- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Chromatographic Analysis for Targeted Metabolomics of Antioxidant and Flavor-Related Metabolites in Tomato
Published: Vol 11, Iss 5, Mar 5, 2021 DOI: 10.21769/BioProtoc.3929 Views: 5580
Reviewed by: Carlos Ignacio LescanoAvinash Chandra PandeyAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Step-by-step Protocol for Crossing and Marker-Assisted Breeding of Asian and African Rice Varieties
Yugander Arra [...] Wolf B. Frommer
Sep 20, 2024 2333 Views

Vegetative Propagation of Cannabis sativa and Resin Obtained From its Female Inflorescences
Sebastián D´Ippolito [...] Silvana L. Colman
Feb 20, 2025 1736 Views

A New Approach to Detect and Semi-quantify All Molecular Species and Classes of Anionic Phospholipids Simultaneously in Plant Samples
Manon Genva [...] Laetitia Fouillen
Apr 20, 2025 1719 Views
Abstract
Targeted metabolomics is a useful approach to evaluate crop breeding studies. Antioxidant and flavor-related traits are of increasing interest and are considered quality traits in tomato breeding. The present study presents chromatographic methods to study antioxidants (carotenoids, vitamin C, vitamin E, phenolic compounds, and glutathione) and flavor-related characters (sugars and organic acids) in tomato. Two different extraction methods (for polar and apolar entities) were applied to isolate the targeted compounds. The extraction methods developed in this work were time and cost-effective since no further purification was needed. Carotenoids, vitamin C, glutathione, and phenolic acids were analyzed by HPLC-PDA using a RP C18 column at an appropriate wavelength for each compound. Vitamin E and sugars were analyzed by HPLC with RP C18 and NH2 columns and detected by FLD and RI detectors, respectively. In addition, organic acids were analyzed with GC-FID using a Rtx 5DA column after derivatization with MSTFA. As a result, sensitive analytical methods to quantify important plant metabolites were developed and are described herein. These methods are not only applicable in tomato but are also useful to characterize other species for flavor-related and antioxidant compounds. Thus, these protocols can be used to guide selection in crop breeding.
Keywords: HPLCBackground
Metabolomics is an applied biochemical approach which has gained attention for its potential to aid crop breeding studies. In tomato breeding, both antioxidant and flavor-related metabolites are of increasing interest because of consumer preferences for improved crop quality. Humans cannot synthesize antioxidant molecules themselves; therefore, these molecules must be provided by the external daily diet (Lobo et al., 2010). Flavor is a complex trait that also affects the consumer marketability of tomato (Kader, 2008). Thus, the quantification of these traits is important. Because the metabolome is complex and consists of a wide variety of compounds including lipid-soluble metabolites, aqueous polar metabolites, stable and unstable metabolites, as well as acidic and basic metabolites, many methods for both extraction and detection of plant metabolites are available. These methods encompass a range of different chromatographic techniques with different extraction methods depending on the type of metabolite. In the literature, different techniques including ultrasonication (Tan et al., 2021), supercritical CO2 extraction (Pellicanò et al., 2020), water-induced hydrocolloidal complexation (Nagarajan et al., 2020), and solid phase extraction (Figueira et al., 2017) have been used to extract antioxidant molecules or flavor-related metabolites. Most of these extraction methods include many steps, sometimes require special chemicals and equipment, or entail pre-purification procedures. As a result, many of these methods are expensive and time consuming. Similarly, different chromatographic techniques have been used to quantify metabolites. The most commonly used method for both targeted (Dumont et al., 2020, Tohge et al., 2020) and untargeted metabolic profiling (Capanoglu et al., 2008, Treutler et al., 2016) is mass spectrophotometry coupled with either liquid or gas chromatography. The nuclear magnetic resonance technique (NMR) can also be used for metabolomics (Ingallina et al., 2020, Masetti et al., 2020). These methods not only require very expensive equipment but also require expertise. An alternative is the use of spectrophotometric methods which can be applied to evaluate lycopene content (Migalatev, 2017), total antioxidant capacity (Martínez et al., 2020), total phenolic acids and flavonoid content (Alenazi et al., 2020). Although spectrophotometric methods are easy and cheap, they are not as sensitive as chromatographic methods. Moreover, quantification of individual molecules such as specific phenolic acids or sugars is not feasible.
Given the limitations of the available extraction and analysis protocols, the current work was designed to establish rapid, easy and relatively cheap extraction methods for two groups of compounds, polar and apolar metabolites. In addition, cheap, easy and sensitive chromatographic methods (HPLC and GC) were developed to detect and quantify the different types of antioxidant and flavor-related metabolites. Apolar extracts were used for analysis of carotenoids with a HPLC method modified from those described by Ishida et al. (2001) and Serino et al. (2009). Polar extracts were used for the analysis of vitamin C, vitamin E, phenolic acids, reduced and oxidized glutathione, and sugars using HPLC. The HPLC methods for these compounds were modified from those described in previous studies for vitamin C (Li and Chen, 2001a and 2001b), vitamin E (Turner and Burri, 2012; Bakre et al., 2015), phenolic acids (Gómez-Alonso et al., 2007), glutathione (Khan et al., 2011), and sugars (Petkova et al., 2013). Organic acids were also detected from polar extracts using a GC method modified from those described in previous studies (Roessner et al., 2001; Namgung et al., 2010). All protocols were developed using fully ripe fruits (at market stage) from a tomato inbred backcross line population as samples. This interspecific population was derived from S. lycopersicum by S. pimpinellifolium cross (Celik et al., 2017) and provided sufficient variation for the metabolites of interest to test the new protocols.
Materials and Reagents
Plant Material
An interspecific IBL (inbred backcross line) population (BC2F6) derived from the cross S. lycopersicum cv. Tueza (recurrent parent) × S. pimpinellifolium (LA1589) (donor parent) was used as plant material. This population was developed in previous studies and is described more fully in the literature (Celik et al., 2017; Gürbüz Çolak et al., 2020a and 2020b). Tueza is a cultivated fresh market tomato line with large (150-160 g), red, slightly flattened round fruits. LA1589 is a wild tomato accession with small (0.8-1 g), red, round fruits. The IBL population and parents were grown in the greenhouse in Antalya, Turkey. Ten plants per genotype were grown in double rows with 140 and 30 cm between wide and narrow rows, respectively. Plants were spaced at 40 cm intervals within rows. For basal fertilization, 500 kg 15:15:15 (N:P:K) fertilizer and 50 t of composted manure were applied per ha. Drip irrigation was used with fertigation (1.4 dS m-1 EC value) at each irrigation using 1-2-1 fertilizer until first fruit set, 2-1-1 fertilizer until first fruit ripening and 1-1-2 fertilizer after first fruit ripening.
Major Ingredients
C18 column (GL Sciences, catalog number: 5020-03946 )
NH2 column (GL Sciences, catalog number: 5020-05546 )
RTx 5DA column (Restek, catalog number: 10523 )
Syringe (BD, Emerald, catalog number: 307742 )
Syringe filter (Millipore, catalog number: Z227412 )
Polyamide membrane filter (45 µm, 47 mm) (Sartorius, catalog number: 25006-47-N )
Acetonitrile (VWR, catalog number: 97065 )
Ammonium dihydrogen phosphate (Merck, catalog number: 1.01126.0500 )
Butylated hydroxytoluene (BHT) (Sigma-Aldrich, catalog number: W218405 )
Chloroform (VWR, catalog number: JT9175 )
Dichloromethane (VWR, catalog number: BDH23373 )
Ethyl acetate (VWR, catalog number: JT9282 )
Hexane (VWR, catalog number: BDH24575 )
Methanol (VWR, catalog number: BDH20864 )
Methoxamine hydrochloride (Sigma-Aldrich, catalog number: M6524 )
N-Methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) (Sigma-Aldrich, catalog number: 69479 )
Ortho-phosphoric acid (Merck, catalog number: 100573 )
Potassium dihydrogen phosphate KH2PO4 (Riedel-de Haën, catalog number: 04243)
Pyridine (Sigma-Aldrich, catalog number: 270970 )
Triethylamine (Merck, catalog number: 8083520500 )
Trifluoroacetic acid (Sigma-Aldrich, catalog number: T6508 )
0.1 M KH2PO4, pH = 7.0 (see Recipes)
0.05% Trifluoroacetic acid(aq) (see Recipes)
Chemicals used as control molecules
β-carotene (Carl Roth, catalog number: 5669.1 )
Apigenin (Applichem, catalog number: A3641 )
Caffeic acid (Sigma-Aldrich, catalog number: C0625 )
Catechin (Applichem, catalog number: A4325 )
Chlorogenic acid (Sigma-Aldrich, catalog number: 00500590)
Chrysin (Fluka, catalog number: 9582 )
Cinnamic acid (Sigma-Aldrich, catalog number: W228826 )
Citric acid (Carl Roth, catalog number: X863.1 )
Coumaric acid (Carl Roth, catalog number: 9906.1 )
Cyanidine (Carl Roth, catalog number: 4545.1 )
Epicatechin (Applichem, catalog number: A3424 )
Epigallocatechin (Applichem, catalog number: A2010 )
Ferulic acid (Sigma-Aldrich, catalog number: 128708 )
Fructose (Sigma-Aldrich, catalog number: F0127 )
Fumaric acid (Sigma-Aldrich, catalog number: 47910 )
Gallic acid (Sigma-Aldrich, catalog number: 91215 )
Glucose (Sigma-Aldrich, catalog number: G8270 )
Hydroxybenzoic acid (Sigma-Aldrich, catalog number: W398608 )
Lactic acid (Sigma-Aldrich, catalog number: L1750 )
Lutein (Applichem, catalog number: A1283 )
Luteolin (Applichem, catalog number: A3424 )
Lycopene (Carl Roth, catalog number: 5670.1 )
Malic acid (Carl Roth, catalog number: 8684.1 )
Malvidin (Applichem, catalog number: A8720 )
Myricetin (Carl Roth, catalog number: 6461.1 )
Oxidized glutathione (Sigma-Aldrich, catalog number: G4376 )
Pelargonidin (Carl Roth, catalog number: 4540.1 )
Peonidin (R&D, catalog number: 0 942 )
Pterostilben (Sigma-Aldrich, catalog number: P1499 )
Quercetin (Applichem, catalog number: A3415 )
Reduced glutathione (Sigma-Aldrich, catalog number: G4251 )
Resveratrol (Sigma-Aldrich, catalog number: R5010 )
Salicyclic acid (Sigma-Aldrich, catalog number: W398500 )
Shikimic acid (Carl Roth, catalog number: 7305.2 )
Sinapic acid (Carl Roth, catalog number: 5317.1 )
Sucrose (Sigma-Aldrich, catalog number: S5016 )
Syringic acid (Carl Roth, catalog number: 5361.1 )
Tartaric acid (Carl Roth, catalog number: K302.1 )
Vanillic acid (Carl Roth, catalog number: 3685.1 )
Vitamin C (Sigma-Aldrich, catalog number: A5960 )
Vitamin E (Sigma-Aldrich, catalog number: T3251 )
Zeaxanthin (Carl Roth, catalog number: 5672.2 )
Extraction solvent 1 (see Recipes)
Extraction solvent 2: Chloroform: methanol: water (1:3:1, v:v:v) (see Recipes)
Equipment
-80 °C freezer
Analytical balance (Mettler Toledo, model: AB54-S )
Centrifuge (Beckman Coulter, model: Allegra X-15R )
Centrifuge (Thermo Scientific, model: SL 16 )
GC/FID (Shimadzu, model: GC 2010 plus ) with RTx 5DA column (0.25 mm × 0.25 mm × 30 m) (Restek; catalog number: 10523 )
HPLC (Shimadzu, model: LC20-AT ; refractive index (RI) detector model: RID 10A; photodiode array (PDA) detector model: SPD M20A; fluorescence detector model: RF 20A) with C18 column (5 µm-25 × 4.6 mm) (GL Sciences, catalog number: 5020-03946 ) and NH2 column (5 µm-25 × 4.6 mm) GL Sciences; catalog number: 5020-05546 )
Knife grinder (Retsch, model: GM200 )
Lyophilizer (Christ, model: Epsilon 1-4 LSC )
Orbital shaker (IKA, model: KS260 )
Vacuum evaporator (Labconco, model: Centrivap )
Vacuum filtration (Do-Chrom, model: FB01 )
Software
LC Solution (Shimadzu, https://www.shimadzu.com/an/lcms/opensolution/opensol4.html)
GC Solution (Shimadzu, https://www.shimadzu.com/an/gc/advflowtech/sw-dl.html)
Procedure
Extraction of Metabolites
Collect 10 ripe tomato fruits (average fresh weight was 65 g), bulk and dice. Take 100 g sample and lyophilize for two days (make sure they are completely dry). Grind the dried samples with a knife grinder and obtain a fine powder.
Weigh 1 g of dried sample and add 5 ml of extraction solvent 1 (see Recipes) to extract the apolar metabolites. Extract the metabolites by shaking on an orbital shaker at 400 rpm at 18 °C overnight.
Centrifuge the samples at 3,724 × g at 4 °C for 20 min. Save the supernatant and do a second overnight extraction with the pellet using the same solvent.
Centrifuge the samples again and combine the extracts. Aliquot the extracts (1 ml) and store at -80 °C until the analysis.
To extract the polar metabolites, use the pellet obtained in Step A4 and follow the same steps 1-4 using extraction solvent 2 (see Recipes). Aliquot the extracts (1 ml) and store at -80 °C until the analysis.
See Note 1.
HPLC Analysis of Metabolites
Carotenoids
Two different methods were used for the analysis of carotenoids: method 1 for the analysis of lycopene and β-carotene; method 2 for the analysis of lutein and zeaxanthin.
Method 1:
Prepare standard solutions of lycopene in methanol:acetone (1:1, v:v) at 1, 5, 10, 25, 50 and 100 ppm concentrations, and β-carotene in dichloromethane at 1, 5, 10, 25, 50, 100 ppm concentrations.
Add 0.05% trimethylamine to ethyl acetate and acetonitrile. Filter methanol, ethyl acetate and acetonitrile through membrane filter using vacuum filtration.
Use methanol:ethyl acetate:acetonitrile at 50:40:10 (v:v:v) ratio as the mobile phase for isocratic elution.
Use RP column and set the column temperature at 30 °C. Set the flow rate as 1.5 ml/min.
Filter the sample and the standard solutions through syringe filter. Inject 20 µl of the sample or the standard solution. Detect lycopene and β-carotene at 469 nm.
See Notes 2 and 3.
A chromatogram for 100 ppm standard molecules (Figure 1), standard curves (Figure 2) and the sample chromatogram for tomato (Figure 3) are shown.
Prepare standard solutions of lutein and zeaxanthin in dichloromethane containing 0.01% BHT at 0.5, 1, 5, 12.5, 50, 100, 200 and 300 ppm concentrations
Add 0.05% trimethylamine to acetonitrile. Filter methanol and acetonitrile through membrane filter using vacuum filtration.
Use methanol:acetonitrile at 90:10 (v:v) ratio as the mobile phase for isocratic elution.
Use a RP column and set the column temperature at 30 °C. Set the flow rate as 1 ml/min.
Filter the sample and the standard solutions through syringe filter. Inject 20 µl of the sample or the standard solution. Detect lutein and zeaxanthin at 469 nm.
See Notes 2 and 3.
Prepare standard solutions of vitamin C in ultrapure water at 10, 50, 100, 150, 200 and 250 ppm concentrations.
Filter methanol and phosphate buffer through membrane filter using vacuum filtration.
Use methanol: KH2PO4 10:90 (v:v) ratio as the mobile phase for isocratic elution.
Use RP column and set the column temperature at 40 °C. Set the flow rate as 1 ml/min.
Filter the sample and the standard solutions through syringe filter. Inject 20 µl of the sample or the standard solution. Detect vitamin C at 265 nm.
See Note 2.
Prepare standard solutions of vitamin E in acetonitrile:methanol (80:20, v:v) at 10, 50, 100, 150, 200, 250, 300 and 500 ppm concentrations.
Filter methanol and acetonitrile through membrane filter using vacuum filtration.
Use methanol:acetonitrile 25:75 (v:v) ratio as the mobile phase for isocratic elution.
Use RP column and set the column temperature at 40 °C. Set the flow rate as 1.5 ml/min.
Filter the sample and the standard solutions through a syringe filter. Inject 20 µl of the sample or the standard solution. Detect vitamin E with the fluorescence detector at 300 nm excitation and 360 nm emission.
See Note 2.
Prepare standard solutions of phenolic acids in methanol at 1, 5, 10, 20, 30, 40, and 50 ppm concentrations.
Filter NH4H2PO4, acetonitrile and H3PO4 through membrane filter using vacuum filtration.
Use NH4H2PO4 (Mobile phase A), acetonitrile (Mobile phase B), and H3PO4 (Mobile phase C) as the mobile phases for gradient elution. See Table 1 for gradient elution parameters.
Table 1. Gradient elution parameters
Time (min) Flow rate (ml/dk) Mobile Phase A % Mobile Phase B % Mobile Phase C % 0 1 100 0 0 2 1 100 0 0 5 1 92 8 0 17 1 0 14 86 22 1 0 18 82 29.5 1 0 21 70 55 1 0 33 67 70 1 0 50 50 75 1 0 50 50 78 1 20 80 0 81 1 20 80 0 86 1 100 0 0 Use RP column and set the column temperature at 35 °C. Set the flow rate as 1 ml/min.
Filter the sample and the standard solutions through a syringe filter. Inject 20 µl of the sample or the standard solution. Detect phenolic acids with PDA detector at different wavelengths. Epigallocatechin, epicatechin and chrysin are detected at 204 nm. Hydroxybenzoic acid, vanillic acid, myricetin and quercetin are detected at 254 nm. Gallic acid, catechin, syringic acid and cinnamic acid are detected at 280 nm. Chlorogenic acid, caffeic acid, coumaric acid, ferulic acid, sinapic acid, resveratrol, apigenin and pterostilben are detected at 320 nm. Luteolin is detected at 360 nm, while cyaniding, pelargonidin and peonidin are detected at 520 nm.
See Note 2.
Prepare standard solutions of phenolic acids in methanol at 10, 50, 100, 150, 200, 250 ppm concentrations.
Filter trifluoroacetic acid and methanol through membrane filter using vacuum filtration.
Use trifluoroacetic acid:methanol (97:3, v:v) as the mobile phase for isocratic elution.
Use RP column and set the column temperature at 35 °C. Set the flow rate as 0.2 ml/min.
Filter the sample and the standard solutions through a syringe filter. Inject 20 µl of the sample or the standard solution. Detect oxidized and reduced form of glutathione with PDA detector at 208 nm.
See Note 2.
Prepare standard solutions of sugars (glucose, fructose and sucrose) in ultrapure water at 100, 150, 200, 250, 300, 500, 750, 1,000, 1,500, 2,000, 2,500 ppm concentrations.
Filter trifluoroacetic acid and methanol through membrane filter using vacuum filtration.
Use acetonitrile: water (90:10, v:v) as the mobile phase for isocratic elution.
Use RP column and set the column temperature at 40 °C. Set the flow rate as 1 ml/min.
Filter the sample and the standard solutions through a syringe filter. Inject 20 µl of the sample or the standard solution. Detect sugars with RI detector using positive mode.
See Notes 2 and 4.
GC Analysis of Metabolites
- Organic acids
Weigh standard molecules and dissolve them in methoxamine hydrochloride (40 µl, 20 mg/ml in pyridine) at 0.25, 0.5, 1, 2.5, 5, 7.5, 10 mg/ml concentrations.
Take 100 µl of the sample. Evaporate the organic solvent of the sample using a vacuum evaporator at 30 °C until completely dry.
Dissolve the tomato sample in methoxamine hydrochloride (40 µl, 20 mg/ml in pyridine) in an ultrasonic bath for 5 min.
Derivatize both the samples and standard molecules at 37 °C for 90 min.
Perform the second derivatization with MSTFA. Add 60 µl MSTFA to the samples and the standard molecules, and incubate at 37 °C for 30 min.
Centrifuge the samples at 25,830 × g, 5 min.
Inject the supernatant into GC-FID.
Analyze the organic acids on an Rtx 5DA column with a thermogradient program. Set the column temperature from 100 °C (1 min hold) to 150 °C at a rate of 5 °C min-1, from 150 °C (1 min hold) to 280 °C at a rate of 5 °C min-1, 2 min hold at the final temperature.
Hold the injection port temperature at 250 °C, and detector temperature at 300 °C. Use nitrogen as the carrier gas, and adjust the split ratio 1/25.
See Notes 5 and 6.
Chromatogram for 1 mg/ml standard molecules (Figure 22), standard curves (Figure 23) and the sample chromatogram for tomato (Figure 24) are given below.
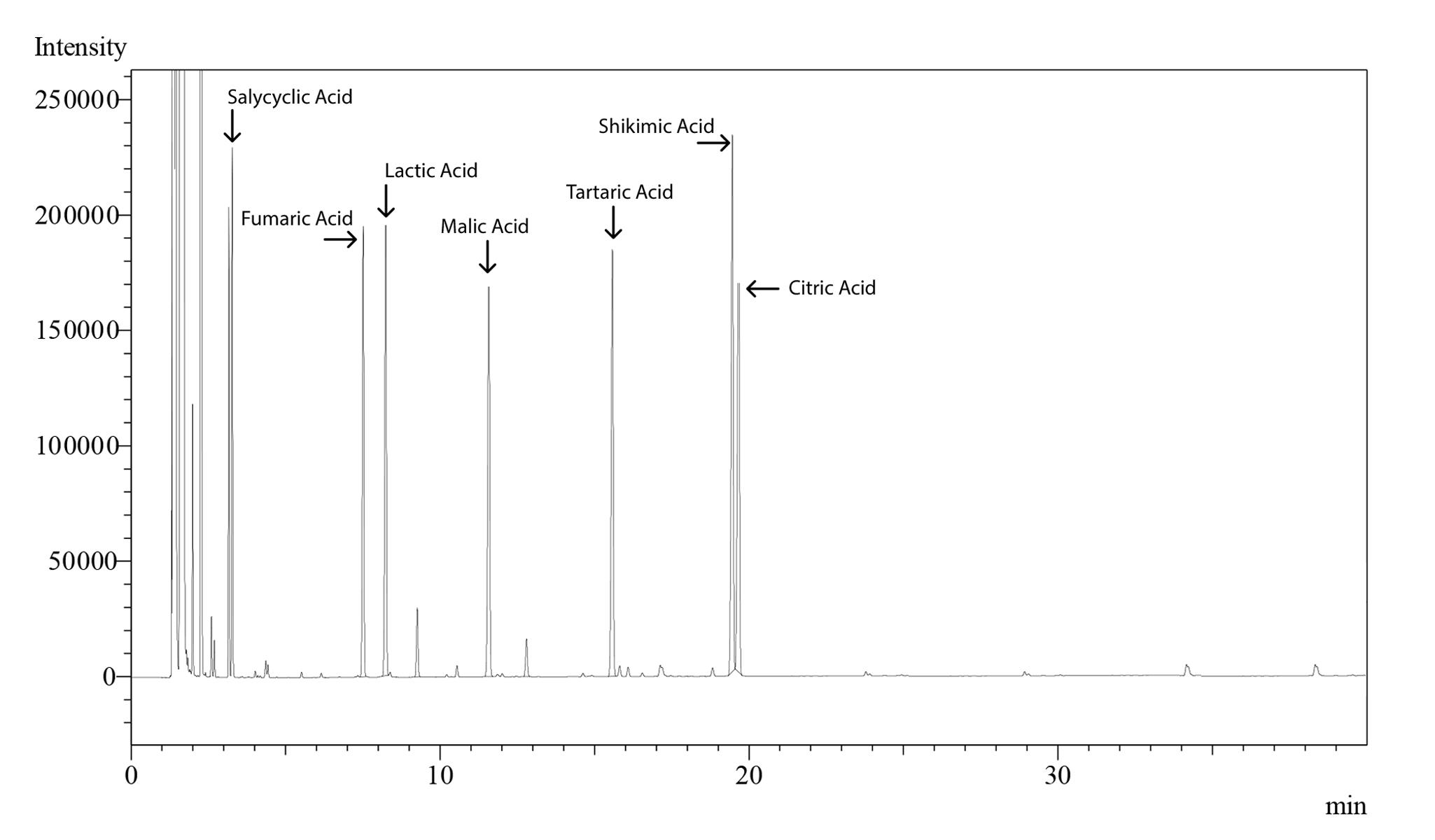
Figure 22. Chromatogram of organic acid standards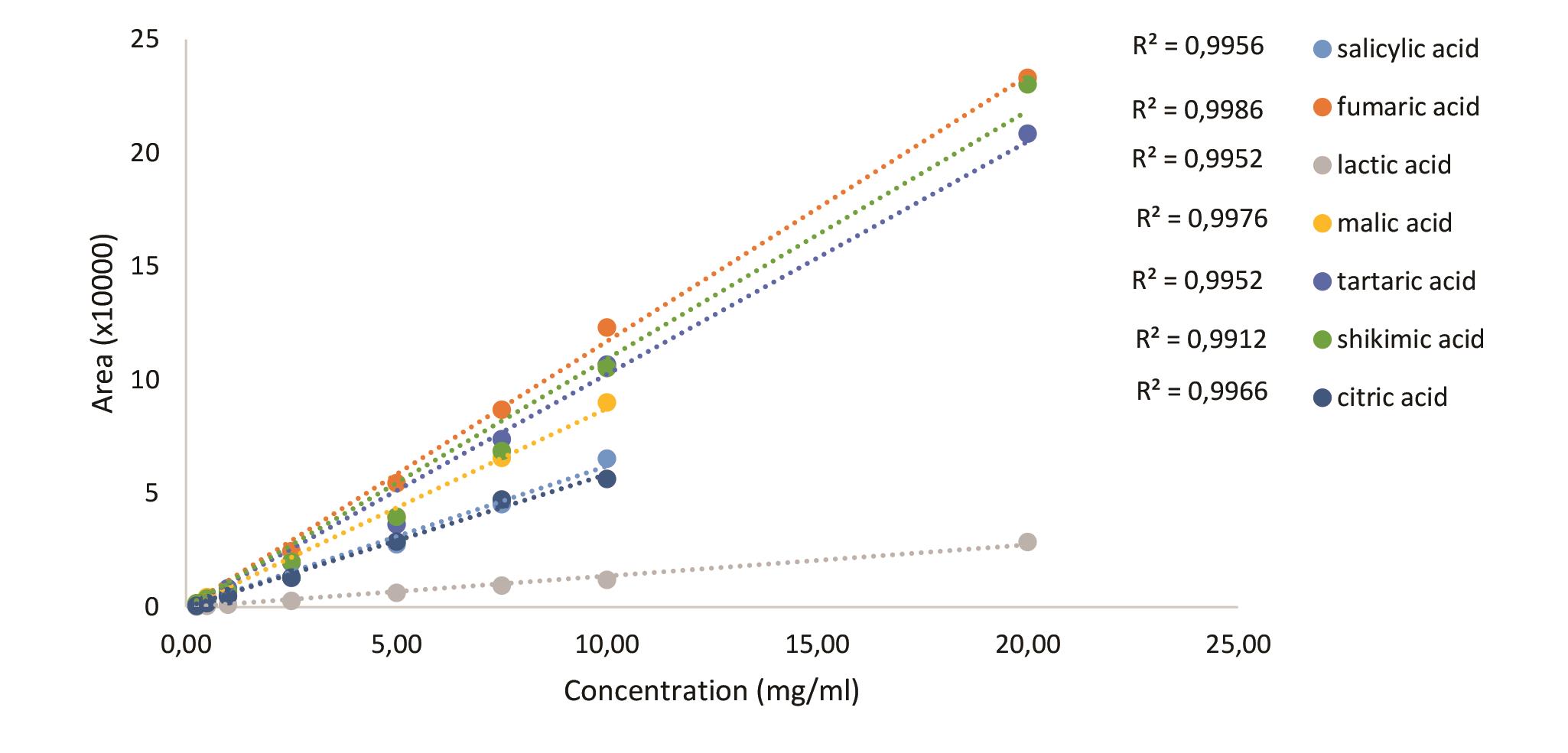
Figure 23. Standard curves of organic acids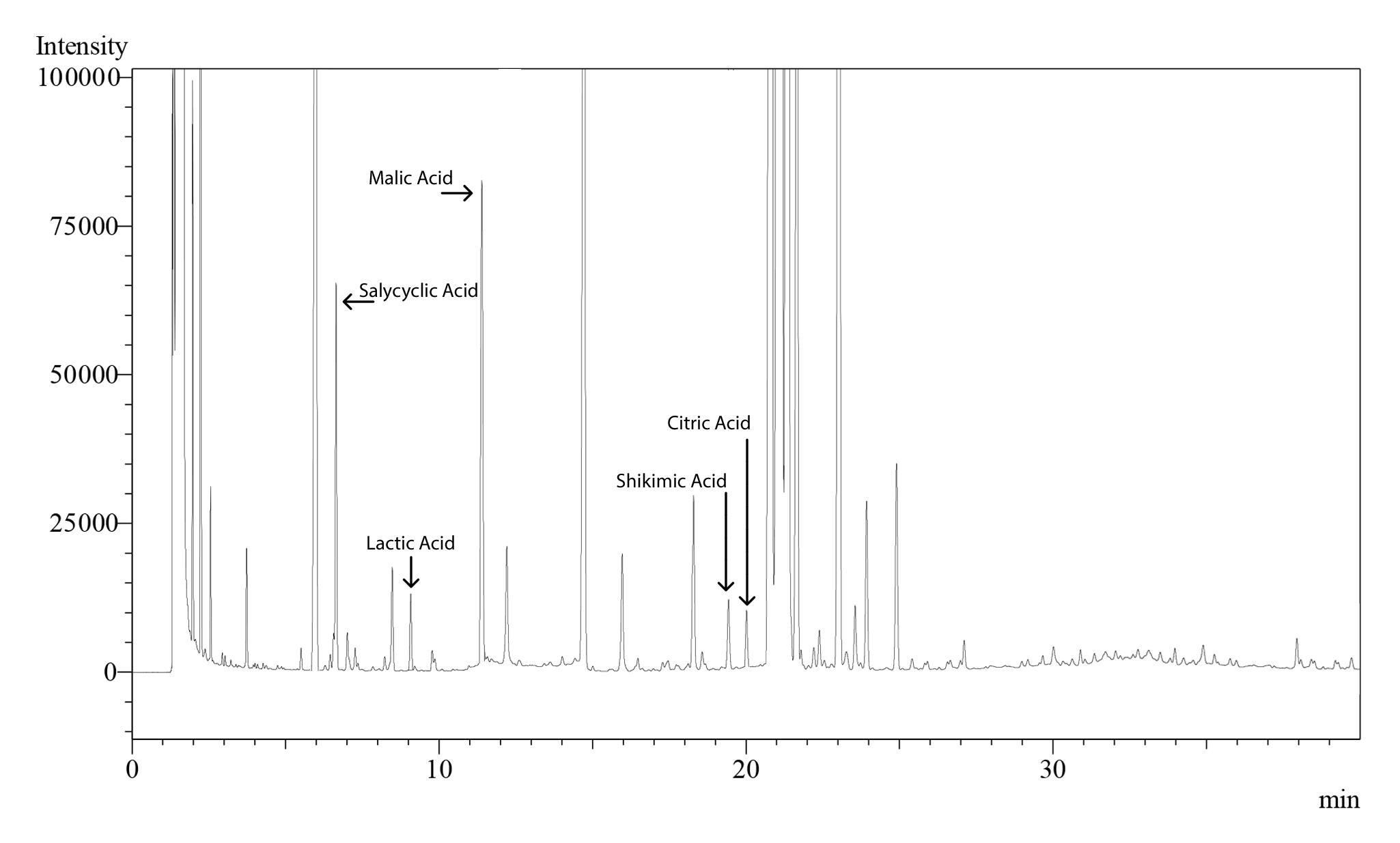
Figure 24. Sample chromatogram for tomato showing peaks for organic acids
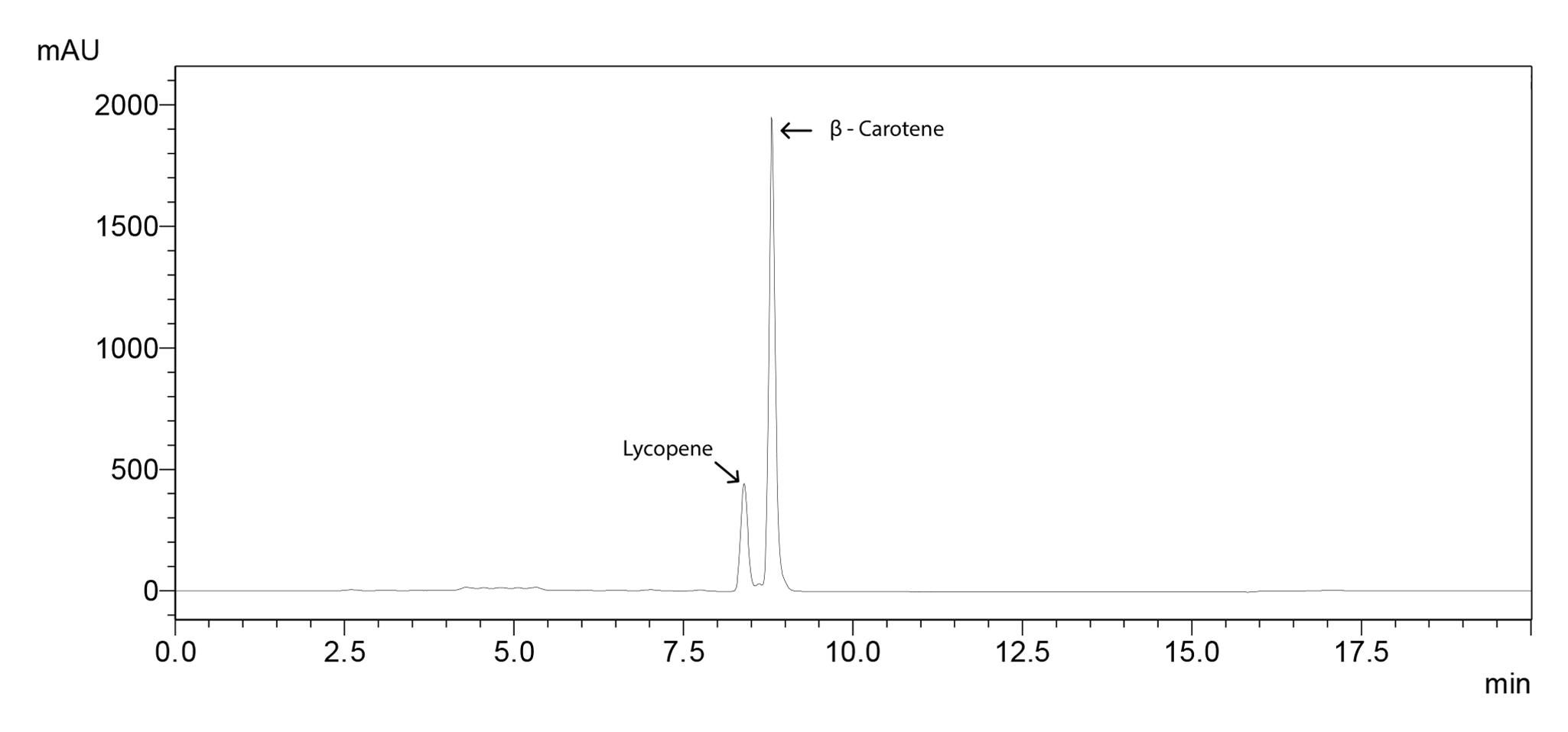
Figure 1. Chromatogram of lycopene and β-carotene standards
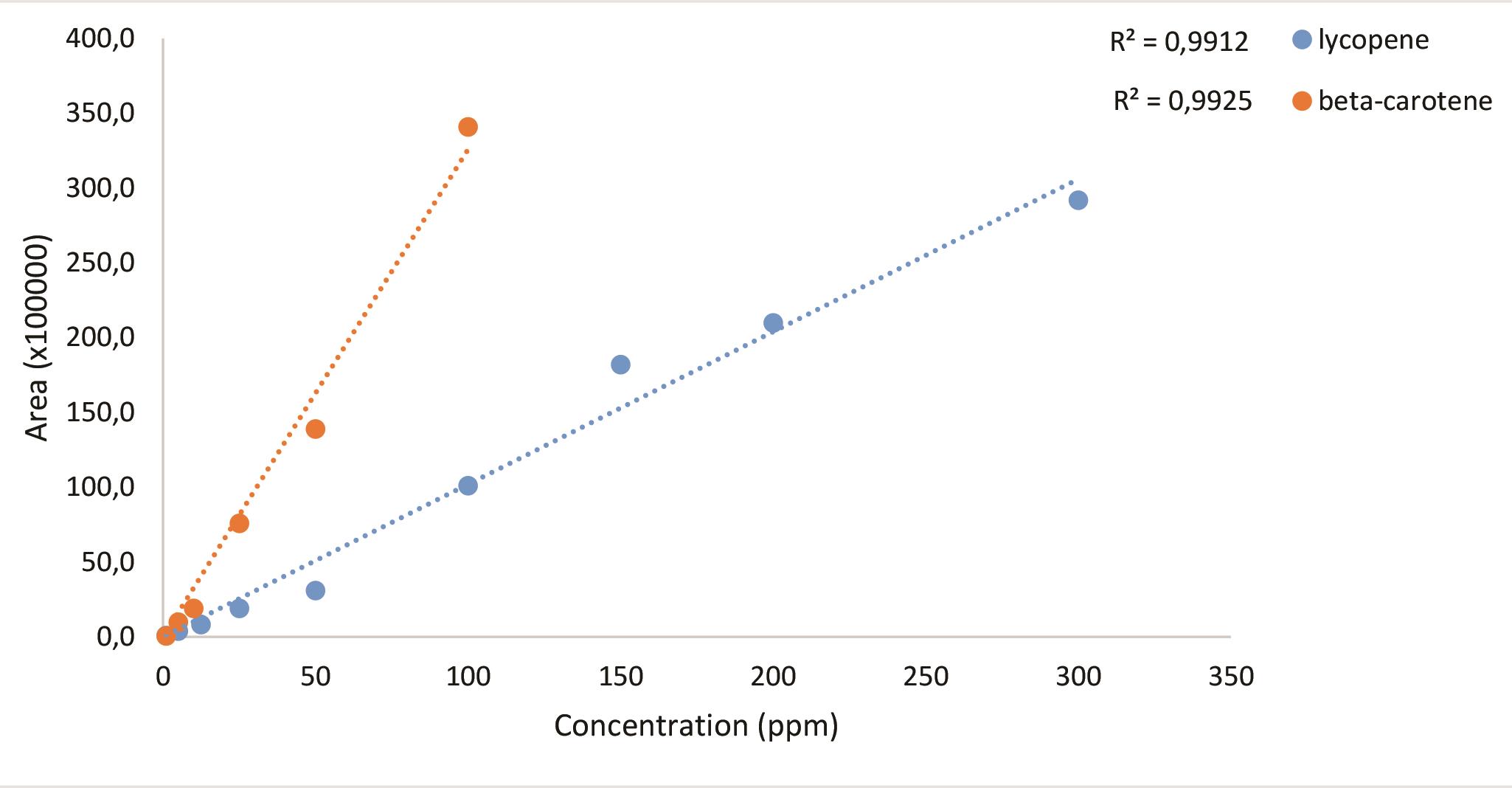
Figure 2. Standard curves of the carotenoids
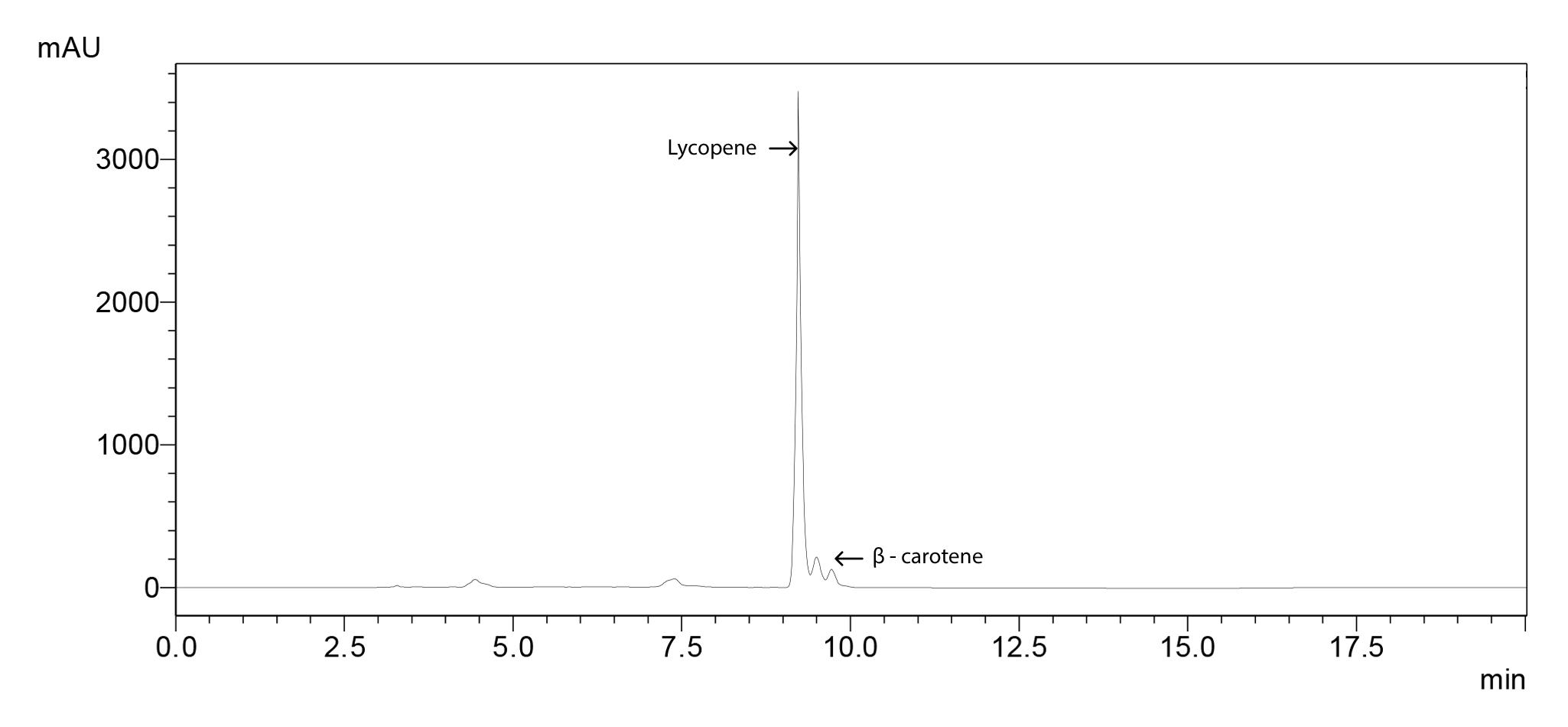
Figure 3. Sample chromatogram for tomato sample showing lycopene and β-carotene peaks
Method 2:
Chromatogram for 50 ppm standard molecules (Figure 4), standard curves (Figure 5) and the sample chromatogram for tomato (Figure 6) are shown.
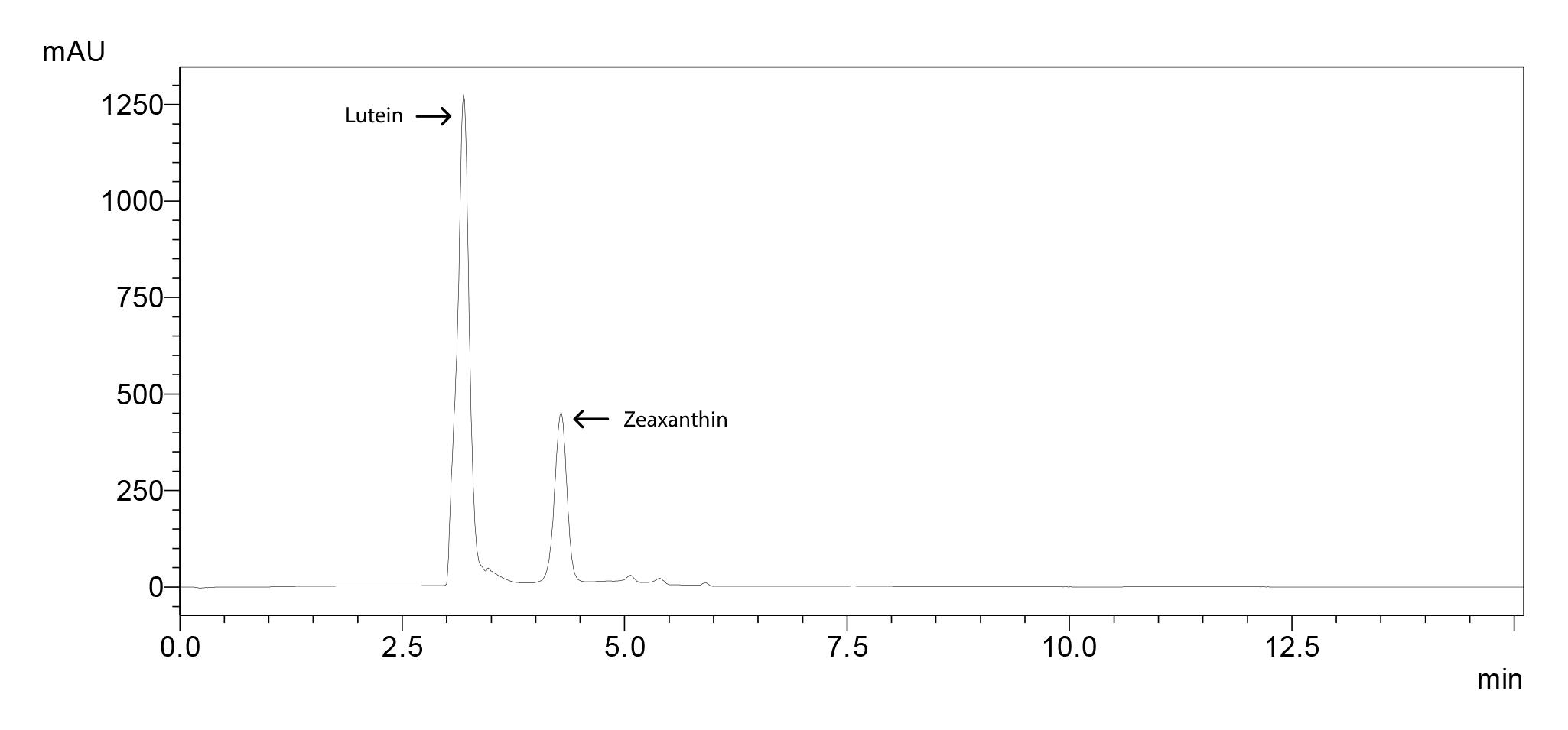
Figure 4. Chromatogram of lutein and zeaxanthin standards
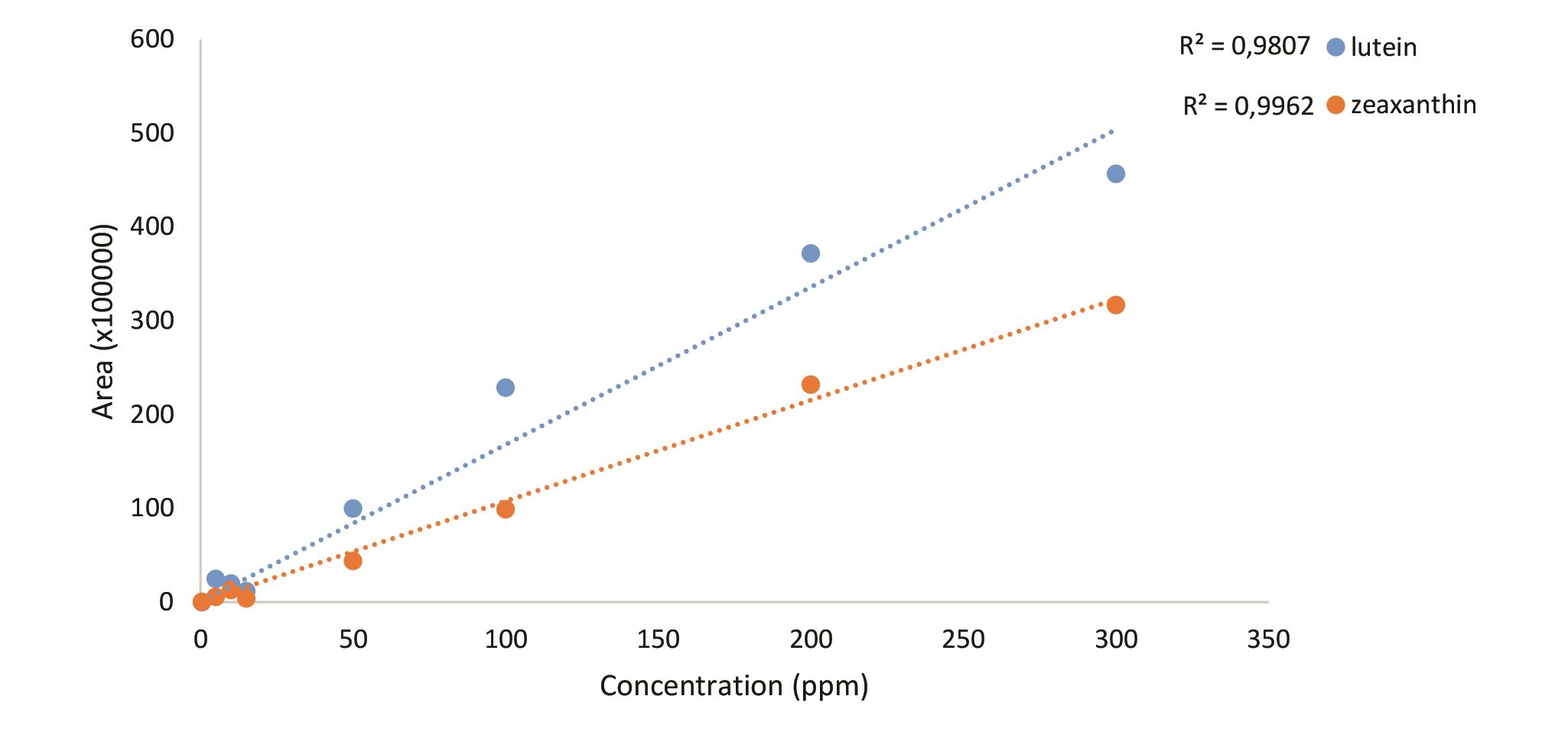
Figure 5. Standard curves of the carotenoids
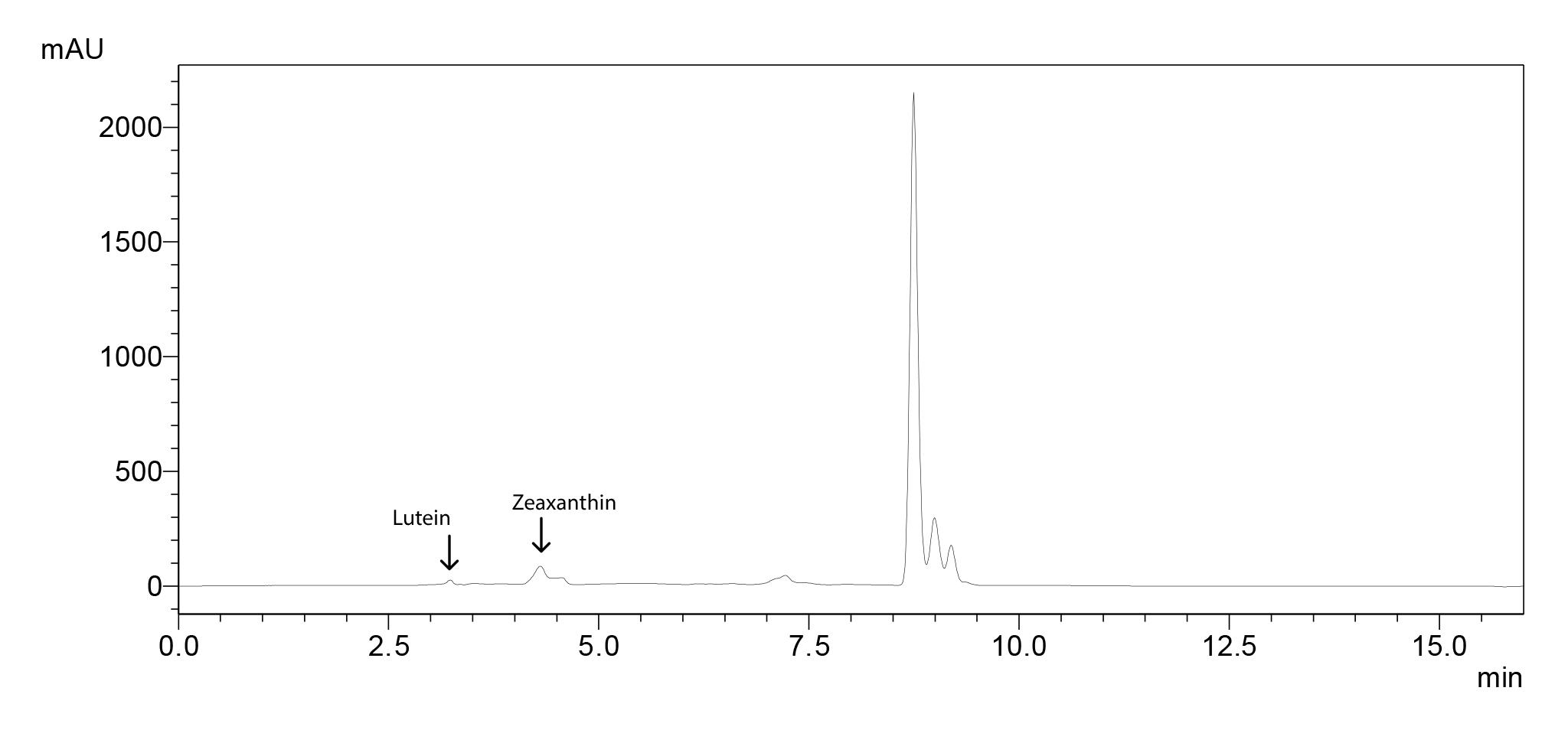
Figure 6. Sample chromatogram for tomato with lutein and zeaxanthin peaks
Vitamin C
Chromatogram for 100 ppm standard molecule (Figure 7), standard curve (Figure 8) and the sample chromatogram for tomato (Figure 9) are shown.
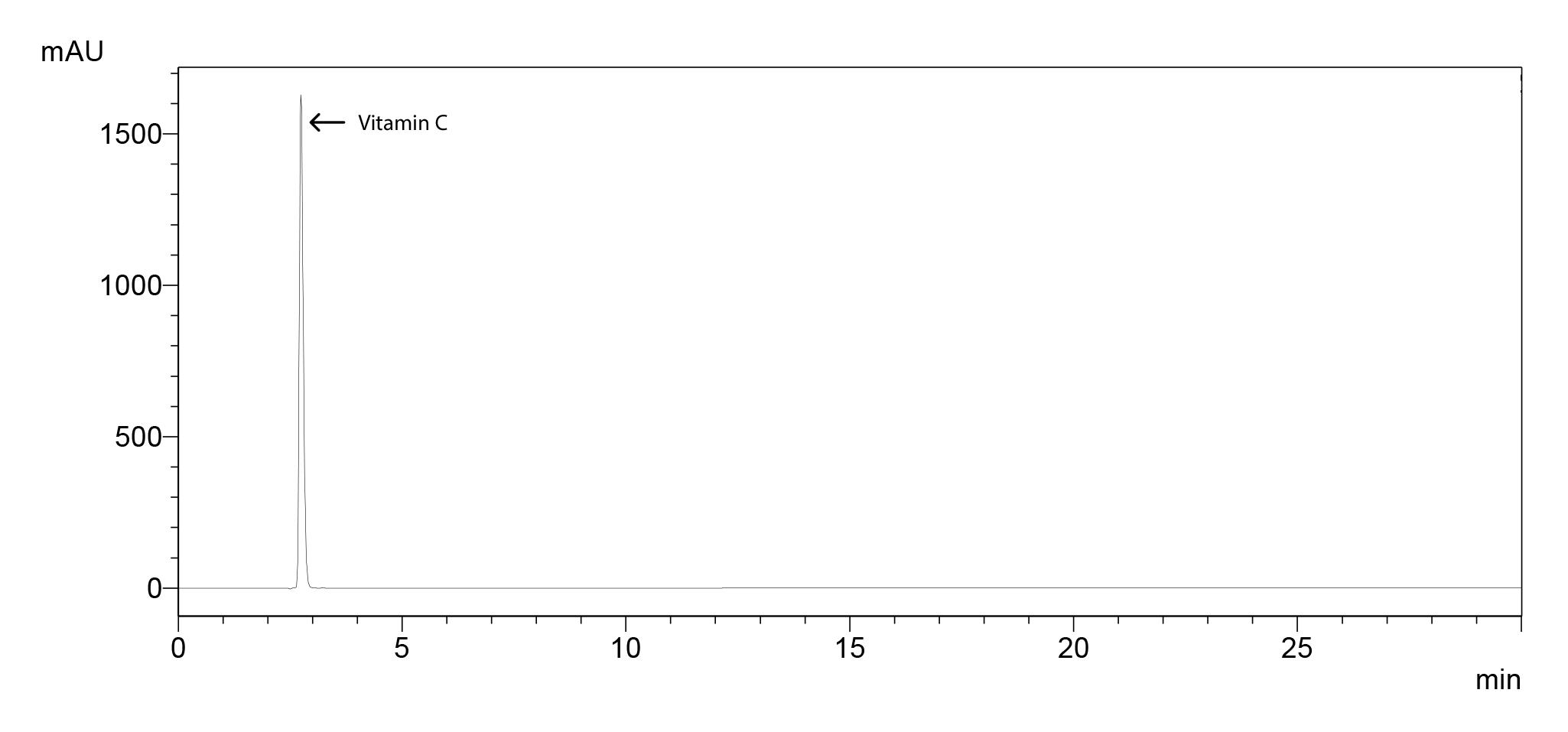
Figure 7. Chromatogram of vitamin C standard
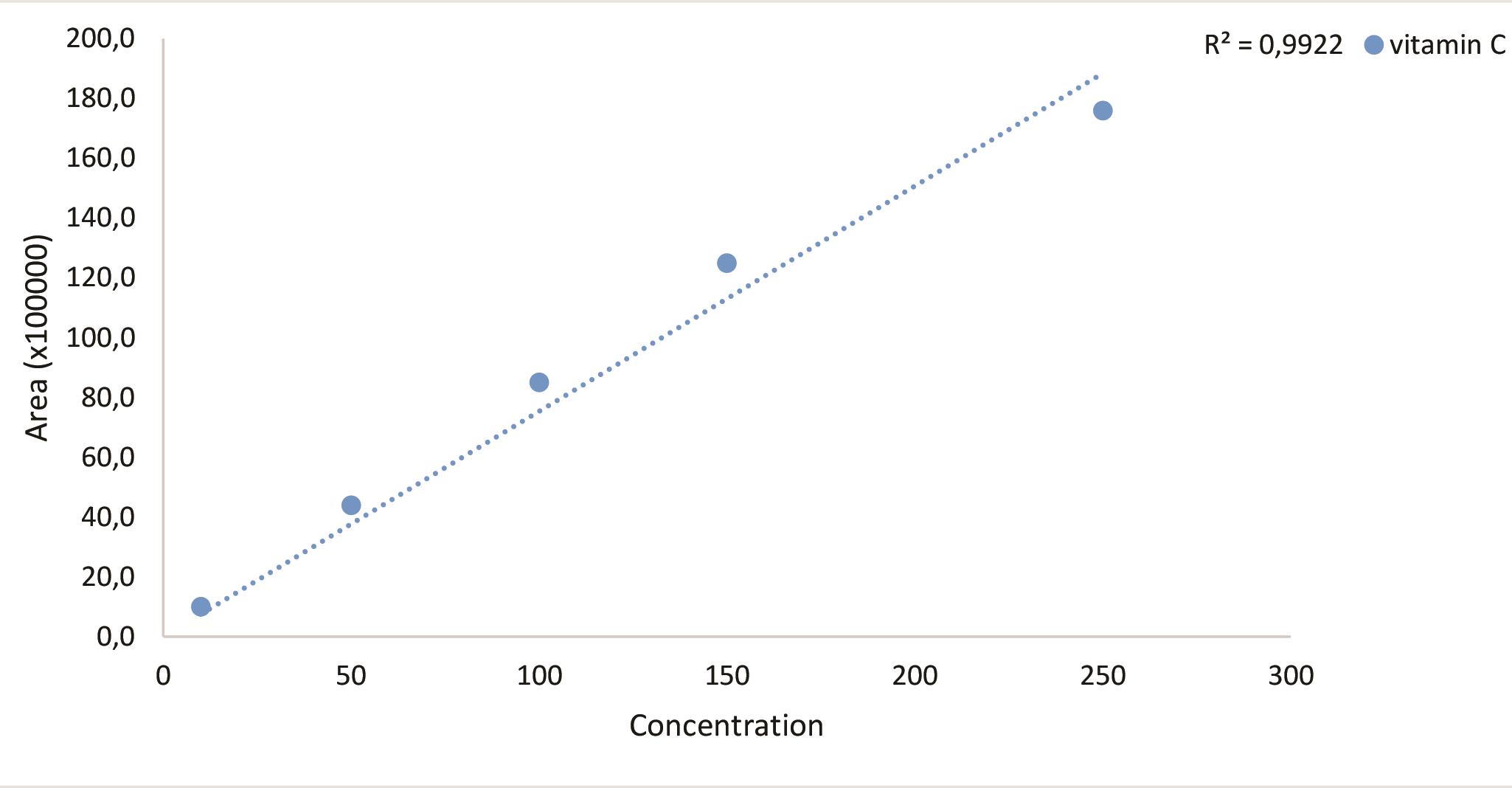
Figure 8. Standard curve for vitamin C
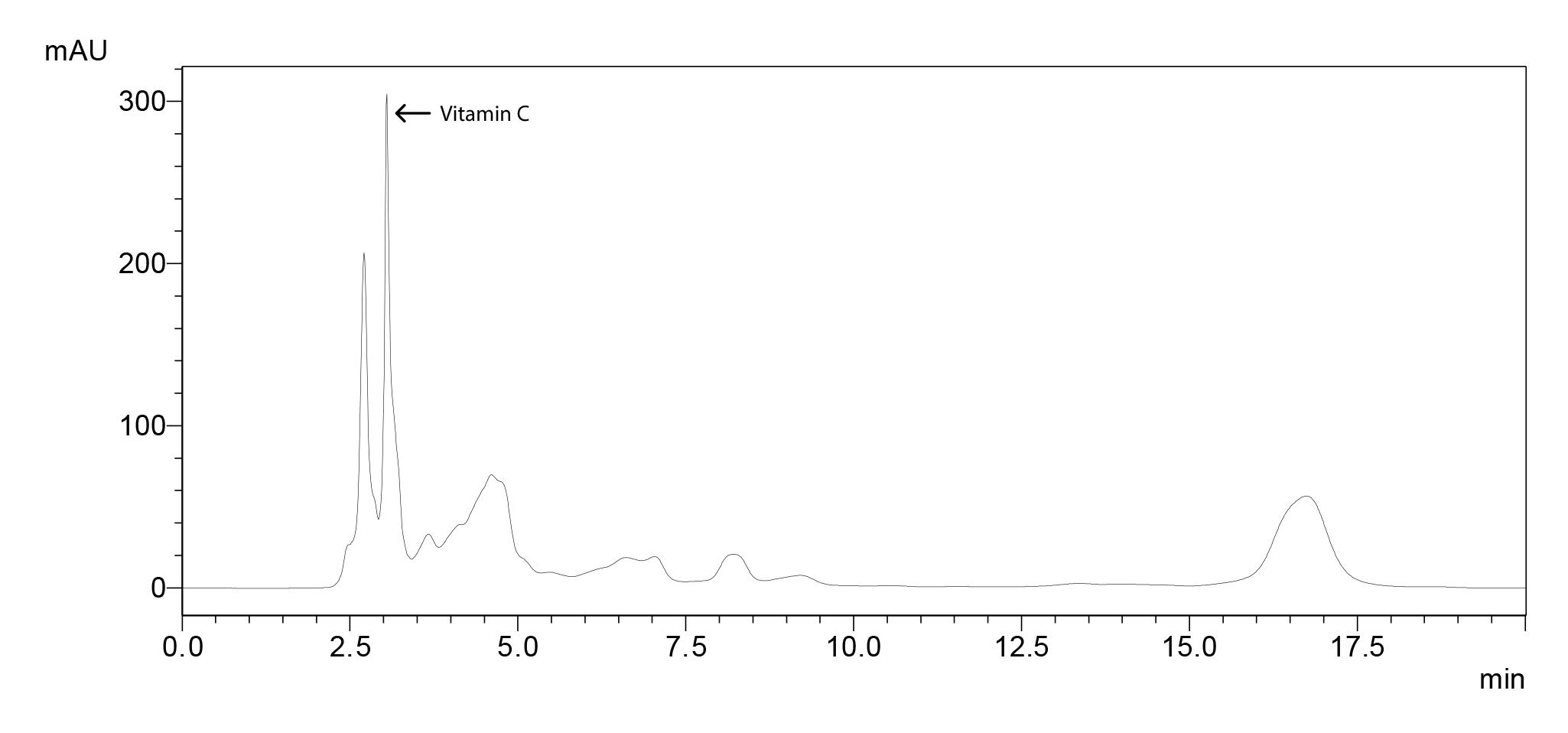
Figure 9. Sample chromatogram for tomato with vitamin C peak
Vitamin E
Chromatogram for 500 ppm standard molecule (Figure 10), standard curve (Figure 11) and the sample chromatogram for tomato (Figure 12) are shown.
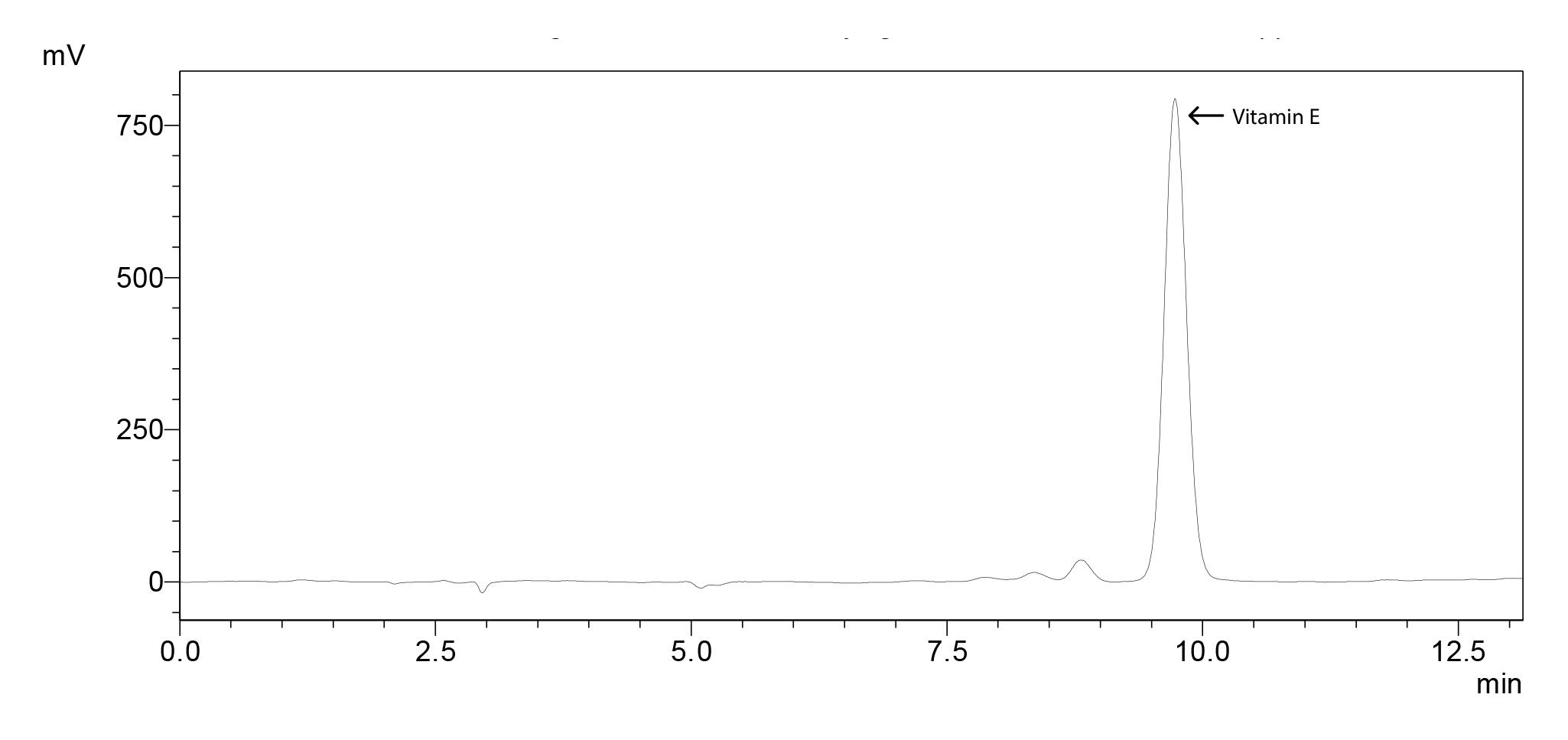
Figure 10. Chromatogram of vitamin E standard
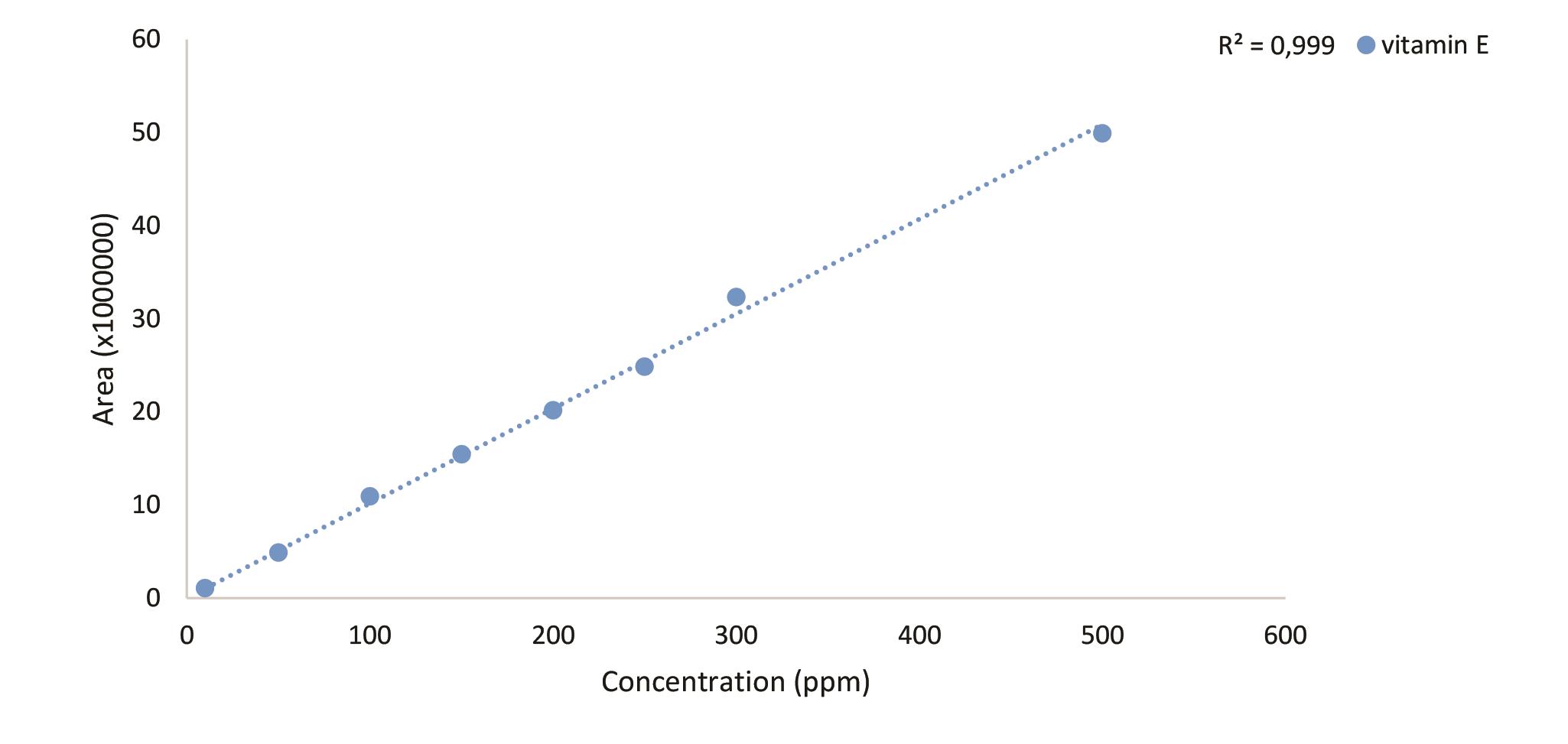
Figure 11. Standard curve of vitamin E
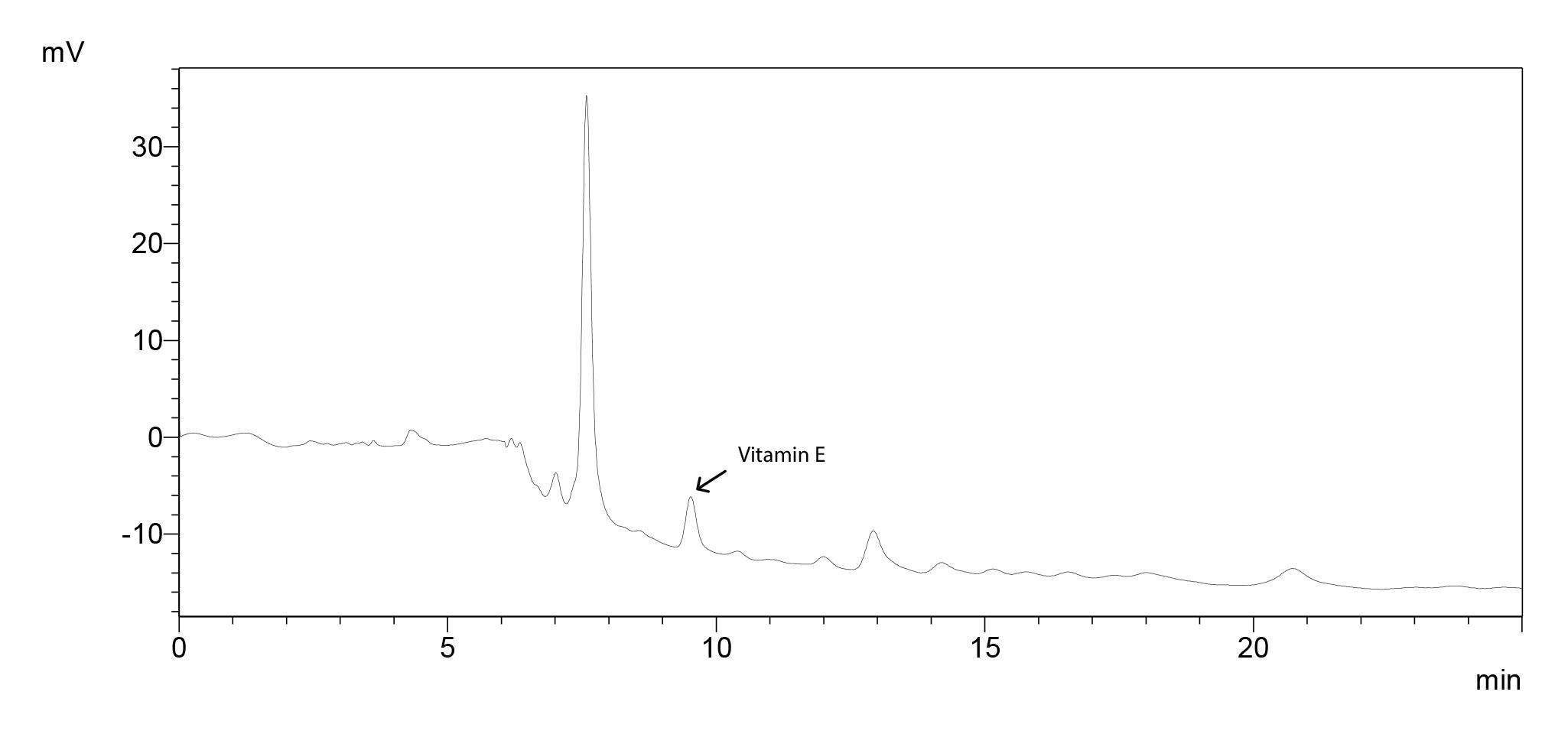
Figure 12. Sample chromatogram for tomato with vitamin E peak
Phenolic acids
Chromatogram for 50 ppm standard molecules (Figure 13), standard curves (Figure 14) and the sample chromatogram for tomato (Figure 15) are shown.
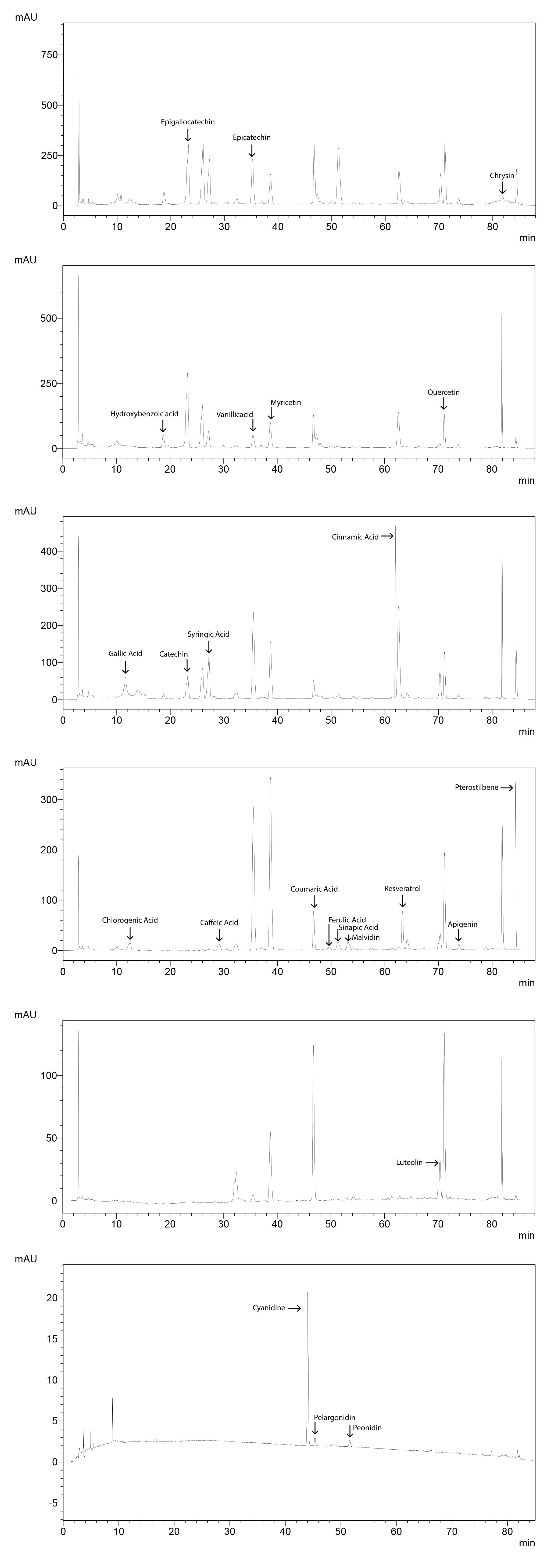
Figure 13. Chromatograms of standard molecules
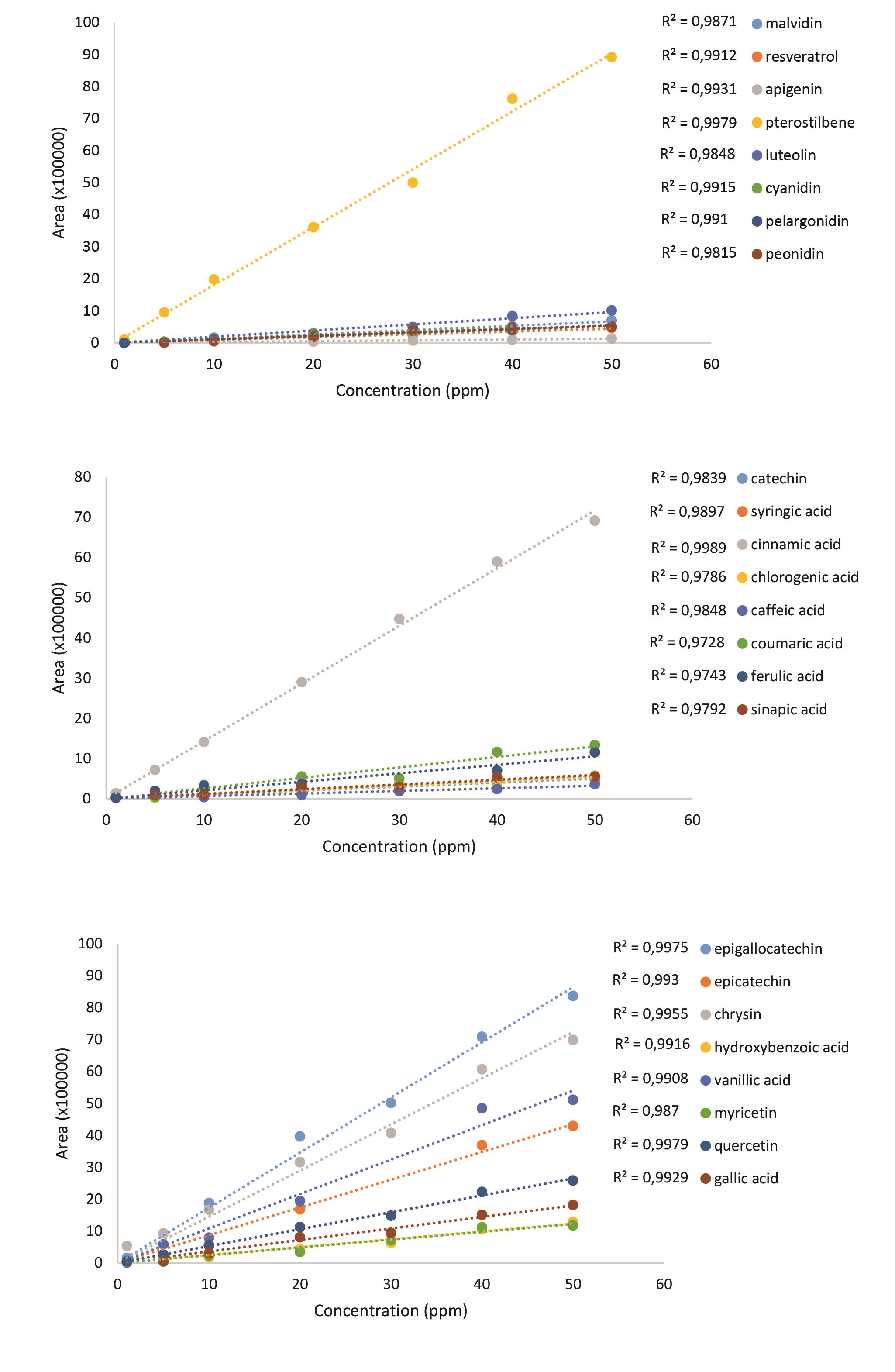
Figure 14. Standard curves of phenolic acids
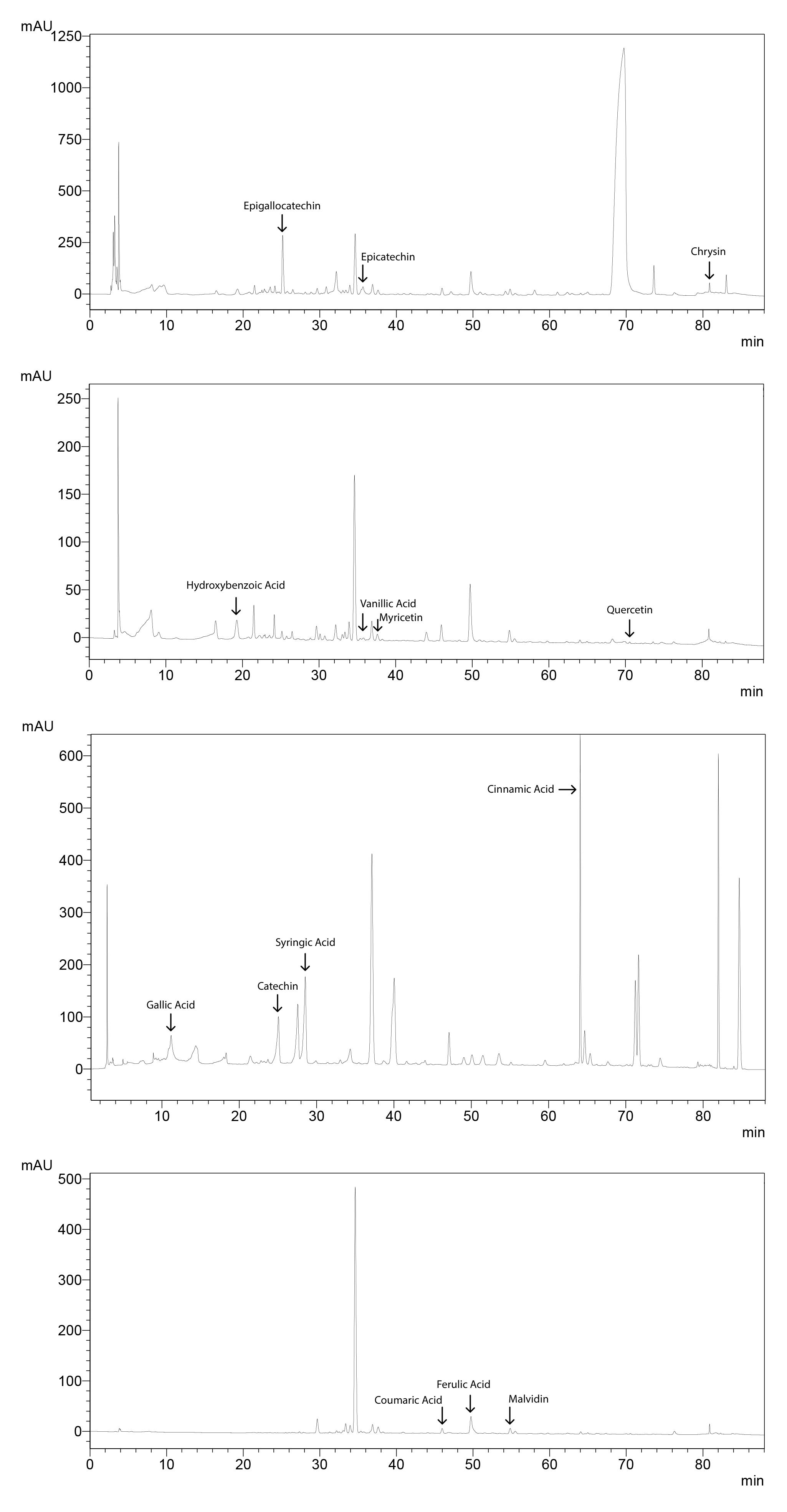
Figure 15. Sample chromatogram for tomato showing peaks for phenolic acids
Glutathione
Chromatogram for 50 ppm standard molecules (Figure 16), standard curves (Figure 17) and the sample chromatogram for tomato (Figure 18) are given below.
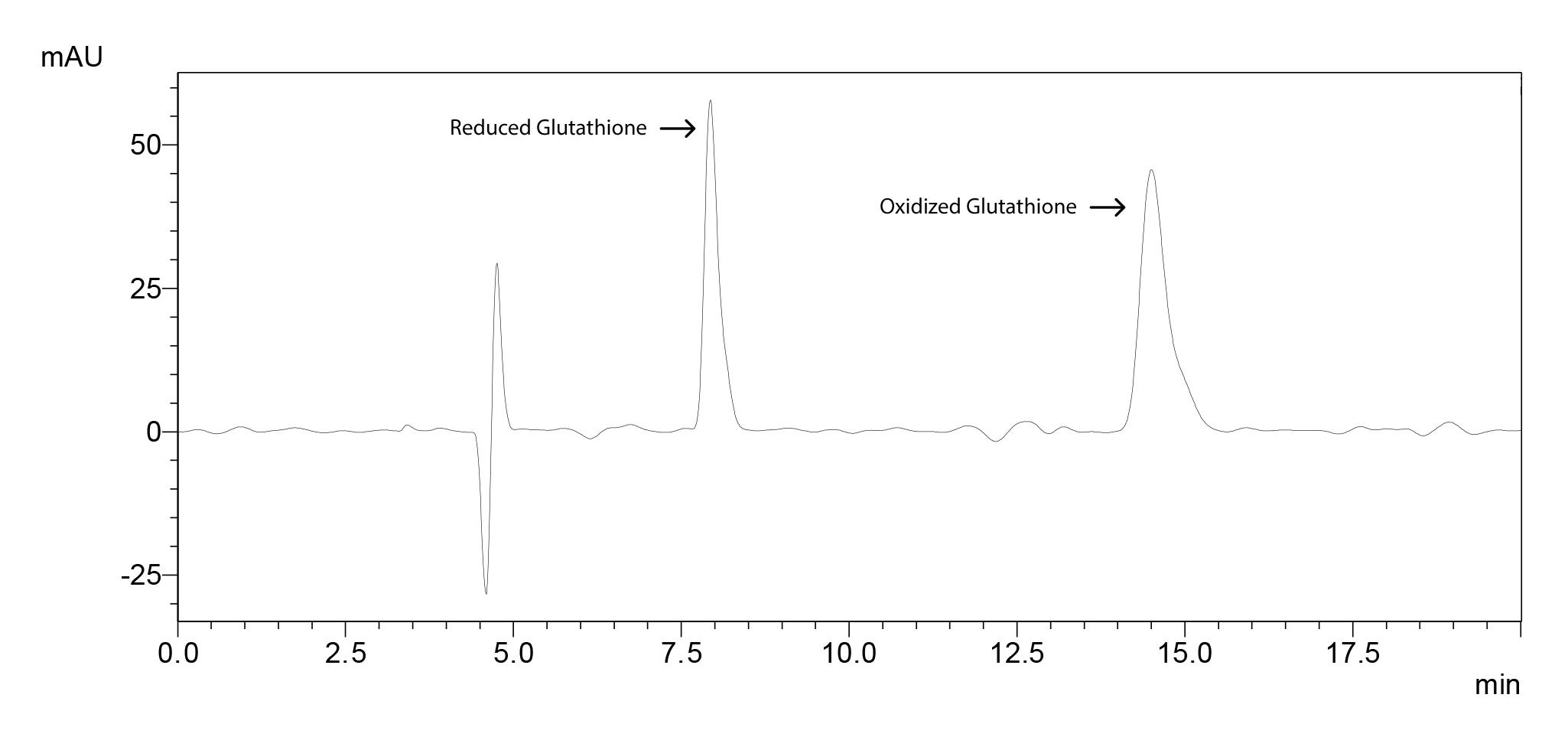
Figure 16. Chromatogram of reduced and oxidized glutathione standards
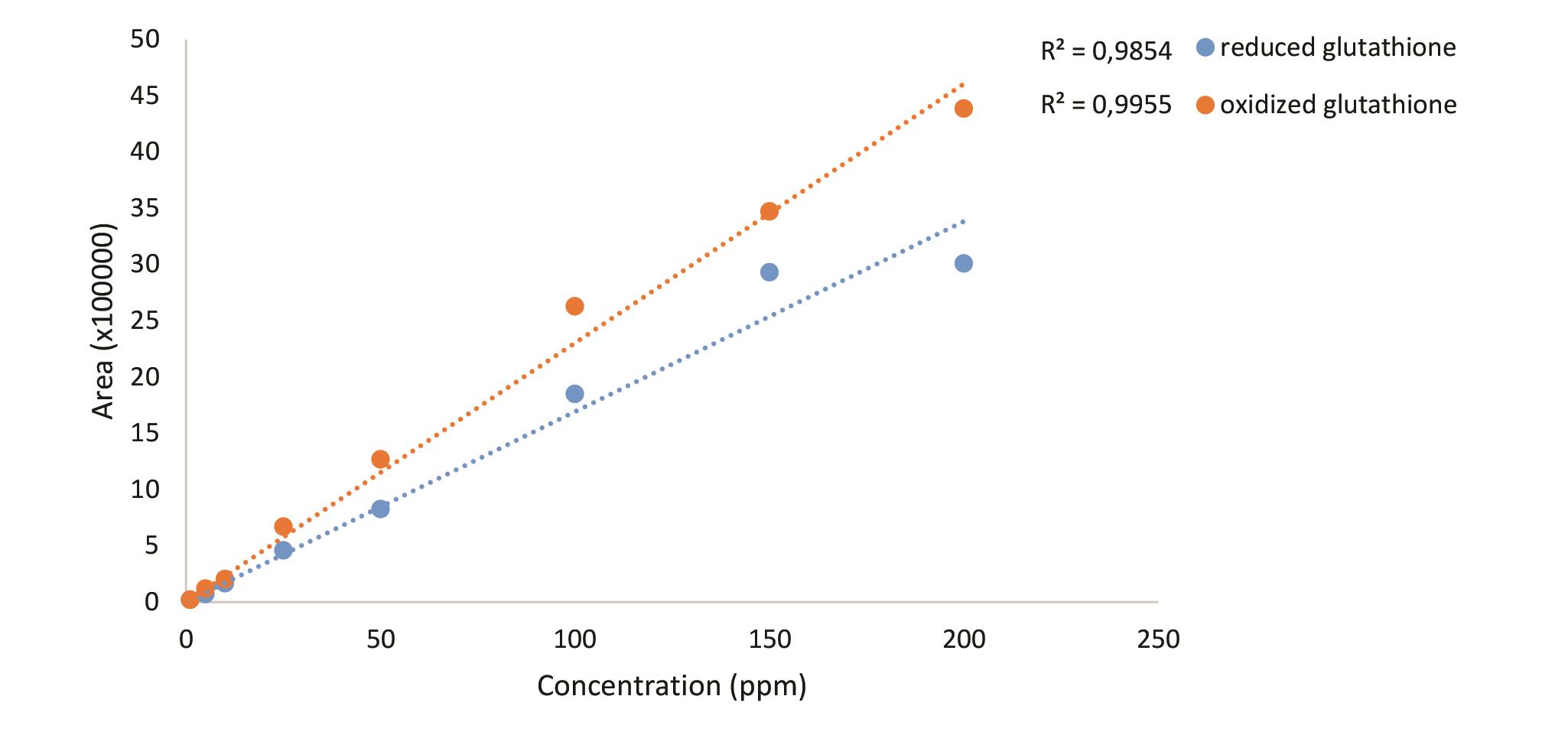
Figure 17. Standard curves reduced and oxidized glutathione
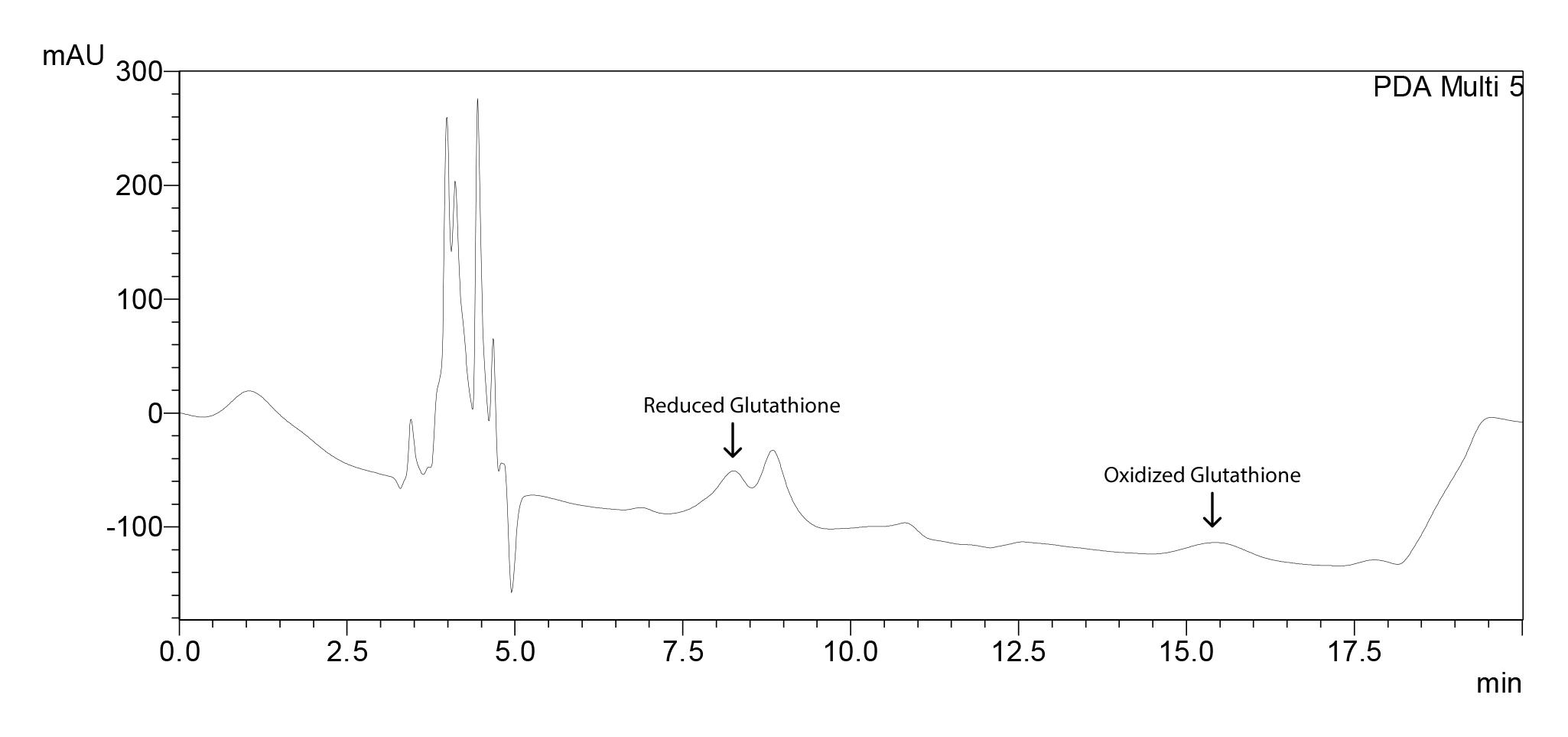
Figure 18. Sample chromatogram for tomato with glutathione results
Sugars
Chromatogram for 50 ppm standard molecules (Figure 19), standard curves (Figure 20) and the sample chromatogram for tomato (Figure 21) are given below.
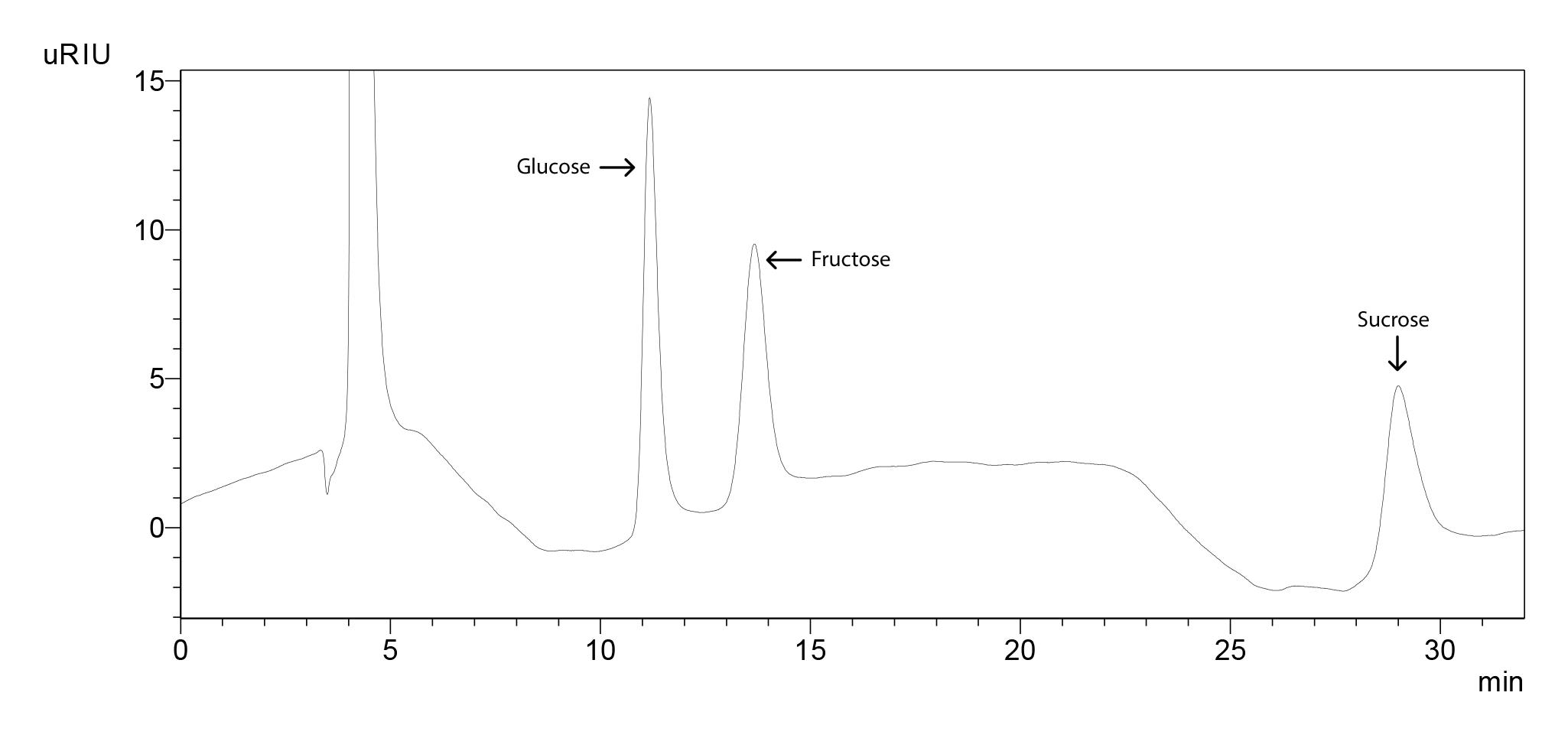
Figure 19. Chromatogram of glucose, fructose and sucrose standards
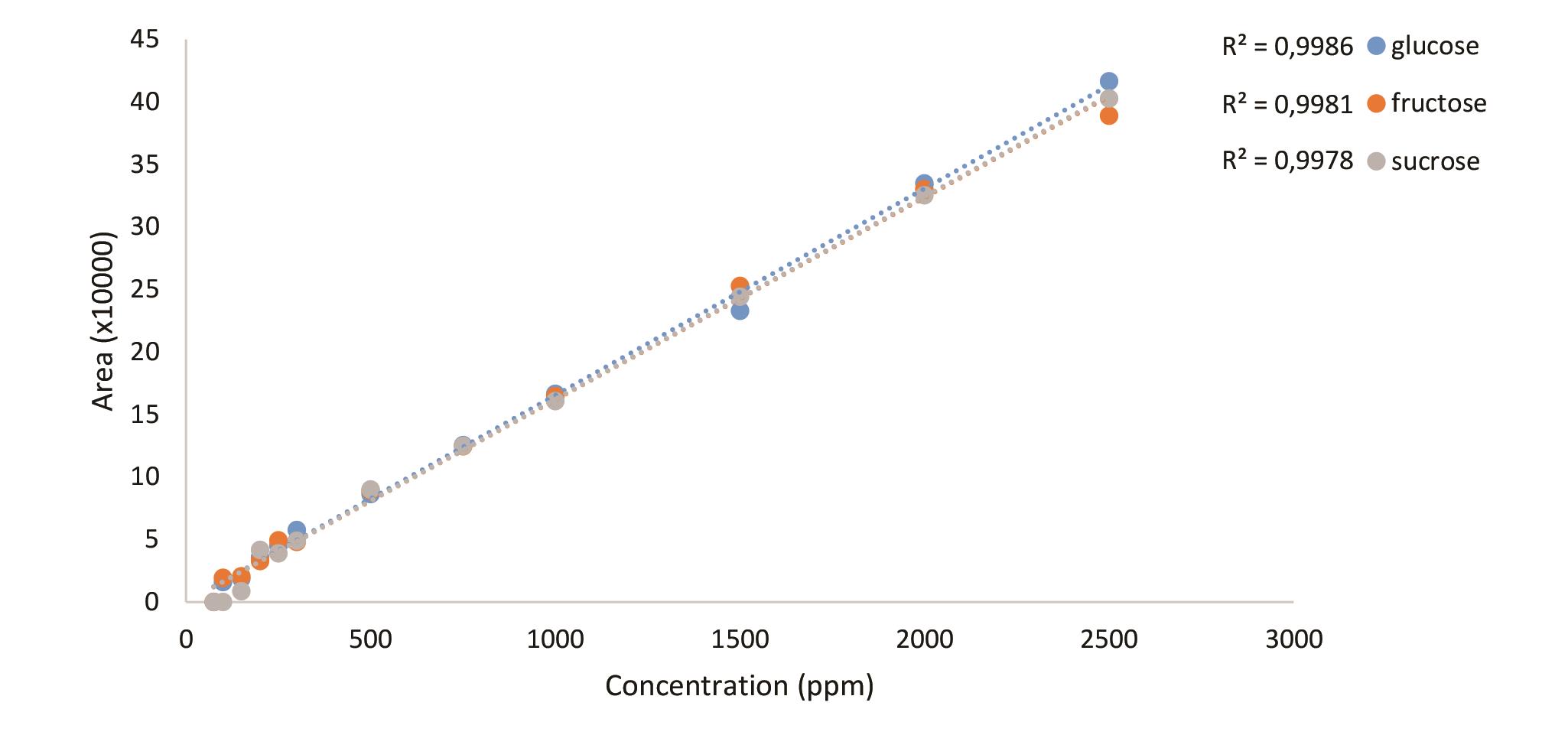
Figure 20. Standard curves of sugars
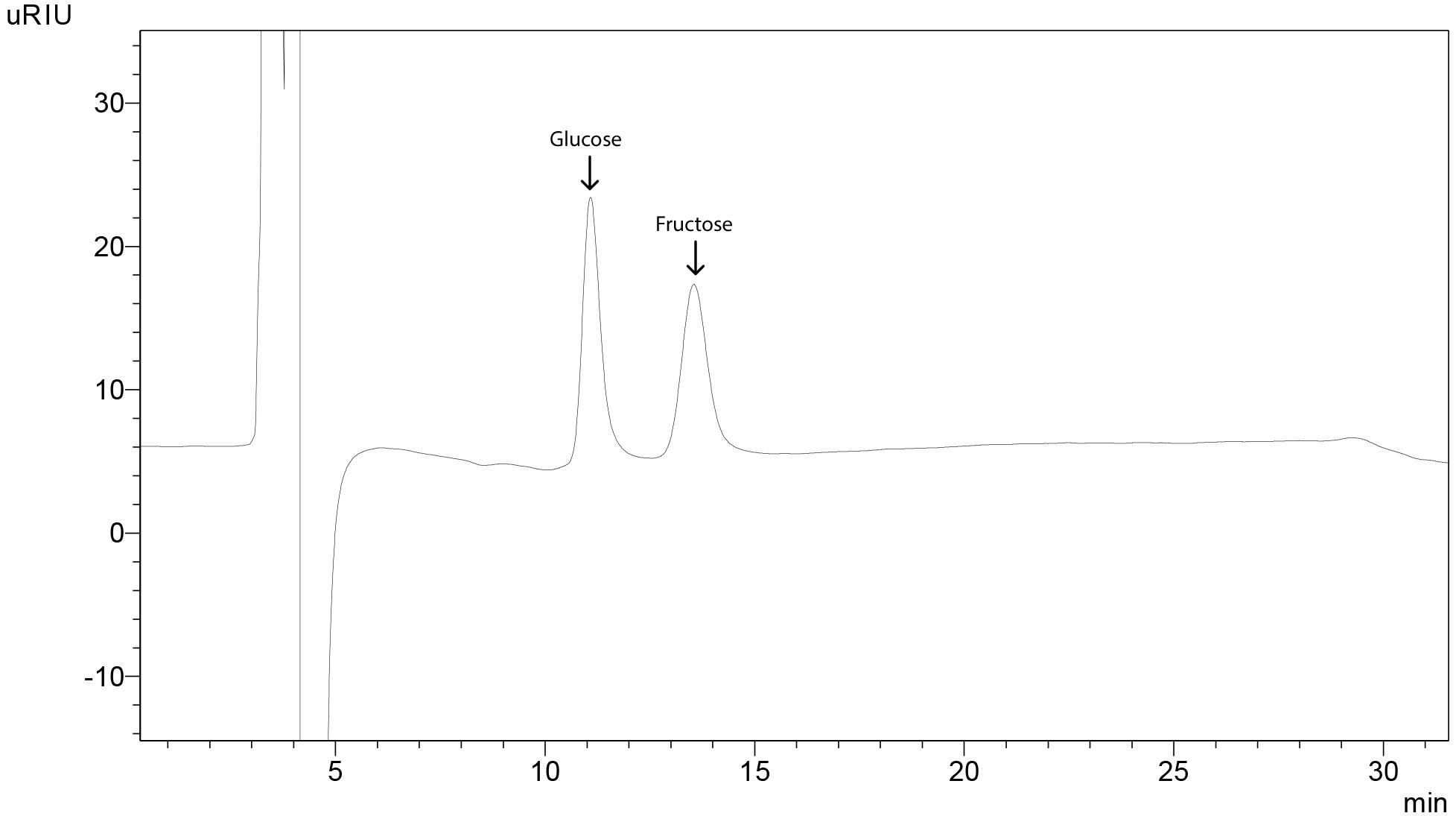
Figure 21. Sample chromatogram for tomato showing peaks for sugars. Sucrose was not detected in fully ripe fruit.
Data analysis
Experiments were repeated and data recorded for three replicates for control molecules. Draw standard curves using LC solution for HPLC data and GC Solution for GC data. These software programs calculate the average value of the triplicate analysis and plot the curves based on the area and concentrations automatically. Also, the software programs calculate metabolite concentrations in the samples automatically using the standard curves. Average metabolite contents of the parental lines and population are shown in Table 2.
Table 2. Mean metabolite content measured in the IBL population and parents
| Metabolites | S. lycopersicum | S. pimpinellifolium | Population |
| Carotenoids | |||
| Lycopene | 16141.58 | 26733.95 | 16919.46 |
| β-carotene | 56.62 | 36.06 | 45.98 |
| Lutein | 3.25 | 5.06 | 3.83 |
| Zeaxanthin | 3.26 | 2.78 | 3.60 |
| Vitamins | |||
| Vitamin C | 19.11 | 20.17 | 17.84 |
| Vitamin E | 3.61 | 20.28 | 20.40 |
| Phenolic acids | |||
| Epigallocatechin | 1.92 | 2.19 | 2.61 |
| Epicatechin | 0.24 | 5.89 | 2.45 |
| Chrysin | 0.42 | 2.07 | 80.12 |
| Hydroxybenzoic acid | 6.60 | 1.37 | 46.80 |
| Vanillic acid | 12.8 | 2.05 | 61.12 |
| Myricetin | 10.00 | 40.00 | 3.06 |
| Quercetin | 1.88 | 1.67 | 1.025 |
| Gallic acid | 31.46 | 1.06 | 5.07 |
| Catechin | 0.37 | 0.59 | 26.53 |
| Syringic acid | 3.96 | 20.40 | 89.46 |
| Cinnamic acid | 0.06 | 0.11 | 0.98 |
| Chlorogenic acid | 19.20 | 0.83 | 0.53 |
| Caffeic acid | 9.54 | 3.32 | 3.99 |
| Coumaric acid | 0.24 | 2.13 | 1.87 |
| Ferulic acid | 2.08 | 31.00 | 3.29 |
| Sinapic acid | 1.47 | 1.29 | 1.60 |
| Malvidin | 0.76 | 0.75 | 3.48 |
| Resveratrol | - | - | - |
| Apigenin | 2.18 | 1.24 | 13.04 |
| Pterostilbene | - | - | - |
| Luteolin | 0.62 | 0.20 | 0.72 |
| Cyanidin | - | - | - |
| Pelargonidin | - | - | - |
| Peonidin | - | ||
| Glutathione | |||
| Reduced glutathione | 17.75 | 10.79 | 52.17 |
| Oxidized glutathione | 5.18 | 0.09 | 56.14 |
| Sugars | |||
| Glucose | 8738.04 | 4153.29 | 6596.45 |
| Fructose | 8401.38 | 3967.70 | 5839.52 |
| Sucrose | - | - | - |
| Organic acids | |||
| Salicylic acid | 0.01 | 0.02 | 0.03 |
| Fumaric acid | - | - | - |
| Lactic acid | 0.31 | 0.04 | 0.18 |
| Malic acid | 6.19 | 0.86 | 3.58 |
| Tartaric acid | - | - | - |
| Shikimic acid | 0.94 | 0.00 | 1.17 |
| Citric acid | 10.40 | 8.51 | 7.59 |
* Quantities are given as mg 100 g-1 DW.
** Resveratrol, pterostilbene, cyanidin, pelargonidin, peonidin, sucrose, fumaric acid and tartaric acid were not detected in the tomato samples.
Notes
It is better to do extraction in the dark and at low temperatures (+4 °C) since some metabolites are affected by light and high temperatures.
It is better to centrifuge the sample (25,830 × g, 1-2 min) once more before syringe filtration to extend column life.
A RP C30 column can provide better resolution but is more expensive than a RP C18 column.
It is better to wait for conditioning of the RI detector before analysis. Sometimes it is better to leave the detector overnight with mobile phase at low flow rate (e.g., 0.1 ml/min-0.5 ml/min).
Helium can be used instead of nitrogen as the carrier gas.
Freshly prepare methoxamine hydrochloride in pyridine.
Use only ultrapure water for extraction and analysis.
Recipes
Extraction solvent 1
Dichloromethane: hexane (1:1, v:v) containing 0.01% BHT to avoid oxidation reactions while extracting the metabolites
Measure 50 ml of dichloromethane
Add 0.01 g BHT into the dichloromethane and mix until BHT dissolves
Add 50 ml of hexane and shake well
Extraction solvent 2: Chloroform: methanol: water (1:3:1, v:v:v)
Measure 20 ml of chloroform
Add 60 ml of methanol and mix well
Place the solution on a shaker and add water slowly into the solution to avoid phase separation
0.1 M KH2PO4, pH = 7.0
Weigh 13.609 g KH2PO4 in a beaker
Add 800 ml of water and mix until KH2PO4 dissolves
Bring the pH to 7.0
Make up volume to 1,000 ml with water
0.05% Trifluoroacetic acid(aq)
Measure 950 ml of water into a volumetric flask
Add 0.5 ml of trifluoroacetic acid and bring the volume to 1,000 ml with water
Acknowledgments
This work was supported by funds from the Scientific and Technological Research Council of Turkey (TÜBİTAK, project number 114Z116) and the Republic of Turkey Ministry of Agriculture and Forestry, General Directorate of Agricultural Research Institute (TAGEM-16/AR-GE/03). This protocol was derived from Gürbüz Çolak et al. (2020a and 2020b) manuscripts.
Competing interests
The authors declare no financial and non-financial competing interests.
Ethics
No human and/or animal subjects were used in these protocols.
References
- Alenazi, M. M., Shafiq, M., Alsadon, A. A., Alhelal, I. M., Alhamdan, A. M., Solieman, T. H. I., Ibrahim, A. A., Shady, M. R. and Al-Selwey, W. A. (2020). Improved functional and nutritional properties of tomato fruit during cold storage. Saudi J Biol Sci 27(6): 1467-1474.
- Bakre, S. M., Gadmale, D. K., Toche, R. B. and Gaikwad, V. B. (2015). Rapid determination of α- tocopherol in olive oil adulterated with sunflower oil by reversed phase high-performance liquid chromatography. J Food Sci Technol 52(5): 3093-3098.
- Capanoglu, E., Beekwilder, J., Boyacioglu, D., Hall, R. and de Vos, R. (2008). Changes in antioxidant and metabolite profiles during production of tomato paste. J Agric Food Chem 56(3): 964-973.
- Celik, I., Gurbuz, N., Uncu, A. T., Frary, A. and Doganlar, S. (2017). 942 3" target="_blank">Genome-wide SNP discovery and QTL mapping for fruit quality traits in inbred backcross lines(IBLs) of Solanum pimpinellifolium using genotyping by sequencing. BMC Genomics 18(1): 1. doi 10.1186/s12864-016-3406-7.
- Dumont, D., Danielato, G., Chastellier, A., L., Hibrand Saint Oyant, Fanciullino, A. L. and Lugan, R. (2020). Multi-targeted metabolic profiling of carotenoids, phenolic compounds and primary metabolites in goji(Lycium spp.) berry and tomato(Solanum lycopersicum) reveals inter and intra genus biomarkers. Metabolites 10(10). doi:10.3390/metabo10100422.
- Figueira, J. A., Pereira, J. A. M. and Camara, J. S. (2017). Quantification of δ-, γ- and α-tocopherol in tomatoes using an improved liquid-dispersive solid-phase extraction combined with ultrahigh pressure liquid chromatography. Food Anal Methods 10(7): 2507-2517.
- Gómez-Alonso, S., García-Romero, E. and Hermosỉn-Gutiẻrrez, I. (2007). HPLC analysis of diverse grape and wine phenolics using direct injection and multidetection by DAD and fluorescence. J Food Compos Anal 20(7): 618-626.
- Gürbüz Çolak, N., Eken, N. T., Ulger, M., Frary, A. and Doganlar, S. (2020). Mapping of quantitative trait loci for antioxidant molecules in tomato fruit: Carotenoids, vitamins C and E, glutathione and phenolic acids. Plant Sci 292: 110393.
- Çolak, Gürbüz, N., Tek Eken, Ülger, N., Frary, M. and Doğanlar, A. (2020). Exploring wild alleles from Solanum pimpinellifolium with the potential to improve tomato flavor compounds. Plant Sci 298: 110567.
- Ingallina, C., Sobolev, A. P., Circi, S., Spano, M., Giusti, A. M. and Mannina, L. (2020). New hybrid tomato cultivars: An NMR-based chemical characterization. Appl Sci 10: 1887. doi:10.3390/app10051887.
- Ishida, B. K., Ma, J. and Chan, B. (2001). A simple, rapid method for HPLC analysis of lycopene isomers. Phytochem Anal 12(3): 194-198.
- Kader, A. A., (2008). Flavor quality of fruits and vegetables. J Sci Food Agric 88(11): 1863-1868.
- Khan, A., Khan, M. I., Iqbal, Z., Shah, Y., Ahmad, L., Nazir, S., Watson, D. G., Khan, J. A., Nasir, F. and Khan, A. (2011). A new HPLC method for the simultaneous determination of ascorbic acid and aminothiols in human plasma and erythrocytes using electrochemical detection. Talanta 84(3): 789-801.
- Li, H. B. and Chen, F. (2001). Simultaneous determination of twelve water- and fat-soluble vitamins by high-performance liquid chromatography with diode array detection. Chromatographia 54: 270-273.
- Li, H. B. and Chen, F. (2001). Simultaneous determination of nine water-soluble vitamins in pharmaceutical preparations by high- performance liquid chromatography with diode array detection. J Sep Sci 24(4): 271-274.
- Lobo, V., Patil, A., Phatak, A. and Chandra, N. (2010). Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn Rev 4(8): 118-126.
- Martínez, J. P., Fuentes, R., Farías, K., Lİzana, C., Alfaro, J. F., Fuentes, L., Calabrese, N., Bigot, S., Quinet, M. and Lutts, S. (2020). Effects of salt stress on fruit antioxidant capacity of wild(Solanum chilense) and domesticated(Solanum lycopersicum var. cerasiforme) tomatoes. Agronomy 10: 1481. doi:10.3390/agronomy10101481.
- Masetti, O., Nisini, L., Ciampa, A. and Dell’Abate, M. T. (2020). 1H NMR spectroscopy coupled with multivariate analysis was applied to investigate Italian cherry tomatoes metabolic profile. J Chemom 3191.
- Migalatev, O., (2017). Optimisation of operating parameters for supercritical carbon dioxide extraction of lycopene from industrial tomato waste. Ukr Food J 6(4): 698-716.
- Nagarajan, J., Kay, H. P., Krishnamurthy, N. P., Ramakrishnan, N. R., Aldawoud, T. M. S., Galanakis, C. M. and Wei, O. C. (2020). Extraction of carotenoids from tomato pomace via water-induced hydrocolloidal complexation. Biomolecules 10(7): 1019.
- Namgung, H. J., Park, H. J., Cho, I. H., Choi, H. K., Kwon, D. Y., Shim, S. M. and Kim, Y. S. (2010). Metabolite profiling of doenjang, fermented soybean paste, during fermentation. J Sci Food Agric 90(11): 1926-1935.
- Pellicanò, T. M., Sicari, V., Loizzo, M. R., Leporini, M., Falco, T. and Poiana, M. (2019). Optimizing the supercritical fluid extraction process of bioactive compounds from processed tomato skin by-products. Food Sci Technol 40(3): 692-697.
- Petkova, T. N., Pascal, B.A., Annick, M. and Pantele, D. P. (2013). HPLC analysis of mono- and disaccharides in food products. Presentation at the meeting of the Scientific Works Volume LX, Food Science, Engineering and Technology, Plovdiv, Bulgaria, 761-765.
- Roessner, U., Wagner, C., Kopka, J., Trethewey, R. N. and Willmitzer, L. (2000). Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23(1): 131-142.
- Serino, S., Gomez, L., Costagliola, G. and Gautier, H. (2009). HPLC assay of tomato carotenoids: validation of a rapid microextraction technique. J Agric Food Chem 57(19): 8753-8760.
- Tan, S., Ke, Z., Chai, D., Miao, Y., Luo, K. and Li, W. (2021). Lycopene, polyphenols and antioxidant activities of three characteristic tomato cultivars subjected to two drying methods. Food Chem 338: 128062.
- Tohge, T., Scossa, F., Wendenburg, R., Frasse, P., Balbo, I., Watanabe, M., Alseekh, S., Jadhav, S. S., Delfin, J. C., Lohse, M., Giavalisco, P., Usadel, B., Zhang, Y., Luo, J., Bouzayen, M. and Fernie, A. R. (2020). Exploiting natural variation in tomato to define pathway structure and metabolic regulation of fruit polyphenolics in the lycopersicum complex. Mol Plant 13(7): 1027-1046.
- Treutler, H., Tsugawa, H., Porzel, A., Gorzolka, K., Tissier, A., Neumann, S. and Balcke, G. U. (2016). Discovering regulated metabolite families in untargeted metabolomics studies. Anal Chem 88(16): 8082-8090.
- Turner, T. and Burri, B. J. (2012). Rapid isocratic HPLC method and sample extraction procedures for measuring carotenoid, retinoid, and tocopherol concentrations in human blood and breast milk for intervention studies. Chromatographia 75: 241-252.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Gürbüz Çolak, N., Eken, N. T., Frary, A. and Doganlar, S. (2021). Chromatographic Analysis for Targeted Metabolomics of Antioxidant and Flavor-Related Metabolites in Tomato. Bio-protocol 11(5): e3929. DOI: 10.21769/BioProtoc.3929.
Category
Plant Science > Plant biochemistry > Metabolite
Plant Science > Plant breeding
Biochemistry > Carbohydrate
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









