- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Molecular and Phenotypic Characterization Following RNAi Mediated Knockdown in Drosophila
(*contributed equally to this work) Published: Vol 11, Iss 4, Feb 20, 2021 DOI: 10.21769/BioProtoc.3924 Views: 5641
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Immunofluorescent Staining Assay of 3D Cell Culture of Colonoids Isolated from Mice Colon
Trisha Mehrotra [...] Didier Merlin
Mar 5, 2024 2486 Views

Intraepidermal Nerve Fiber Quantification of the Mouse Hind Paw Footpads: A Detailed and Simplified Protocol
Anastasia Yerushkin [...] Amir Dori
Dec 5, 2025 1227 Views
Abstract
Loss of function studies shed significant light on the involvement of a gene or gene product in different cellular processes. Short hairpin RNA (shRNA) mediated RNA interference (RNAi) is a classical yet straightforward technique frequently used to knock down a gene for assessing its function. Similar perturbations in gene expression can be achieved by siRNA, microRNA, or CRISPR-Cas9 methods also. In Drosophila genetics, the UAS-GAL4 system is utilized to express RNAi and make ubiquitous and tissue-specific knockdowns possible. The UAS-GAL4 system borrows genetic components of S. cerevisiae, hence rule out the possibility of accidental expression of the system. In particular, this technique uses a target-specific shRNA, and the expression of the same is governed by the upstream activating sequence (UAS). Controlled expression of GAL4, regulated by specific promoters, can drive the interfering RNA expression ubiquitously or in a tissue-specific manner. The knockdown efficiency is measured by RNA isolation and semiquantitative RT-PCR reaction followed by agarose gel electrophoresis. We have employed immunostaining procedure also to assess knockdown efficiency.
RNAi provides researchers with an option to decrease the gene product levels (equivalent to hypomorph condition) and study the outcomes. UAS-GAL4 based RNAi method provides spatio-temporal regulation of gene expression and helps deduce the function of a gene required during early developmental stages also.
Background
Drosophila melanogaster (fruit fly) is a versatile model organism frequently used in research laboratories. Fruit flies are easy to handle, propagate, and maintain. Moreover, the elaborate yet short life span with high fecundity is an added advantage of Drosophila. The facile nature of Drosophila genetics tools helps develop a comprehensive understanding of a gene function. Since 60% of the Drosophila genes are homologous to human genes, and other advantages mentioned earlier, Drosophila is an obvious model organism of choice to study in vivo gene functions.
The UAS-GAL4 system was used for in vivo activation of transcription of genes. Moreover, it was suggested that antisense RNAs could be expressed using the UAS-GAL4 system to achieve significant inhibition of gene expression (Brand and Perrimon, 1993). For using the UAS-GAL4 system in Drosophila, transgenic fly lines carrying the upstream activating sequence (UAS) capable of regulating RNAi expression are generated. Genome-scale shRNA-dependent RNAi resource was generated for stage-specific effective knockdown of Drosophila genes (Ni et al., 2011). RNAi lines are publicly available from Vienna Drosophila Resource Center (VDRC, maintaining GD and KK RNAi lines) and Drosophila RNAi Screening Center (TRiP and shRNA lines were generated at DRSC and are currently maintained and available from Bloomington Drosophila Stock Center, BDSC). The detailed methodology for the generation of different RNAi lines can be found at the VDRC stock center (https://stockcenter.vdrc.at/control/main) and DRSC (https://fgr.hms.harvard.edu/) websites.
When the RNAi fly line is crossed with the GAL4 driver fly line, the progenies can have both components of the UAS-GAL4 system expressed. The upstream promoter guides GAL4 expression, and wherever the GAL4 is expressed, it binds to the UAS sequence with high affinity, allowing expression of the RNAi component (Figure 1). In the absence of GAL4 expression, the transgenic RNAi flies behave like wild-type due to lack of dsRNA expression. This method allows the knockdown of the desired gene in a spatiotemporal manner. Classically, X-ray mutagenesis, and P-element mobilization have been used to achieve the loss of gene expression. Further, the EMS mutagenesis screens have been utilized to induce point mutation, occasionally leading to truncations affecting gene expression. The RNAi mediated knockdown is rather facile and less labor-intensive.
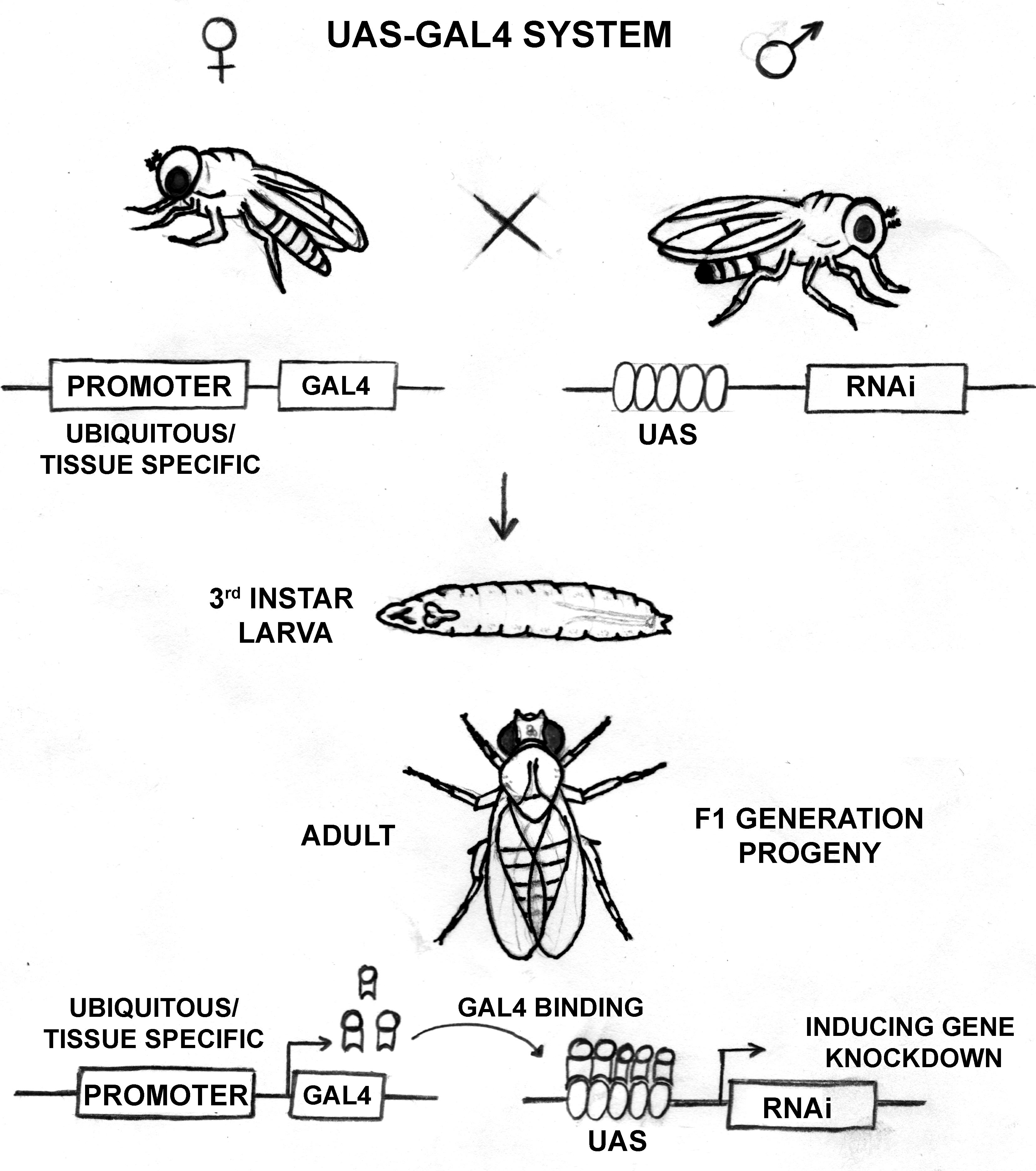
Figure 1. Schematic diagram detailing the functioning of the UAS-GAL4 system in Drosophila Due to varying GAL4 expression degrees in different tissues, a gradient in the knockdown can be seen in different tissues. Besides, temperature-dependent regulation on the GAL4 function provides an advantage of regulating knockdown levels. Together, these handles on gene expression regulation using the UAS-GAL4 system provide more information regarding the functions of our gene of interest. Importantly, this method provides a controlled spatiotemporal regulation on RNAi mediated knockdown and can help study functions of developmentally essential genes, genes with housekeeping functions, and mutations in genes associated with harmful consequences. This method also helps in the functional characterization of a new gene like dElys (Mehta et al., 2020).
A typical qRT-PCR or semiquantitative PCR validates the reduction in gene expression levels upon RNAi mediated knockdown. The knockdown efficiencies can be further assessed by detecting the desired gene product levels by immunostaining and Western blotting methods. We have utilized the RNAi mediated knockdown in salivary glands and assessed its effectiveness using immunostaining or assessing mRNA levels by semiquantitative PCR. We successfully depleted nucleoporins in eyes and wings using the RNAi-mediated knockdown method and reported the importance of nucleoporins in tissue and organism development.
Materials and Reagents
1.5 ml microcentrifuge tubes (Genaxy, catalog number: GEN-MT-150-C-S )
Barrier tips
0.5-10 µl tips (Axygen Scientific, catalog number: TF-300-L-R-S )
1-20 µl tips (Axygen Scientific, catalog number: TF-20-L-R-S )
1-200 µl tips (Axygen Scientific, catalog number: TF-200-L-R-S )
100-1,000 µl tips (Axygen Scientific, catalog number: TF-1000-L-R-S )
Glass vials and bottles
SYLGARD 184 Silicone elastomer (DOW, catalog number: 1673921 )
Micropipette tips (2, 20, 200, 1,000 µl)
0.5-10 µl tips (Axygen Scientific, catalog number: T-300-L-R-S )
1-200 µl tips (Axygen Scientific, catalog number: TR-222-C-L-R-S )
100-1,000 µl tips (Axygen Scientific, catalog number: T-1000-C-L-R-S )
3 cm dish (Eppendorf, catalog number: 00 30700112 )
Food bottles
CO2 pads
Cotton plugs
Corn flour
Table sugar
Yeast extract (HiMedia, catalog number: RM027 )
Agar (Merck, catalog number: A5306 )
Dextrose (HiMedia, catalog number: GRM077 )
Methyl-4-Hydroxybenzoate (Sigma-Aldrich, catalog number: H5501 )
Ortho-Phosphoric Acid (Merck, catalog number: 100573 )
Propionic Acid (HiMedia, catalog number: GRM3658 )
Ethanol (Changshu Hongsheng Fine Chemical CHN01 )
Liquid Nitrogen
RNA isolation kit (Favorgen, catalog number: FATRK001 )
RNaseZAP (Thermo Scientific, catalog number: AM9780 )
DNase (Thermo Scientific, catalog number: EN0523 )
Diethyl pyrocarbonate (HiMedia, catalog number: MB076 )
Ethanol (MP Biomedicals, catalog number: 180077 )
Formamide (Sigma-Aldrich, catalog number: F7503 )
6× DNA loading dye purple (New England Biolabs, catalog number: B7024S )
cDNA synthesis kit (iScript, Bio-Rad, catalog number: 1708891 )
Agarose (Invitrogen, catalog number: 16500500 )
Ethidium bromide (EtBr) (MP Biomedicals, catalog number: 193993 )
Custom Primers (Integrated DNA Technology)
G9-Taq DNA polymerase (GCC Biotech, catalog number: G7115A )
dNTPs solution mix (New England Biolabs, catalog number: N0447L )
Sodium Chloride (Sigma-Aldrich, catalog number: S3014 )
Potassium Chloride (Sigma-Aldrich, catalog number: P5405 )
di-Sodium hydrogen phosphate heptahydrate (Merck, catalog number: 1065751000 )
Potassium dihydrogen phosphate (Sigma-Aldrich, catalog number: P9791 )
Tris-base (Sigma-Aldrich, catalog number: T6066 )
Glacial acetic acid (Merck EMPLURA, catalog number: 1.93402.2521 )
Carbon conductive tape (TED PELLLA Inc. catalog number: 16073-1 )
Aluminum stub (TED PELLLA Inc. catalog number: 16111-9 )
Glutaraldehyde (Sigma-Aldrich, catalog number: G5882 )
Gold target (Quorum Technology inc, Catalog number: SC502-314A )
EDTA (Merck EMPARTA, catalog number: 1.93312.1021 )
Paraformaldehyde (Sigma-Aldrich, catalog number: 158127 )
Triton X-100 (Sigma-Aldrich, catalog number: X100 )
Glass Slide (Borosil, catalog number: 9100P02 )
Coverslips (Borosil, catalog number: 9115S01 )
Bovine serum albumin (Merck, catalog number: 621650500501730 )
Fluoroshield with DAPI (Sigma-Aldrich, catalog number: F6057 )
mAb414 (Biolegend 902901) - mAb414 recognizes FG-repeat rich four distinct nucleoporins of nuclear pores
Anti-dElys (Mehta et al, 2020)
Alexa Fluor-488 (Invitrogen, catalog number: A-11029 )
Alexa Fluor-568 (Invitrogen, catalog number: A-11036 )
Coverslip sealant (Transparent nail polish)
Diethyl ether (Merck, catalog number: 107026 )
Phosphate Buffer Saline (1×) (see Recipes)
PBS-T (see Recipes)
Fly food ingredients (see Recipes)
RNA loading dye (see Recipes)
DEPC treated water (see Recipes)
TAE Buffer (50×) (see Recipes)
4% Paraformaldehyde (see Recipes)
Control Wild type (w1118)
Driver lines (in house generated combinations)
+/+; Actin5C-GAL4/CyO-GFP; UAS-Dicer/UAS-Dicer
+/+; wingless-GAL4/wingless-GAL4; UAS-Dicer/UAS-Dicer
+/+; eyeless-GAL4/eyeless-GAL4; UAS-Dicer/UAS-Dicer
+/+; v103547/v103547; UAS-Dicer/UAS-Dicer (dElys RNAi KK line combined with UAS-Dicer, in house generation)
dElys RNAi KK line (VDRC, VDRC ID: 103547)
Nup160 RNAi GD line (VDRC, VDRC ID: 21937)
Sec13 RNAi KK line (VDRC, VDRC ID: 110428)
Nup107 RNAi GD line (VDRC, VDRC ID: 22407)
Equipment
Micropipette (Nichiriyo, Gilson, Corning)
BOD Incubators
Millipore water purification unit
Genova Nanodrop (Bibby Scientific, model: 737501 )
Forceps Dumont 5 (Fine Science Tools, catalog number: 11295-10 )
Forceps Dumont 55 (Fine Science Tools, catalog number: 11295-51 )
Scissors (Fine Science Tools, catalog number: 91500-09 )
Leica DM2500
Fluorescent stereomicroscope (Leica, model: M205 FA )
Confocal Laser Scanning Microscope (Zeiss, model: LSM780 )
Stereomicroscope (Leica, model: S6E )
Scanning Electron microscope (Zeiss, model: Gemini II Ultra plus )<
Tabletop centrifuge (Eppendorf, model: 5424, Thermo Scientific MicroCL 21R )
PCR machine (Applied Biosystems, model: 2720 Thermal cycler )
Water bath (Grant-Bio, model: PSU-10i )
Gel running apparatus (CBS Scientific, model: GCMGU-202T )
Power packs (CBS Scientific, model: EPS 300 )
UVP MultiDoc-It Digital Imaging System (UVP, catalog number: 97-0192-02 )
Software
ZEN 3.2 Blue edition (Zeiss)
ImageJ/Fiji (Free, NIH)
Procedure
The UAS-GAL4 dependent and RNAi mediated knockdown of genes in Drosophila requires several steps. It includes steps like rearing the organism, setting up genetic crosses, harvesting larvae of desired genetic combination, RNA isolation and knockdown assessment, dissections, immunostaining, and phenotypic analysis. Mentioned below is the general scheme of tissue preparation and processing (Figure 2).
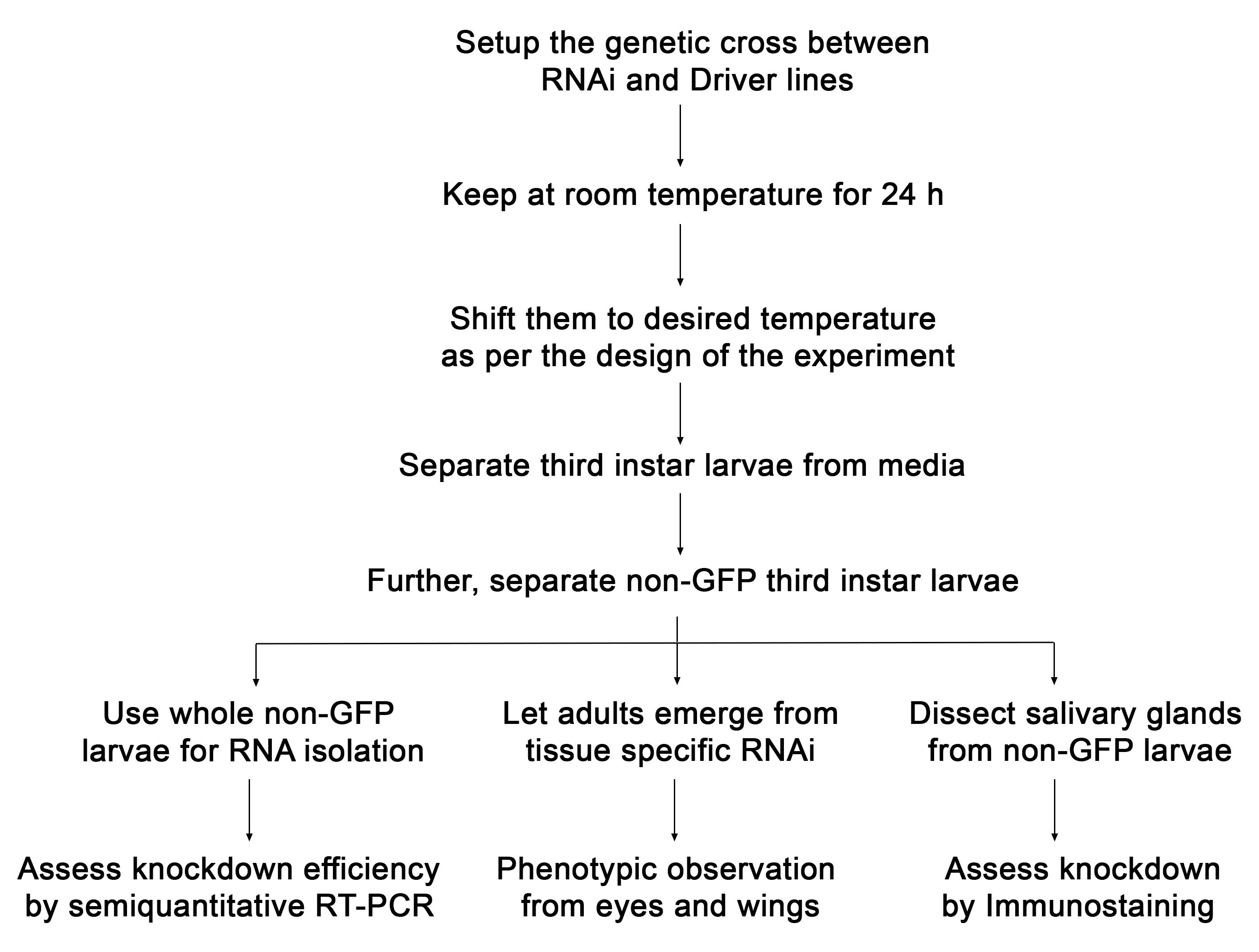
Figure 2. A flow chart with details of the protocol
Genetic crosses for RNAi mediated knockdown
Grow +/+; Actin5C-GAL4/CyO-GFP; UAS-Dicer/UAS-Dicer flies in large numbers in vials containing fly media.
Collect virgin females having the GAL4 driver in a separate media vial.
Grow RNAi and wild type (w1118) flies in separate media vials to get sufficient male flies.
Place 6-7 virgin driver females and 2-3 UAS-RNAi males of the same age (3-4 days old) inside a fresh media vial to establish a mating cross.
For the control cross, use w1118 males instead of UAS-RNAi males and set up the cross with virgin females as in Step A4.
Incubate vials with flies for the genetic cross at room temperature for 24 h.
According to the experimental design, shift them to 28 °C, 25 °C, or 18 °C.
Note: Incubation at a higher temperature allows higher GAL4 activity inducing increased RNAi expression.
Keep crosses in an incubator set at the desired temperature until the third instar larvae are visible.
Remove adult flies and put 2-3 ml water or 2% sucrose solution to harvest larvae from the media's top layer.
Note: Steps A8 to A13 are in Figure 3.
Using a paintbrush, mix the upper layer of food and water and collect it in a clean plastic dish.
Separate third instar larvae in a clean distilled water/phosphate buffer saline containing plate.
Wash collected larvae 2-3 times with water to clean them.
Segregate non-GFP larvae (+/+; Actin5C/RNAi; UAS-Dicer/+, indicating the desired genetic combination of Driver and RNAi) from GFP (+/+; RNAi/CyoGFP; UAS-Dicer/+) larvae for subsequent processing.
For the control, use non-GFP larvae from the control cross as described in Step A5.
Dissect the salivary gland or the head complex of Actin5C-GAL4 driven RNAi from the non-GFP third instar larvae in PBS.
Note: Refer to Figure 3 below and Steps 1-5 mentioned in Part-C for details of salivary gland dissection.
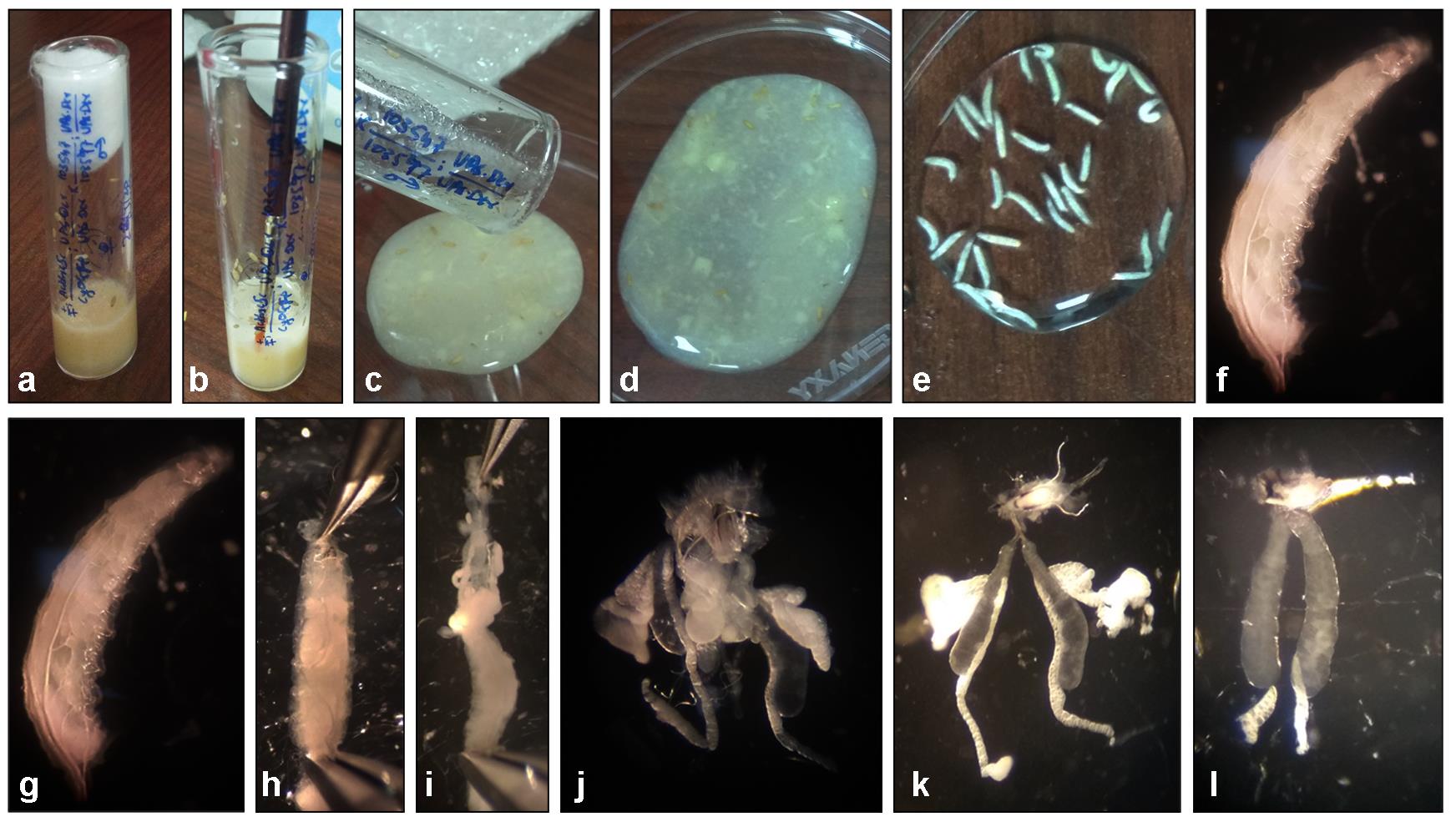
Figure 3. Steps in the isolation of Salivary glands from Drosophila third instar larva. Upper panels (a-f), Steps in larva separation and cleaning; (a) Larvae and Pupae in food vial, (b) Squirting water and suspending the top layer of food and larva with a brush, (c) Decanting suspension in a petri-plate, (d) Fly food and third instar larva suspension, (e) Cleaned larva collected in distilled water, (f) A third instar larva. Lower panels (g-l) Steps in larval dissection and salivary gland isolation (g) A third instar larva, (h) Holding a third instar larva with forceps, (i) Pulled out salivary glands and abdominal parts of the third instar larva, (j) Head complex having the brain, imaginal disc, and salivary gland (k) Salivary gland with associated fat bodies, (l) Dissected and cleaned up pair of salivary glands with an attached mouth hook.
RNA isolation
The most critical step of this method is RNA isolation. RNAi mediated knockdown reduces the specific RNA levels to affect the gene function. RNA digesting enzyme, RNaseA is a very stable and active protein; thus, taking proper precaution and using RNase inactivating solution like RNaseZAP is necessary.
Prepare DEPC treated water for use (see Recipes).
Make sure to use barrier tips, RNase-free microcentrifuge tube (MCT), and glassware.
Put larvae inside a clean RNase-free MCT, flash freeze, and store for RNA isolation.
Dissect sufficient salivary glands from larvae and flash freeze glands inside liquid nitrogen.
Crush the salivary gland/larva using RNase free pestle and follow the manufacturer's protocol for RNA extraction given in the kit.
Crush flash-frozen tissues directly using a pestle or first add lysis solution from the RNA isolation kit and then crush the tissue or larva in the solution.
If adding the lysis solution before crushing, add half of the lysis buffer's final volume, and properly crush the tissue/larva. Subsequently, add the remaining lysis buffer.
Check the purity of RNA using a UV-VIS spectrophotometer/nanodrop. A260/280 and A260/230 ratio equal to 2.0-2.2 suggests pure RNA. Lower values of these ratios indicate the presence of contaminants like proteins, carbohydrates, or phenol.
In an MCT, heat the purified RNA sample at 65 °C for 10 min in a 25% formamide containing RNA loading dye.
Keep RNA sample tubes on ice for 2 min and load immediately on the EtBr containing 1% agarose gel made in Tris-Acetate EDTA (TAE) buffer and run the gel to check the integrity of the isolated RNA.
Prepare cDNA as per instruction given with the BioRad cDNA synthesis kit and store at -20 °C until further use.
A typical cDNA synthesis reaction in 20 µl final volume contains 1 µg purified RNA, 1× iScript reaction mixture, 1 µl of iScript reverse transcriptase enzyme.
Reaction steps include 5 min of priming at 25 °C, 20 min of reverse transcription at 46 °C, and 1 min of inactivation at 95 °C.
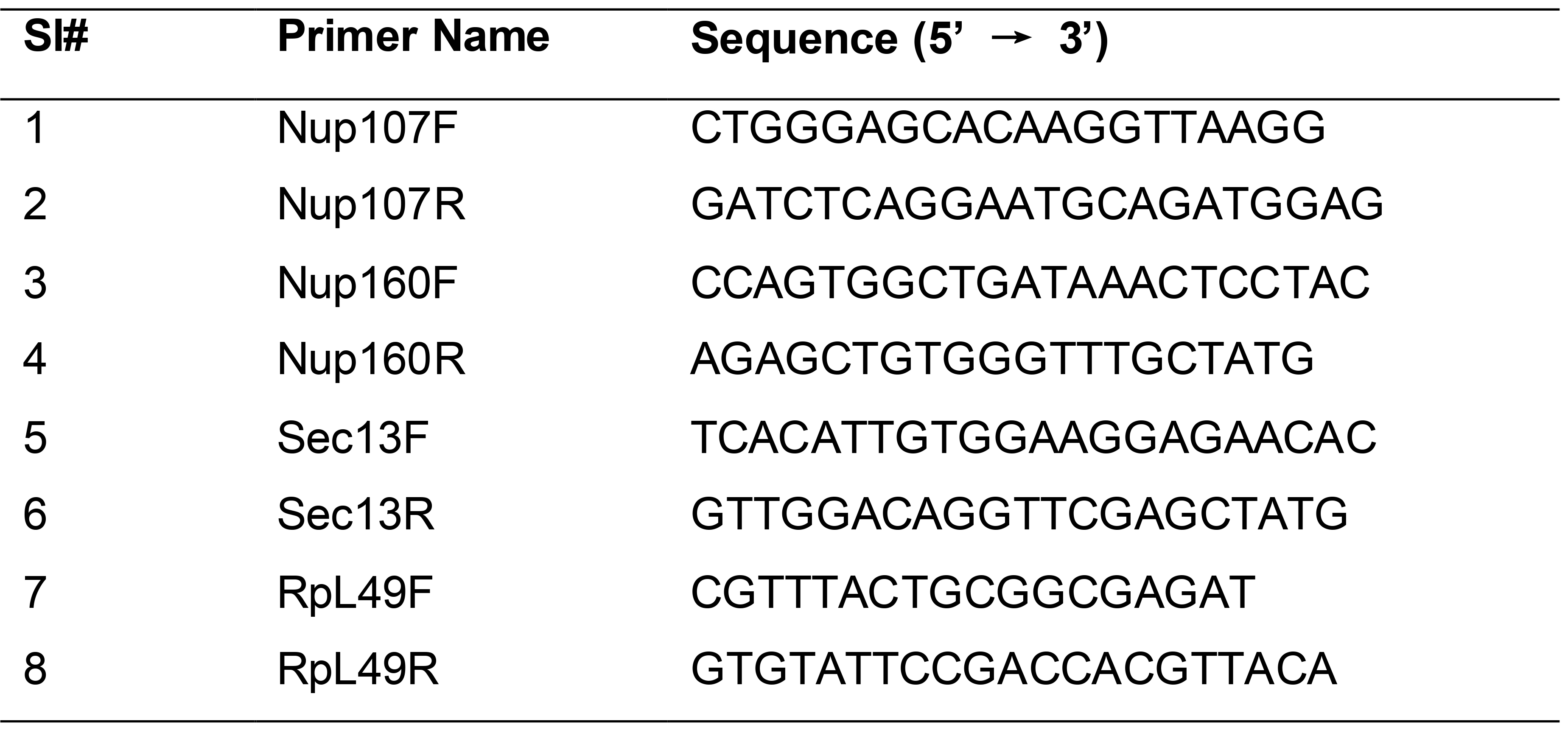
Use gene-specific primers (200 nM) on prepared cDNA and run a typical PCR reaction (see Table 1 for primer sequences).
Note: We used a standard annealing temperature (Ta) of 55 °C for all semiquantitative PCR.
Use RpL49 primers as an internal control gene in the PCR reaction (see Table 1 for primer sequences).
Table 1. List of primers used for assessing the knockdown levels
Load 50% of PCR product on EtBr containing 1% Agarose gel to assess the extent of knockdown (Figure 4).
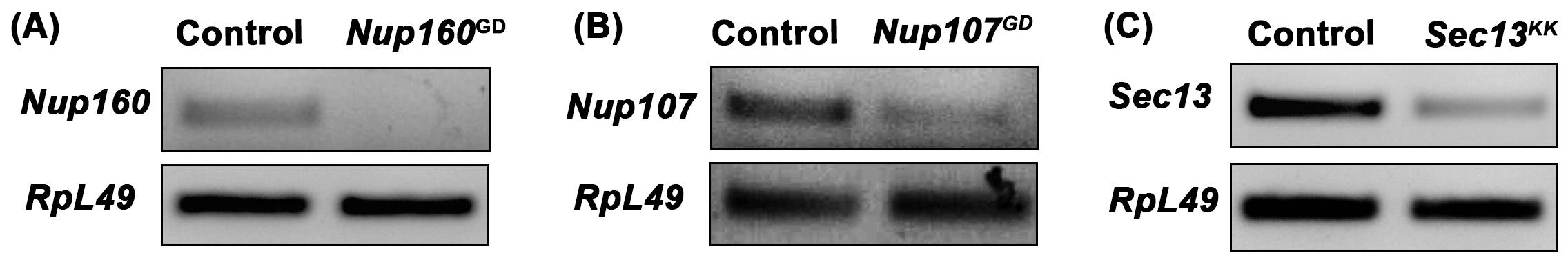
Figure 4. Semiquantitative PCR based assessment of knockdown. cDNAs prepared from Control and nucleoporin knockdown used in a PCR reaction. Upper panels show levels of Nup160 (A), Nup107 (B), and Sec13 (C) from control and knockdown tissues. RpL49 served as an internal loading control between control and knockdown samples.
Dissection and Staining of Salivary gland
Salivary gland is post-mitotic tissue and carries large nuclei. It possesses polytene chromosomes, which serve as excellent model tissue in several cytological studies and gene expression analyses. The loss of nucleoporins and consequent perturbations in nuclear pore assembly and functions can be studied using immunostaining. dElys was knocked down in the salivary glands using RNAi. Subsequent steps in the dissection of salivary glands in immunostaining are the following.
Dissect out salivary glands in PBS on a silicone elastomer plate from 20 third instar larvae.
Note: Replace PBS solution after every 2-3 dissections.
Grab the larva approximately at the middle portion of its size using forceps.
Use Dumont 5 forceps to grab the mouth-hook and pull out the head complex.
Simultaneously remove the gut region and other carcass tissues using Dumont 55 forceps.
Use pointed Dumont 5 forceps to grab the head complex by mouth-hook all this while.
Clean the dissected salivary gland by removing the brain, imaginal discs, and fat tissue and ensure that the mouth-hook remains attached.
After dissecting each salivary gland, immediately transfer it to an MCT containing 1 ml of 4% paraformaldehyde.
After all the salivary glands are dissected and collected, keep the MCT on an orbital shaker with gentle agitation for 40 min at room temperature.
Note: Fixation should not go beyond 1 h; otherwise, the tissue becomes fragile, and staining is affected.
Remove paraformaldehyde and wash tissues three times with PBS-T for 10 min each.
Note: Do all the wash steps carefully to avoid the loss of salivary glands. It applies to all subsequent steps.
Dissecting the salivary gland with the mouth hook attached to it gives an additional advantage that we can easily see the salivary gland, and thus their loss will be restricted.
Block salivary glands in 3% normal goat serum containing PBS-T or 3% Bovine Serum Albumin (BSA) in PBS-T.
Keep at room temperature for 1 h on an orbital shaker with 110 rotations per minute (rpm).
Note: Perform all incubations/wash on an orbital shaker set at 110 rpm.
Add 200 µl of anti-dElys and mAb414 primary antibodies containing solution (1:800 dilution of each of the two antibodies prepared in PBS-T).
Incubate overnight at 4 °C on a shaker.
Wash samples three times with 200 µl of PBS-T for 10 min each at room temperature (25 °C).
Add 200 µl of Alexa Fluor-488 or Alexa Fluor-568 conjugated secondary antibodies dilution (1:800 dilution prepared in PBS-T) and incubate at room temperature (25 °C) for 1.5-2.0 h.
Wash samples three times with 200 µl of PBS-T, 10 min each at room temperature (25 °C).
Put 15 µl of Fluoroshield with DAPI mounting medium.
Pick all salivary glands using a cut 200 µl pipette tip.
Note: Cut the sample inlet end of the fine 200 µl pipette tip to avoid any tissue damage.
Place salivary glands on a previously cleaned slide and spread the tissue gently using Dumont 55 forceps.
Note: Do not damage the tissue while spreading. Use the mouth hook to grab the tissue, only if required.
Carefully place a clean coverslip on the tissue.
Avoid trapping air bubbles between the slide and coverslip.
Gently remove the excess mounting medium using tissue paper and seal the coverslip edges with transparent nail polish.
Observe processed tissue under a fluorescent Leica microscope.
On Zeiss LSM780 , use the control slide to adjust settings and focus in a field with a clear view of cells.
Scan the field having salivary gland nuclei under 63× objective in a Confocal Laser Scanning Microscope LSM780 .
Capture images from different visual fields of the control slide (Figure 5).
Reuse the same settings for the knockdown samples also.
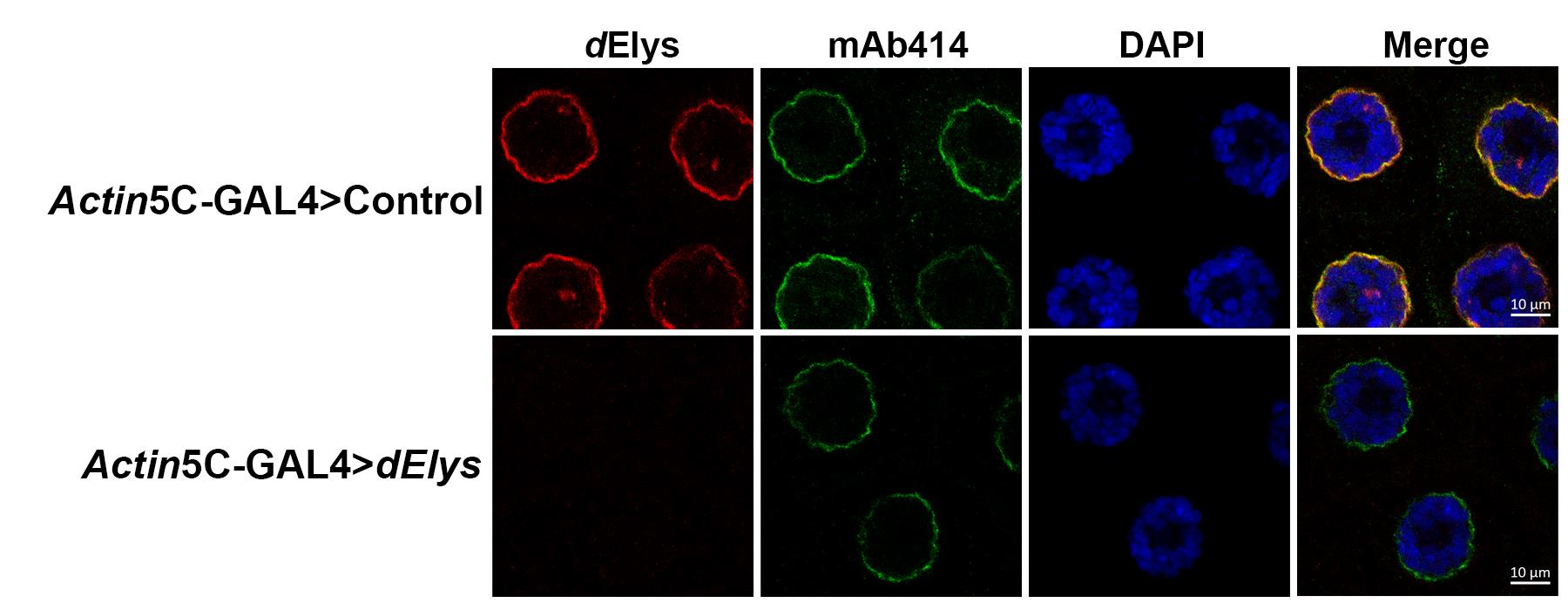
Figure 5. RNAi mediated dElys knockdown depleted dElys signals from salivary gland nuclei. The salivary glands dissected from Control RNAi (upper panels) and dElys RNAi (lower panels) third instar larvae and stained with dElys (red, first panels), mAb414 (green, second panels), DAPI (blue, third panels). The fourth panels represent the merged images. Scale bars =10 μm.
Wing and Eye specific knockdown
Set up crosses between nup160 RNAi and +/+; Ey-GAL4/Ey-GAL4; UAS-Dicer/UAS-Dicer (for eye-specific knockdown) or nup160 RNAi and +/+; wg-GAL4/Wg-GAL4; UAS-Dicer/UAS-Dicer (for wing-specific knockdown).
Transfer crosses to a lower temperature (18 °C). The lower temperature reduces the GAL4 activity.
Reduced GAL4 activity at 18 °C causes a reduction in knockdown levels and allows adult fly emergence.
Dissection and visualization of eyes and wings
Visualization of eyes
Drosophila compound eyes are complex yet pertinent tissue to observe growth defects upon RNAi mediated knockdown. A definite transcriptional paradigm regulates the arrangement of each ommatidium in the compound eye during development. Any perturbation in the ommatidium arrangement, their smooth morphology, and the presence of a bristle with each ommatidium due to RNAi mediated knockdown serves to indicate the importance of the gene function. After knocking down the Nup160 using RNAi, process wings in the following manner. If the gene of interest induces embryonic to larval lethality, eye-specific knockdown helps assess the gene function. After knocking down the Nup160 using RNAi, process compound eyes in the following manner.
Collect adult flies in an empty vial.
Apply 100-200 µl of diethyl ether on cotton and plug the fly containing vial to anesthetize them.
Orient them properly (compound eye facing up) under Stereomicroscope Leica S6E .
Fix the position of the fly using carbon conductive tape for steady observation of eyes.
Observe eye morphology using Leica fluorescent stereomicroscope M205 FA at ×123 magnification (left panels in Figure 6A).
SEM analysis of eye phenotype
Anesthetize the flies using diethyl ether. Flash freeze them and separate heads from the body.
Fix head tissues with 2.5% Glutaraldehyde in PBS for 2 h at 4 °C.
Wash with PBS and 4% sucrose solution.
Dehydrate tissue using a series of graded ethanol wash (single wash with 25%, 50%, 75% ethanol, and twice with 100% ethanol) for 2 h each at room temperature.
Keep inside a lab desiccator for critical point drying.
Mount dried tissue on aluminum stubs with carbon-conductive tape.
Coat the flies with gold particles in a sputter coating apparatus.
Image using Zeiss Gemini II FESEM with In lens detector (middle and right panels in Figure 6A).
Visualization of wings
Drosophila adult wings serve as an excellent tissue to observe the developmental defects induced by perturbation in a gene function. The changes in bristles, wing veins, and notched wings are distinct phenotypes seen with the knockdown of genes. Wing imaginal discs are present in the third instar larva. If the gene of interest induces post-larval lethality, wing imaginal discs are routinely analyzed. After knocking down the Nup160 using RNAi, process wings for visualization in the following manner.
Follow Steps E1a and E1b of the visualization of eyes procedure.
Cut the wings with scissors and place them on a clean slide.
Put coverslip on top of the wing tissue and seal.
Observe under upright light microscope Leica DM2500 at 10× magnification (panels in Figure 6B).
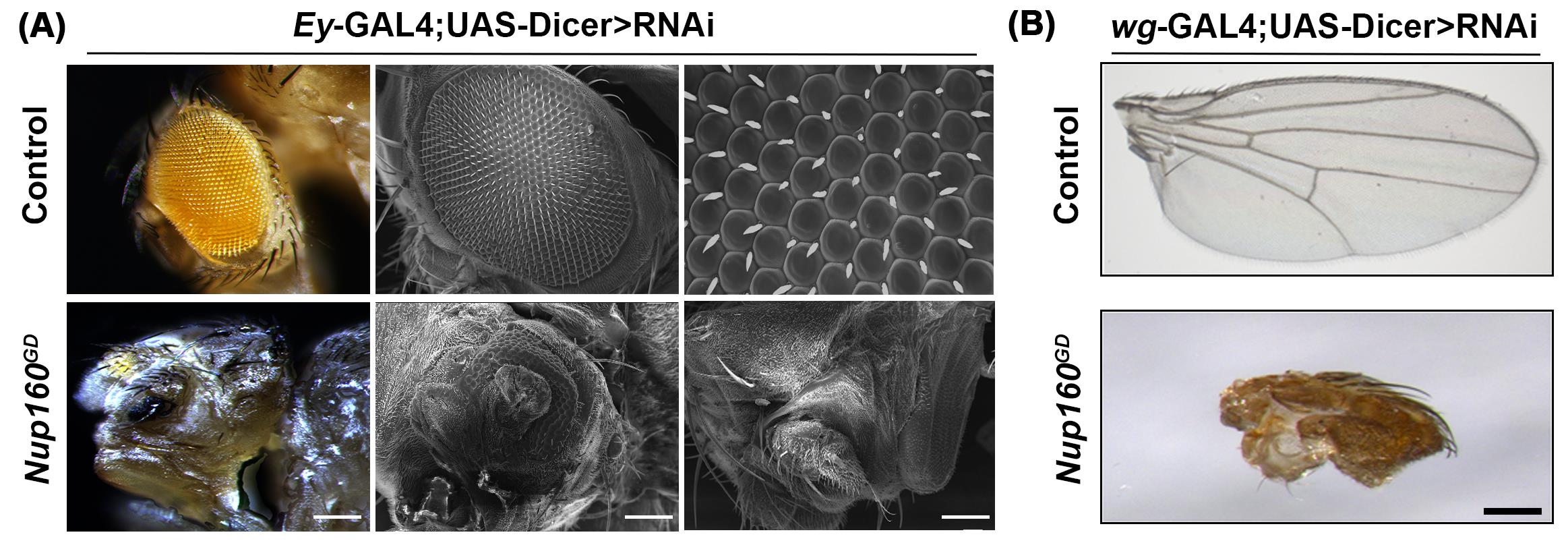
Figure 6. Knockdown of Nup160 in eyes and wings induces developmental defects. (A) Eye specific knockdown of Nup160 using Ey-GAL4 and visualization of the compound eye under bright field light microscope and SEM. (B) Wing specific knockdown of Nup160 using Wg-GAL4 and visualization of wings under bright field light microscope. The control knockdown is for comparison. Scale bars (A) are 200 μm in the left panels, 20 μm in the middle panels, and 2 μm in the right panels. The scale bar in (B) is 200 μm.
Data analysis
We have utilized RNAi mediated knockdown of dElys in Drosophila and have explored the functional significance of dElys in Drosophila development. Observations made regarding dElys function highlight that dElys is important for embryonic development, nuclear pore assembly, nucleocytoplasmic shuttling of the developmentally regulated molecule, Dorsal. The genetic crosses between flies carrying RNAi against dElys, Nup160, Nup107, and Sec13, and promoter-specific GAL4 sequences produced desired progenies where both RNAi and UAS-GAL4 were present in the same fly. Progenies were selected at the third instar larva stage based on the absence of GFP expression (non-GFP larva). In salivary gland specific analysis, we dissected paired salivary glands from non-GFP larvae (Figure 3). Use of Actin5C-GAL4 for knockdown of genes of interest, Nup160, Nup107, and Sec13 successfully achieved significant gene expression knockdown. The semiquantitative RT-PCR analysis established that these genes are efficiently depleted (Figure 4).
We achieved a knockdown ranging from 50-80% using RNAi for different genes. Determination of RpL49 gene levels by PCR helped in making these precise calculations. In conditions where the ubiquitous knockdown appears lethal or harmful, we explored the option of tissue-specific knockdown. Additionally, when a gene's function confines to a particular tissue, the power of tissue-specific promoter mediated knockdown can be utilized. Any potential off-target gene of utilized RNAi must also be analyzed to build a correlation between gene and phenotype. Mehta et al., 2020 reported that two off-targets of dElys RNAi were not perturbed significantly. Simultaneously, a gentle lowering of RNA transcript for a dosage-sensitive protein can also bring about the observable phenotypes.
dElys levels reduced strongly under dElys knockdown conditions (Actin5C>dElys). The ubiquitous knockdown of dElys and subsequent analysis of the salivary gland nuclei indicated that nuclear pore assembly is affected by dElys depletion. Under similar conditions, dElys levels are unperturbed in control knockdown (Actin5C>Control) (Figure 5). mAb414 antibody recognizes four distinct FG-repeat rich nucleoporins. A noticeable reduction in mAb414 staining of nuclear pores is also evident upon dElys RNAi. Based on these observations, we suggest an essential role for dElys in nuclear pore assembly in Drosophila.
We further utilized the UAS-GAL4 system's power for RNAi mediated knockdown of Nup160 in Drosophila eye and wings. Eye-specific Ey-GAL4 and wing-specific wg-GAL4 promoters depleted Nup160 to produce significant perturbation in the development of these two tissues. In Nup160 knockdown animals, the entire compound eye is shrunken and seems absent. The SEM imaging of knockdown eyes shows drastic changes compared with eye-specific control knockdown (Figure 6, panel A). Similarly, the wing-specific knockdown of Nup160 caused a reduction in the wing blade size, and the veins are completely missing (Figure 6, panel B).
The observations made with RNAi mediated knockdown of nucleoporin genes dElys and Nup160 established the robustness of this gene knockdown paradigm. Further, the tissue-specific knockdown and analysis from salivary glands, eyes, and wings highlighted the functional significance of nucleoporins in Drosophila development.
Recipes
Phosphate Buffer Saline (1×)
Sodium Chloride 137 mM
Potassium Chloride 2.7 mM
di-Sodium hydrogen phosphate heptahydrate 10 mM
Potassium dihydrogen phosphate 1.8 mM
PBS-T
Lukewarm the falcon tube containing 1× PBS and add Triton X-100 (100%) to a final concentration of 0.2% with a cut pipette tip.
Fly food ingredients
Ingredient Amount/liter
Corn flour 80 g
Sugar 40 g
Dextrose 20 g
Yeast Extract 15 g
Agar 10 g
Methyl-4-hydroxybenzoate 1 g
Propionic acid 4 ml
Orthophosphoric acid 0.6 ml
Ethanol absolute 10 ml
Weigh corn flour, sugar, dextrose, yeast extract, Agar separately, and add to the flask.
Note: Follow the ratio of different components mentioned in the table above.
Autoclave at 121 °C for 20 min and let it cool for some time till the temperature reaches ~50-55 °C.
Meanwhile, weigh 1 g of methyl-4-hydroxybenzoate and dissolve in 10 ml ethanol.
Once the medium has cooled down to ~50-55 °C (check it with a thermometer), add methyl-4-hydroxybenzoate solution, propionic acid, and orthophosphoric acid to the media.
Mix it thoroughly and pour 8-10 ml into each glass vial.
Note: Avoid the incorporation of bubbles. Do not let it cool below ~50 °C; otherwise, Agar will start to solidify.
Once the media has solidified inside the vial, put a tight cotton plug on each vial, and use it as required.
Note: Keep vials inside a cage for cooling. The cage helps avoid contamination of vials by stray flies in the room.
RNA loading dye
50 µl Formamide
35 µl 6× DNA loading dye
15 µl Ultrapure water (to make up the final volume)
DEPC treated water
Add 1 ml of DEPC to 1,000 ml of ultrapure water (0.1%).
Mix well, cover the bottle and leave it overnight at room temperature.
Autoclave two times, let it cool down to room temperature.
Make aliquots and store them at -20 °C for use.
TAE Buffer (50×)
Add 242 g of Tris base to 700 ml of double-distilled water.
Add 57.1 ml of Glacial acetic acid carefully.
Mix 100 ml of 0.5 M EDTA having pH 8.0 to it.
Adjust volume to 1,000 ml.
Note: The pH should be around 8.5 and need not be adjusted.
Use 20 ml of 50× TAE stock to make a 1 liter of 1× TAE buffer.
1× TAE buffer will have the following final concentrations: 40 mM Tris base, 20 mM Acetic acid, and 1 mM EDTA
4% Paraformaldehyde
Take 50 ml of 1× PBS in a clean glass bottle.
Add 4 g of paraformaldehyde while keeping the bottle on the heating plate at ~60 °C
Note: Make sure that to work inside a ventilated hood as paraformaldehyde is hazardous.
Add 1 N NaOH slowly and dropwise to dissolve all the paraformaldehyde.
When all paraformaldehyde dissolves in PBS, adjust the pH to 7.2-7.4 using 1 N HCl.
Adjust the final volume to 100 ml using 1× PBS.
Filter and store paraformaldehyde as 10 ml aliquots at 0-4 °C.
Note: Do not store the paraformaldehyde solution for more than a month.
Acknowledgments
This protocol is originally published in a research manuscript by Mehta et al. (2020) and was supported by the Science and Engineering Research Board (SERB, EMR/2016/001819). We thank Ms. Jyotsna Kawadkar for the accurate illustration of the UAS-GAL4 system in Drosophila. We also thank IISER Bhopal, the Fly facility at IISER Bhopal, and DST-FIST Live cell imaging and SEM facilities at IISER.
Competing interests
No competing interest to declare.
Ethics
No human subjects or mouse model was used in the study.
For rearing, maintaining, and experimenting with Drosophila, work followed the applicable parts of animal Ethics guidelines of the host Institute.
References
- Brand, A. H. and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118(2): 401-415.
- Mehta, S. J. K., Kumar, V. and Mishra, R. K. (2020). Drosophila ELYS regulates Dorsal dynamics during development. J Biol Chem 295(8): 2421-2437.
- Ni, J.-Q., Zhou, R., Czech, B., Liu, L.-P., Holderbaum, L., Yang-Zhou, D., Shim, H.-S., Tao, R., Handler, D., Karpowicz, P., Binari, R., Booker, M., Brennecke, J., Perkins, L. A., Hannon, G. J. and Perrimon, N. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8(5): 405-407.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Mehta, S. J. K., Joshi, P. A. and Mishra, R. K. (2021). Molecular and Phenotypic Characterization Following RNAi Mediated Knockdown in Drosophila. Bio-protocol 11(4): e3924. DOI: 10.21769/BioProtoc.3924.
- Mehta, S. J. K., Kumar, V. and Mishra, R. K. (2020). Drosophila ELYS regulates Dorsal dynamics during development. J Biol Chem 295(8): 2421-2437.
Category
Developmental Biology > Cell signaling > Apoptosis
Cell Biology > Cell staining > Protein
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









