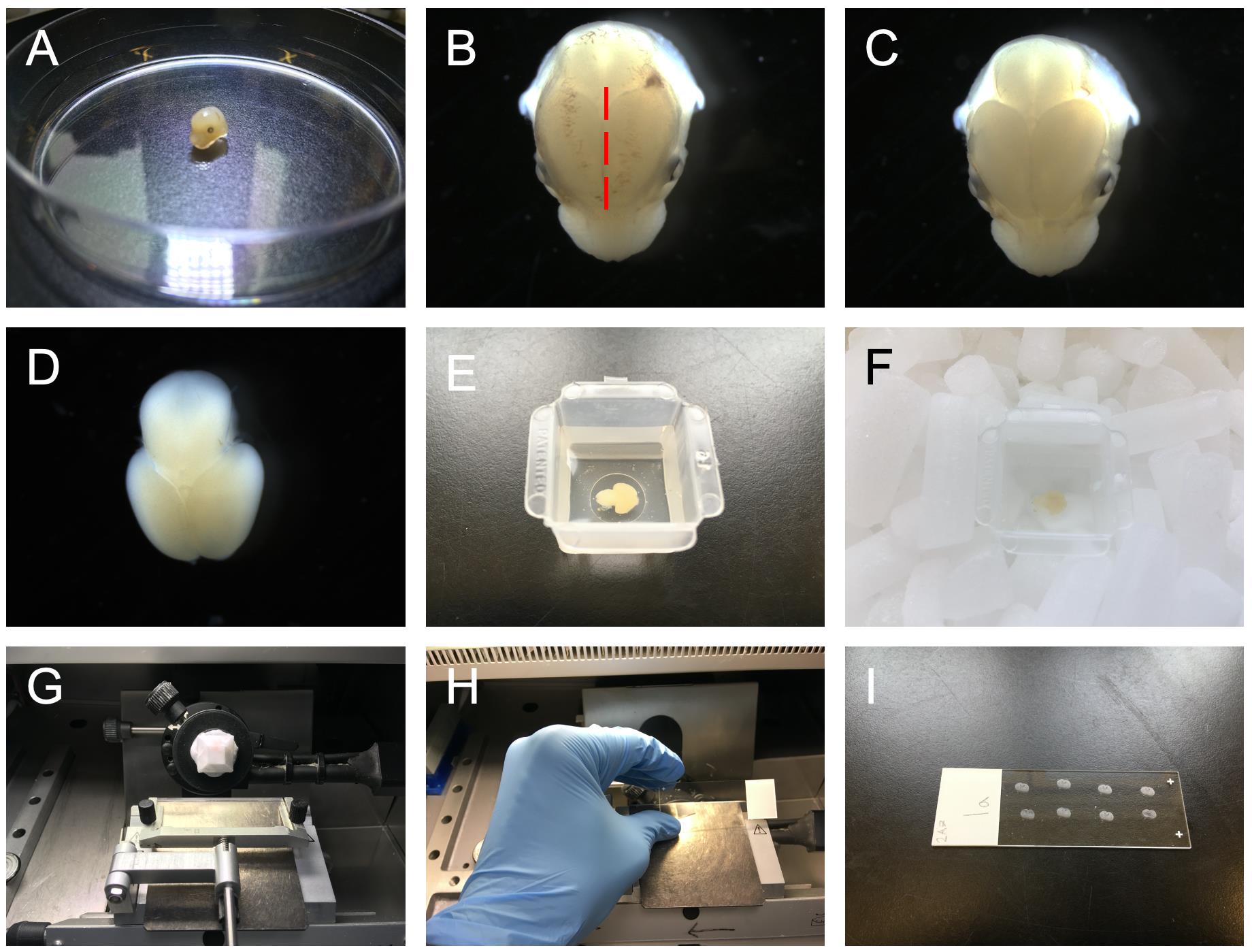
Figure 1. Images of the dissection, embedding and sectioning of embryonic mouse brain tissue. A. The head of the embryo is detached from the body. B. An incision is made along the midline of the skull (dashed red line). C. The skull is peeled away to reveal the brain. D. The brain is detached from the head. E. The brain is placed in an embedding mold and submerged in OCT/sucrose solution. F. Brain tissue is then frozen on dry ice. G-I. Frozen tissue can then be sectioned using a cryostat and sections mounted on superfrost slides.Select slides to be used for the LACE assay, remember to include slides for the heparinase digest and no-ligand controls.
Place slides in 95% ethanol at room temperature for 5 min.
Post fix the sections by placing slides in 4% PFA in PBS at room temperature for 10 min, with shaking
Wash sections in 1× PBS at room temperature with shaking, 2 × 5 min.
Note: All wash steps should be performed at room temperature, placing the slides in a colpin jar, covering with 1× PBS and shaking at 50 rpm.
Incubate sections with 0.05% Sodium borohydride solution in a coplin jar for 5 min at room temperature.
Note: Bubbles of hydrogen gas should form on the tissue sections during incubation, if bubbles do not form make up fresh Sodium borohydride solution and repeat.
Replace with fresh 0.05% Sodium borohydride and incubate sections for a further 10 min.
Wash sections in 1× PBS, 2 × 5 min.
Incubate sections in 0.1 M Glycine in 1× PBS at 4 °C overnight with shaking.
Note: Sodium borohydride/glycine treatment reduces aldehyde fixation-induced autofluorescence, reducing background fluorescence and therefore allowing easier detection of LACE signal.
Wash sections in 1× PBS, 2 × 5 min.
Dilute heparinase in heparinase buffer (0.5 mU/ml working concentration).
Select slides for heparinase digest control and ligand negative control, draw around sections using a hydrophobic barrier pen.
Note: Heparinase digest and ligand negative controls are included to ensure that observed LACE signal is the result of HS:ligand:receptor complex formation. If signal is detected after heparinase digestion (i.e., removing HS) or in the ligand negative control this indicates that the receptor is binding to the tissue directly without complex formation.
Apply heparinase solution to the selected slides, leave untreated slides in 1× PBS.
Place slides in a sealable humidified chamber and incubate at 37 °C for 2 h.
Replace with fresh heparinase solution and incubate at 37 °C for a further 2 h.
Wash treated slides in ddH2O, 1 × 5 min.
Wash all slides in 1× PBS, 1 × 5 min.
Block sections using a blocking solution of 1% bovine serum albumin in 1× TBS for 1 h.
FGFR1/FGF treatment, at 4 °C overnight.
In our experiments we have used FGFR1 and FGFR3 at a concentration of 9 µM and 100 nM respectively. FGF8 and FGF17 were used at a concentration of 30 nM and 3 µM respectively.
Recombinant FGFR and FGF is diluted in 1% bovine serum albumin in 1× TBS.
100 µl FGFR/FGF solution is applied to each slide.
Slides should be placed in a humidified chamber for overnight incubation.
Notes:
The recombinant Fc chimeric fusion proteins used in this protocol are proteins (in our case FGFR1/3) to which the Fc domain of IgG has been genetically linked. This Fc tag allows for easy detection of the protein of interest using antibodies. Fc fusion proteins are commercially available for a wide range of different receptors including FGFRs, VEGFRs, Robos, Neuropilins and DCC.
FGFR/FGF concentrations used here were obtained from the literature which originally described these binding assays (Friedl et al., 2001; Allen and Rapraeger, 2003). In our testing we have always maintained a roughly 3:1 receptor/ligand ratio for the FGFR/FGF combinations we have used. To perform this assay using other receptor/ligand pairs, users would need to titrate these concentrations however we would recommend starting with the concentrations used here and maintaining the 3:1 receptor/ligand ratio.
Wash sections in 1× TBS, 4 × 5 min.
Apply fluorescent conjugated secondary antibody (anti-human IgG, Fc-specific Cy3) diluted 1 in 200 in blocking solution.
Wash sections in 1× TBS, 4 × 5 min.
Counterstain with DAPI diluted at 1 in 1,000 in 1× PBS for 5 min.
Note: DAPI stock solution is at a concentration of 1 mg/ml.
Wash all slides in 1× PBS, 1 × 5 min
Coverslip slides using Vectashield Hardset and leave to dry at room temperature for 1 h.
Once dried, slides can be imaged using epifluorescence or confocal microscopy.
LACE Quantification
Image slides using epifluorescence or confocal microscopy.
Note: All images acquired for quantification must be imaged at constant exposure time and gain using the same objective lens. Care should also be taken to make sure no pixels within the images are over exposed.
LACE signal should be apparent in those tissue sections treated with both ligand and Fc-tagged receptor which have not undergone heparinase digestion (Figures 2A, 2E). Control sections in which the ligand was omitted should be blank (Figures 2D, 2H). Sections which have undergone heparinase digestion should see LACE signal reduced compared to undigested sections (Figures 2C, 2G), this indicates that binding of the ligand/receptor pair is HS dependent.
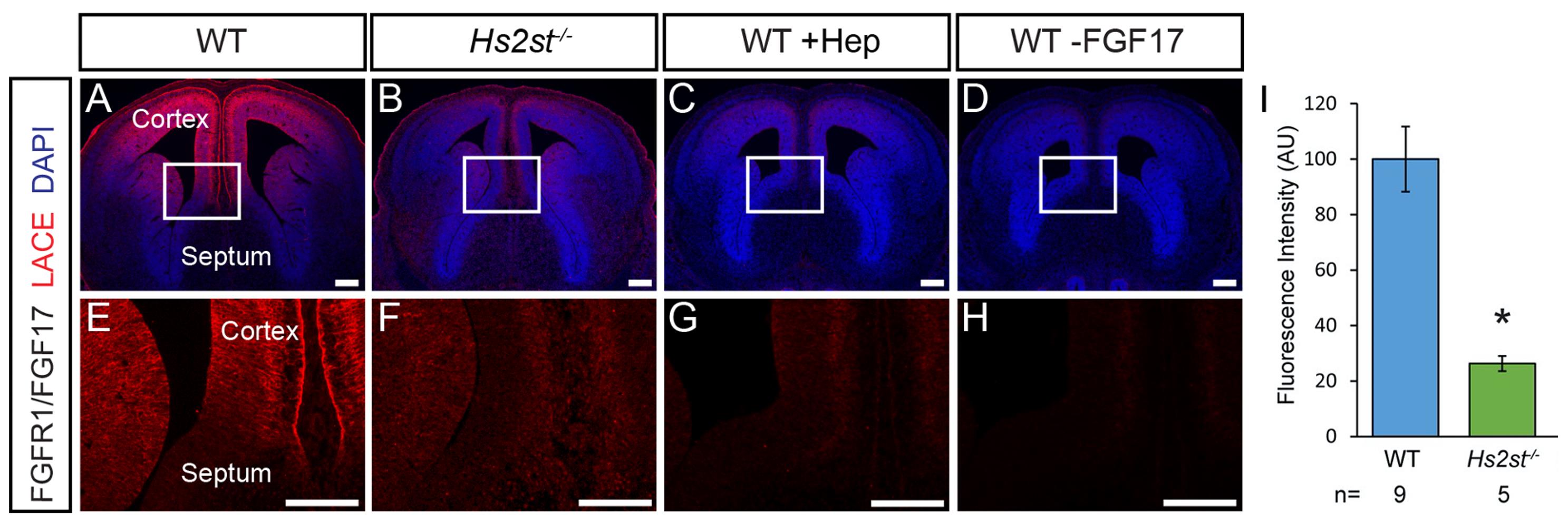
Figure 2. FGFR1/FGF17 LACE assay on E14.5 mouse brain tissue. A, E. LACE signal (red) is visible in the cortex of WT embryos. B, F. LACE signal is greatly reduced in Hs2st-/- embryos. C, G. LACE signal is greatly reduced in WT tissue after heparinase digestion. D, H. LACE signal is completely abolished when FGF17 ligand is omitted from the binding step. I. Quantification of LACE signal in the WT and Hs2st-/- cortex shown in E and F. LACE signal is significantly reduced in Hs2st-/- cortex compared to WT (t-test, P = 0.014). E, F, G, H. are higher magnification images of the boxed regions in A, B, C, D respectively. Scale bars = 200 µm.LACE images should be opened in ImageJ (or FIJI) software (Figure 3A).
Note: Images should not be modified in any way between acquisition and quantification.
Click ‘Analyze’ on the menu bar and select ‘Set Measurements’, in the resulting pop-up window ensure that ‘Mean grey value’ is checked (Figure 3B).
Note: Any other desired measurements can be selected in this window.
Open the ligand-negative control in ImageJ software and using either the rectangle, oval or polygon tool, draw a shape around the region to be quantified, the shape used should be of a known size that can be kept consistent throughout the quantification (Figure 3C).
Click ‘Analyze’ and select ‘Measure’, the resulting pop-up window will give the mean greyscale value of the quantified region under ‘Mean’ (Figure 3D). Repeat this process on at least 2 other ligand-negative control tissue sections and calculate a mean greyscale value. This can then be used as the background fluorescence value for sections within the same LACE experiment.
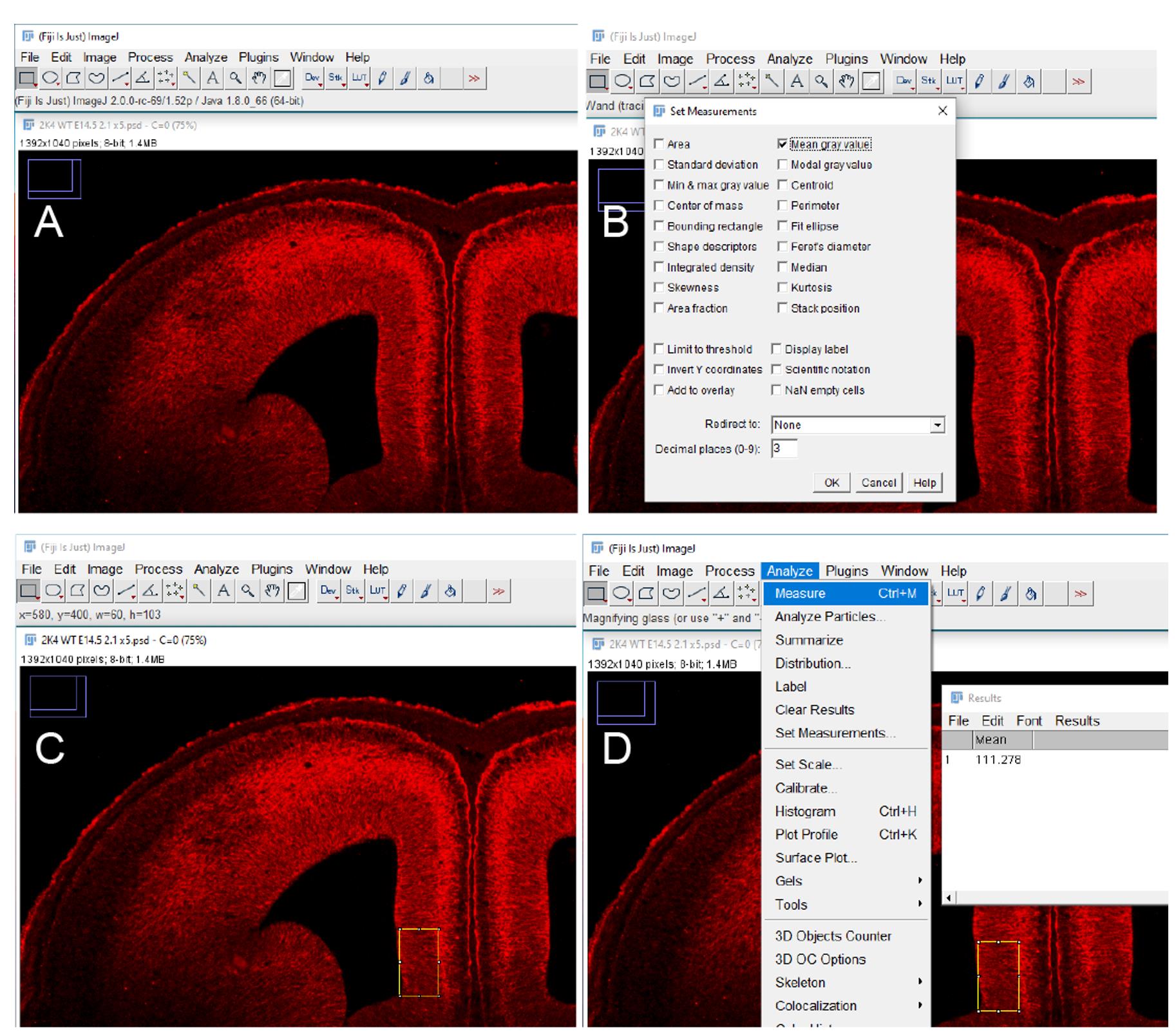
Figure 3. LACE Fluorescence Quantification using FIJI software. A. Open image file in Fiji software. B. Ensure ‘Mean grey value’ is checked in ‘Set Measurements’ window. C. Draw a shape of consistent size at the region to be quantified. D. Click ‘Measure’ and record mean greyscale value.Open experimental image in ImageJ and draw the chosen shape around the region to be quantified as in Step B5. Ensure that the shape is the same size and in the same anatomical position within the tissue section.
Click ‘Analyze’ and select ‘Measure’ as in Step B6 and record the mean greyscale value. Perform background fluorescence subtraction by subtracting the background fluorescence value from the greyscale value recorded for the section.
Repeat Step B8 using at least 2 more tissue sections and calculate a mean greyscale value for that individual animal.
Repeat Steps B8 and B9 for tissue sections from all other animals included in the experiment.
Recipes
Heparinase Buffer
20 mM Tris-HCl
4 mM CaCl2
0.1 mg/ml BSA
Acknowledgments
This work was supported by funding from the Wellcome Trust, the Biotechnology and Biological Sciences Research Council (BB/M00693X/1) and The Simons Initiative for the Developing Brain (SFARI - 529085).
Competing interests
The authors declare no competing financial interests.
Ethics
All mice were bred in-house according to Home Office UK legislation and licenses approved by the University of Edinburgh Ethical Review Committees and Home Office. Animal husbandry was in accordance with UK Animals (Scientific Procedures) Act of 1986 regulations. All work was carried out under Home Office project license number, P1351480E.
References
- Allen, B. L. and Rapraeger, A. C. (2003). Spatial and temporal expression of heparan sulfate in mouse development regulates FGF and FGF receptor assembly. J Cell Biol 163(3): 637-648.
- Allen, B. L., Filla, M. S. and Rapraeger, A. C. (2001). Role of heparan sulfate as a tissue-specific regulator of FGF-4 and FGF receptor recognition. J Cell Biol 155(5): 845-858.
- Clegg, J. M., Parkin, H. M., Mason, J. O. and Pratt, T. (2019). Heparan sulfate sulfation by Hs2st restricts astroglial precursor somal translocation in developing mouse forebrain by a Non-cell-autonomous mechanism. J Neurosci 39(8): 1386-1404.
- Friedl, A., Filla, M. and Rapraeger, A. C. (2001). Tissue-specific binding by FGF and FGF receptors to endogenous heparan sulfates. Methods Mol Biol 171: 535-546.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Clegg, J. M. and Pratt, T. (2021). Ligand and Carbohydrate Engagement (LACE) Assay and Fluorescence Quantification on Murine Neural Tissue. Bio-protocol 11(6): e3952. DOI: 10.21769/BioProtoc.3952.
- Clegg, J. M., Parkin, H. M., Mason, J. O. and Pratt, T. (2019). Heparan sulfate sulfation by Hs2st restricts astroglial precursor somal translocation in developing mouse forebrain by a Non-cell-autonomous mechanism. J Neurosci 39(8): 1386-1404.
Category
Neuroscience > Development
Developmental Biology > Cell signaling
Molecular Biology > Protein
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link