- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Extraction and Quantification of Sphingolipids from Hemiptera Insects by Ultra-Performance Liquid Chromatography Coupled to Tandem Mass Spectrometry
Published: Vol 11, Iss 4, Feb 20, 2021 DOI: 10.21769/BioProtoc.3923 Views: 5229
Reviewed by: Agnieszka ZienkiewiczMarc-Antoine SaniIstvan Stadler

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Identification of R-loop-forming Sequences in Drosophila melanogaster Embryos and Tissue Culture Cells Using DRIP-seq
Célia Alecki and Nicole J. Francis
May 5, 2021 7125 Views

Preparation of Drosophila Larval Blood Cells for Single-cell RNA Sequencing
Sudhir Gopal Tattikota and Norbert Perrimon
Aug 20, 2021 3778 Views

The Centriole Stability Assay: A Method to Investigate Mechanisms Involved in the Maintenance of the Centrosome Structure in Drosophila Cultured Cells
Mariana Lince-Faria [...] Ana Pimenta-Marques
Jun 5, 2025 2201 Views
Abstract
Sphingolipids are major structural components of endomembranes and have also been described as an intracellular second messenger involved in various biological functions in all eukaryotes and a few prokaryotes. Ceramides (Cer), the central molecules of sphingolipids, have been depicted in cell growth arrest, cell differentiation, and apoptosis. With the development of lipidomics, the identification of ceramides has been analyzed in many species, mostly in model insects. However, there is still a lack of research in non-model organisms. Here we describe a relatively simple and sensitive method for the extraction, identification, and quantification of ceramides in Hemiptera Insects (brown planthooper), followed by Ultra-Performance Liquid Chromatography coupled to tandem mass spectrometry (UPLC-MS/MS). C18 is used as the separation column for quantitative detection and analysis on the triple quadruple liquid mass spectrometer. In this protocol, the standard curve method is adopted to confirm the more accurate quantification of ceramides based on the optional detection conditions.
Keywords: Hemipetera InsectsBackground
Sphingolipids are the second largest group of membrane lipids in living organisms and play an important role in many aspects of cell structure, metabolism, and regulation (Lahiri and Futerman, 2007). At first, it was thought that sphingolipids were a complex family of structurally related molecules, but more and more studies have shown that sphingolipids are involved in numerous cellular processes (Mao and Obeid, 2008). Ceramides (Cer) are essential bioactive lipids implicated in various cell biological processes ranging from cell growth regulation to cell death and senescence (Futerman and Hannun, 2004; Hannun and Obeid, 2008) through influencing of multiple signaling pathways. Although the physiological roles of ceramides are widely reported, few studies have described the extraction, identification, and quantification, thus, analysis of ceramides has gained significant interest in investigating the physiological functions of sphingolipid metabolism in Hemiptera Insects.
Currently, various methods have been described for this purpose, such as Diacylglycerol (DAG) Kinase assay (Preiss et al., 1987), Thin-layer chromatography (TLC) (Gorska et al., 2002), Gas chromatography mass spectrum (GC-MS) (Tserng et al., 2003), High-performance liquid chromatography (HPLC) (Yano et al., 1998; Dobrzyn and Gorski, 2002). In the beginning, DAG kinase assay was commonly used for Cer quantitation, but the specificity has been questioned (Watts et al.,1997). Thin layer chromatography was the method of choice, but the resolution, sensibility, and separation were limited, resulting in inefficient separation of similar molecules (Bielawski et al., 2010). Despite the high sensitivity of chromatographic analysis, this method had some limitations, such as the need for standard substances and derivatization (Dobrzyn et al., 2004). High-performance liquid chromatography (HPLC) was introduced to obtain ceramides separation with higher resolution, but complex samples like tissue extracts, therefore, produced many unspecific signals that did not provide any information concerning the metabolism of molecular species by HPLC (Yano et al., 1998; Bode and Graler, 2012).
Given the ultra performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS), it is more efficient, rapid, and sensitive, improving the separation condition of extremely complex samples and reducing matrix interference (Cutignano et al., 2010). Therefore, the current choice method is the analysis of ceramides by Ultra Performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS). This protocol provides a relatively rapid and reproducible method. Moreover, this method can be used to profile ceramides from the extraction of the plant sample. It has evolved as the method of choice to detect sphingolipids metabolites due to its high sensitivity and superior specificity. The content of ceramide is determined and a quantitative system of sphingolipids in Hemiptera Insects is established, which lay a foundation for understanding the metabolic process and elucidating the biological function of each component. This method was described and used successfully to extract other sphingolipids in previously published studies (Bielawski et al., 2010; Shi et al., 2018 and 2019).
Materials and Reagents
1.5 ml Eppendorf tubes (Axygen, catalog number: MCT-150-C )
1.0 mm Ceramic Beads (Nalgene, catalog number: 150010C )
2 ml Micro tube (Sarstedt, catalog number: 72.609 )
Pipette tips (Axygen, catalog numbers: T-300 , T-200-Y , T-1000-B )
Glass Centrifuge Tubes (VWR International, catalog number: 734-4240 )
Nitrogen gas (> 99% Purity) (any brand will suffice)
Isopropyl alcohol (Sangon Biotech, catalog number: A503069 )
Ethyl acetate (Sangon Biotech, catalog number: A507048 )
Liquid nitrogen (any brand will suffice)
HPLC-grade methanol (Sigma-Aldrich, catalog number: 34806 )
Formic acid (Sangon Biotech, catalog number: A503066 )
MilliQ Water (Millipore, catalog number: Direct-Q3 )
Standards (see Table 1)
Note: All Ceramides standards list is shown in Table 1.
Solvent extraction solution A (see Recipes)
Solvent extraction solution B (see Recipes)
Mobile phase A (see Recipes)
Mobile phase B (see Recipes)
Internal standard (Avanti company) (see Recipes)
Table 1. Example of ceramides standards list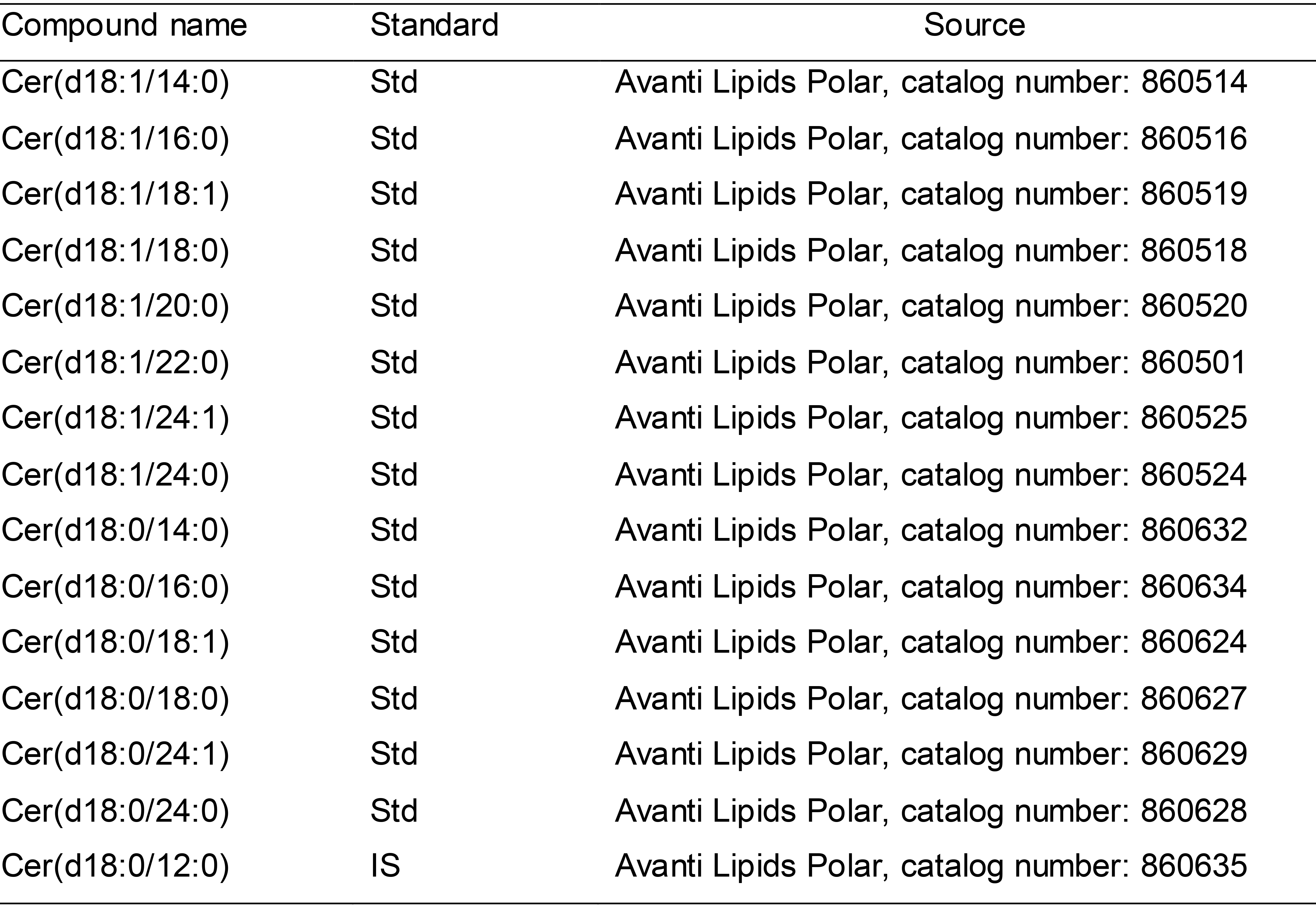
Std: Standard; IS: Internal standard
Equipment
Nitrogen evaporator N-EVAP (Organomation, model: HGC-24A )
Centrifuge (Eppendorf, model: 5430R )
Autoclave (SANYO, model: MLS-3780 )
UHPLC-Q-TOF-MS/MS system (AB SCIEX, Framingham, MA, USA)
UHPLC column (Zorbax sb-C8, 2.1 × 150 mm, 3.5 μm; Agilent, Palo Alto, CA, USA)
Ivory PTFE/red silicone rubber septa (Agilent Technologies, catalog number: 5182-0731 )
2 ml amber screw vial with patch USP 1 expansion (HAMAG Technologies, catalog number: HM-0716H )
Blue open-topped polypropylene cap and white PTFE/red Scilicone septa (HAMAG Technologies, catalog number: HM-0722 )
250 μl clear glass pulled conical-bottom (HAMAG Technologies, catalog number: HM-2085 )
Analytical balance (METTLER TOLEDO, model: XS105 )
Tissue homogenizer (MP Biomedicals, USA, FastPrep-24 )
Oven (Bluepard, model: BPG-9040A )
Vortexer (Germany, IKA, model: vortex 2 )
-80 °C freezer
Software
PeakView (AB Sciex)
Excel software (Microsoft office 2010)
Data Processing software (DPS)
Procedure
Insect samples collection
The laboratory strain of N. lugens (brown planthopper) used in this study originated from a field population in the Huajiachi campus of Zhejiang University, Hangzhou, China. The BPHs (brown planthopper) were reared on susceptible rice seedlings cv.Taichung Native 1 (TN1) at 27 ± 1 °C, 70% relative humidity and a 16:8 h light:dark photoperiod.
About 2 g fresh weight of insects was determined and collected at different development stages (e.g., eggs, first-fifth instar nymphs, female and male adults) in labeled 2 ml tissue grinding tubes. Samples were then stored at -80 °C after quickly freezing in liquid nitrogen. The sample was set for three biological repeats.
Total sphingolipids extraction (Figure 1)
Sphingolipids were extracted from insects according to Bielawski’s method (Bielawski et al., 2010). Details were prepared as follows:
Note: All following steps were performed at room temperature if not stated otherwise.
The samples were taken out from the -80 °C freezer and dissolved the melted samples in 1 ml solvent extraction solution A (see Recipe 1), to which 10 μl Internal standard had been added (see Recipe 5).
Samples were next grounded twice in tissue homogenizer with grinding beads, each time for 20 s.
Note: In the case of plant material, clean and pre-cooled mortars and pestles were used to grind the samples in liquid nitrogen, and the powder was then transferred to a 15 ml glass tube. The sample was homogenized into a fine powder and that powder was kept frozen at every homogenization step. The purpose of the griding bead is to facilitate insect tissue grinding, so it is okay just to cover the bottom layer for the amount of griding beads per mass of sample and total volume.
Powdered tissue was vortexed vigorously for 5 min and centrifuged for 5 min at 1,000 × g.
After centrifugation, the solvent from the upper lipid-containing phase was transferred to a 15 ml sterilized glass bottle.
Note: The vial label was protected with a clear tape to avoid being wiped off by the solution.
The extraction was repeated by adding 1 ml solvent extraction solution A (see Recipe 1) to the remaining aqueous phase, and the organic layers were combined and concentrated under a stream of nitrogen gas.
The dried lipid fractions were re-suspended into 500 μl solvent extraction solution B (see Recipe 2), and immediately sufficient liquid was transferred to glass vials and caped tightly.
The solvent was transferred into a labeled 1.5 ml microcentrifuge tube, centrifuge the samples at 12,000 × g for 5 min at room temperature.
Note: The tube was caped tightly to avoid leakage of liquid during centrifugation.
The reconstitution solution was finally transferred to a mass spectrometer flask with an internal cannula and stored at -20 °C for machine test.
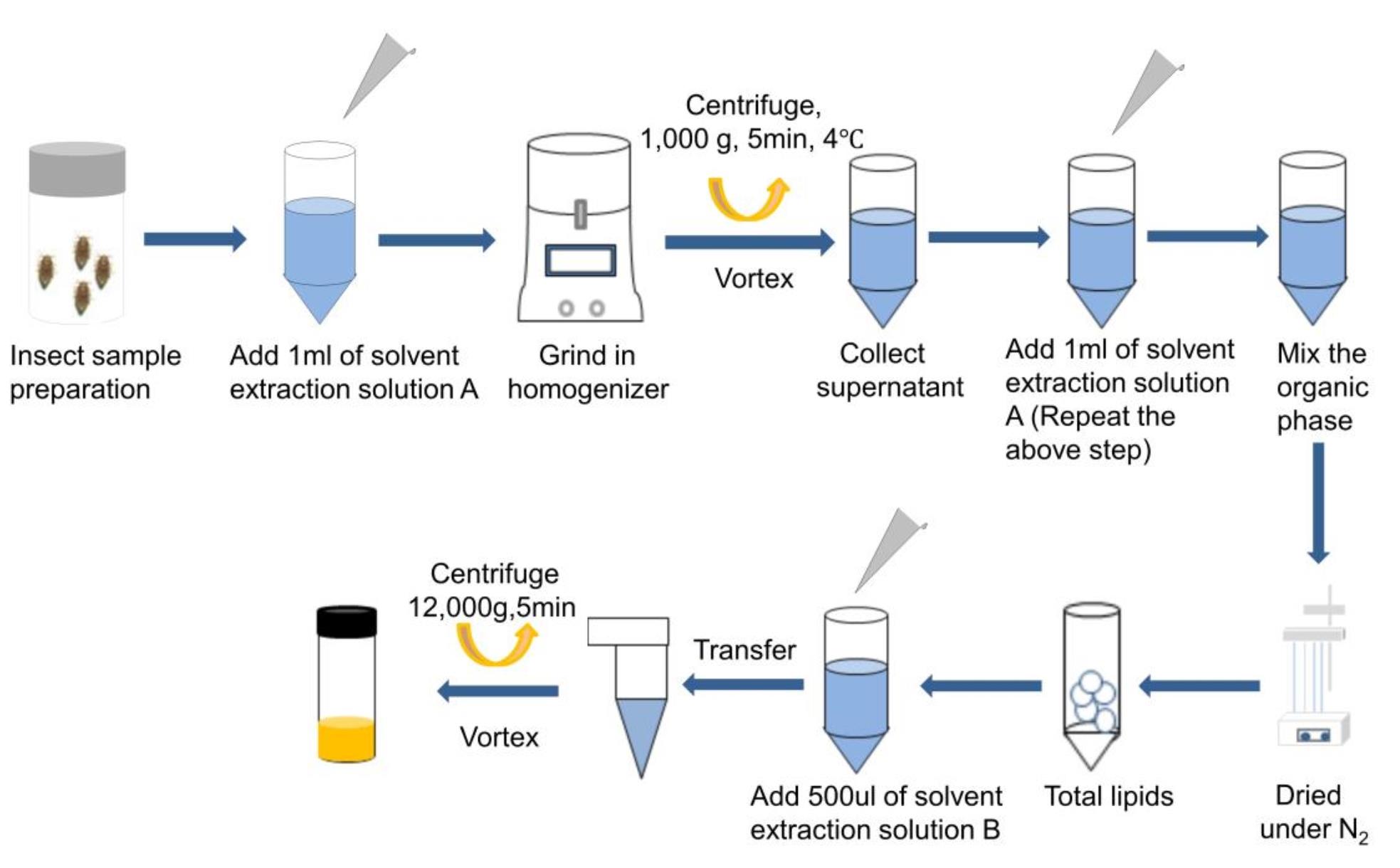
Figure 1. Workflow of sphingolipids extraction from Insect sample
Sphingolipids detection and identification
Sphingolipids were analysed on an AB Seriex 5600+quadrupole Time-of-Flight (Q-TOF) Premier mass spectrometer combined with a Water Acquity Ultra Performance liquid chromatography.
HPLC-program
Solvent A (see Recipe 3)
Solvent B (see Recipe 4)
Constant flow at a rate of 0.3 ml/min stated by solvent A was running in a Waters UPLC (Waters Corp, Milford, MA, USA) coupled with an AB Triple TOF 5600 plus System (AB SCIEX, Framingham, MA, USA).
The changes in gradient were comprised of an increase in solvent B (methanol) from 80 to 99% over 20 min and then from 99 to 100% over 15 min, followed by a reduction back to 80% over 1 min. The percentage of solvent B was then held at 80% for the last 9 min.
The reversed-phase analytical column (Zorbax sb-C8, 2.1 × 150 mm, 3.5 μm; Agilent, Palo Alto, CA, USA) was used to separate ceramides.
10 μl of the samples were applied to the column.
The column was kept at 35 °C during the whole procedure.
The mass spectrum was acquired with an electrospray ionization (ESI) ion source in the positive ionization mode and following settings (Table 2).
Table 2. The instrument settings for sphingolipids analysis
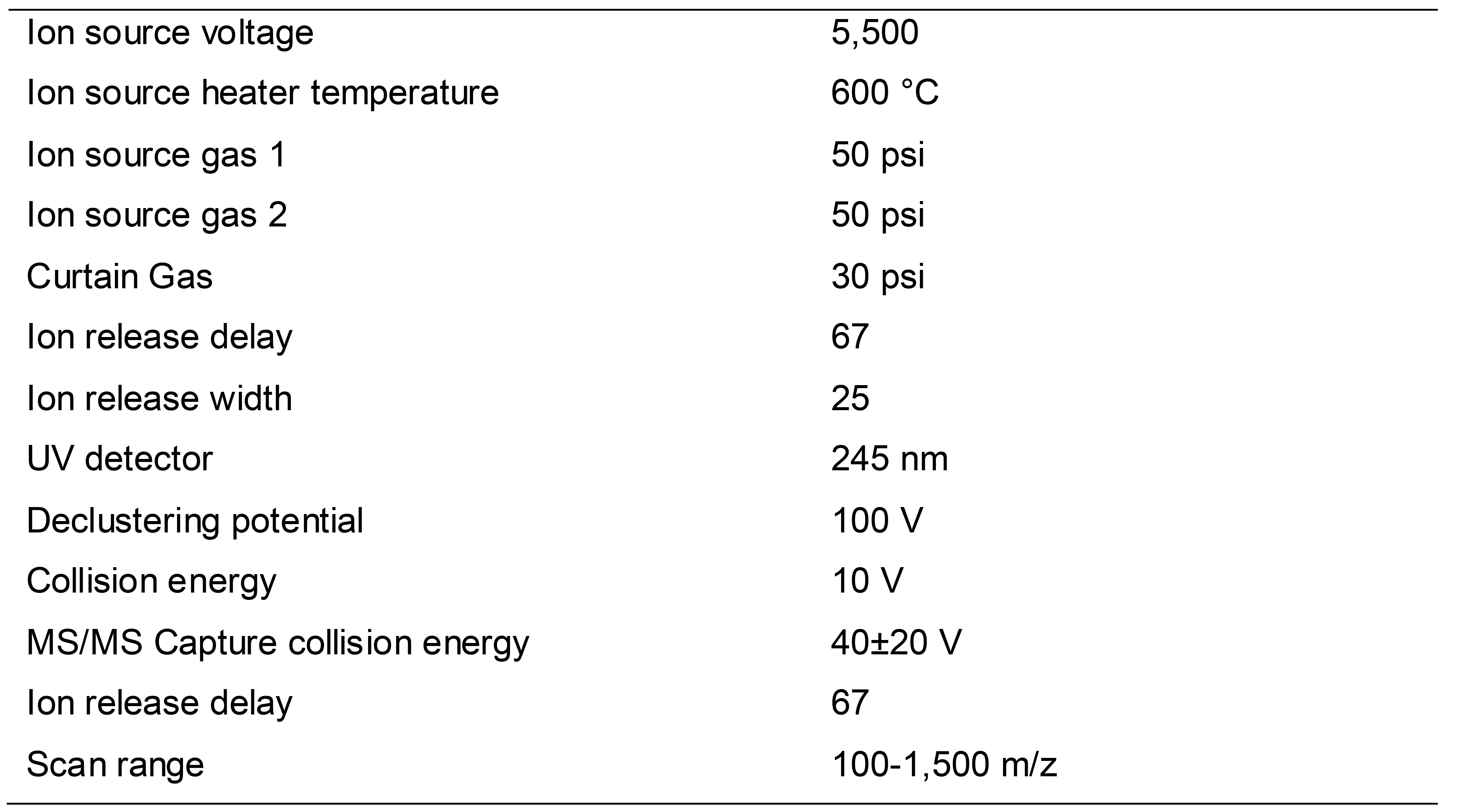
Sphingolipids profiling parameters for detection
The C18 column was used as the separation column on the Aligent 6460+ triple four-pole liquid mass spectrometer to explore the optimal detection conditions of each standard sample. The profiling parameters include precursor ion/targeted ion, retention time, fragmentor, and collision energy. Cer (d18:0/12:0) was used as the internal standard. The scanning parameters for each lipid class were listed in Table 3.
Table 3. Scanning parameters for sphingolipids detection
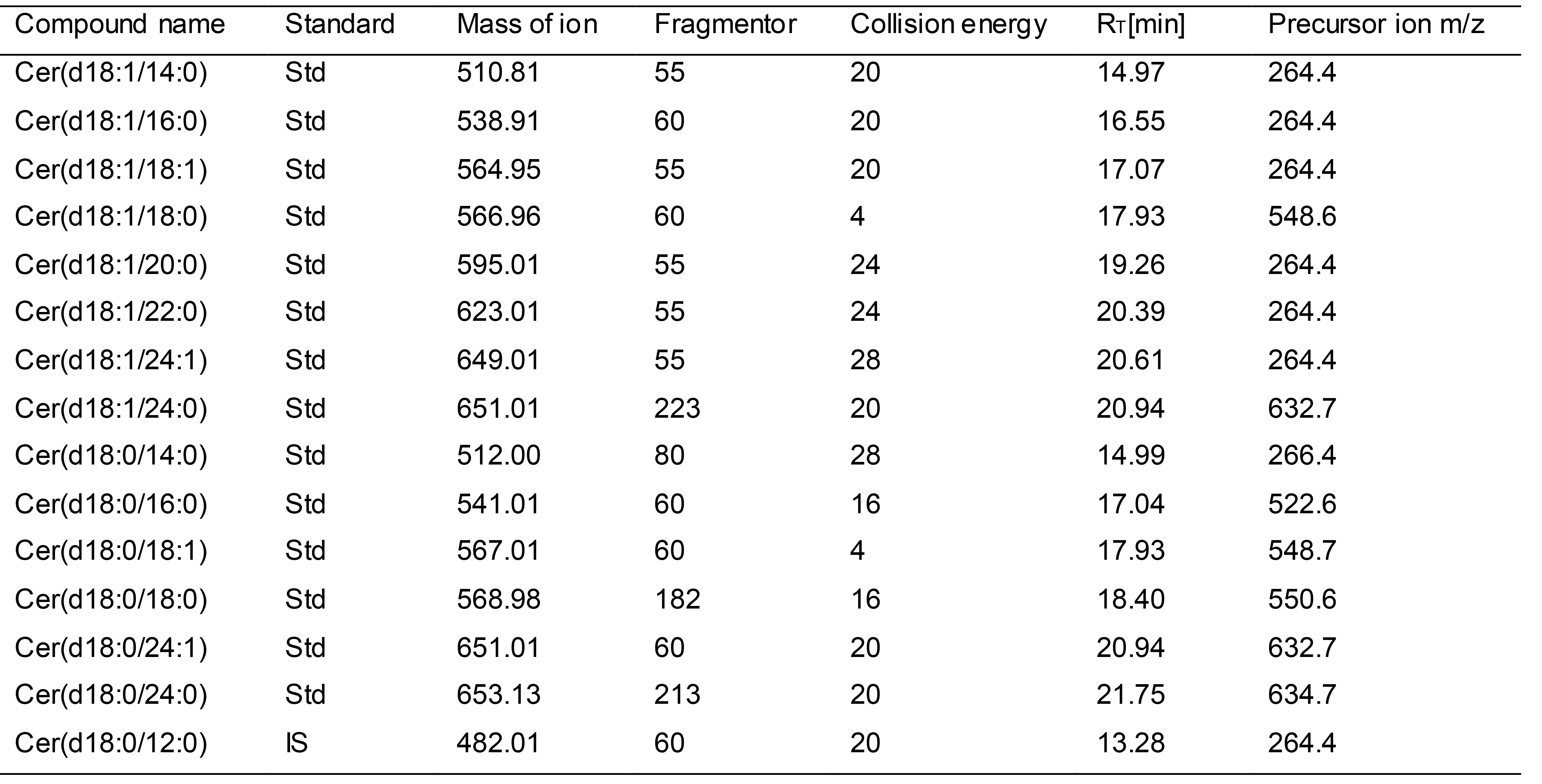
Method optimization notes: For samples containing large amounts of low volatility compounds, the HPLC-program can be made more stringent by increasing the rate flow and by increasing the sampling time. HPLC-program can be adjusted as appropriate for specific analytes and columns by altering holding times. For a few analytes which degrade at high temperatures, it may be desirable to reduce Ion source heater temperatures to 550-580 °C. The timed MS parameters may be used to reduce or eliminate signal from very abundant peaks or contaminants. Those sphingolipids that have different chain lengths, branching, or unsaturation will fragment to yield ions of different m/z. Thus, different precursor ion scans would be necessary to determine the corresponding molecular species. Furthermore, the scan range and collision energy will vary depending on the size and substitution of the various subspecies. All such method adjustments should only be undertaken with the assistance of an experienced MS user.
Data analysis
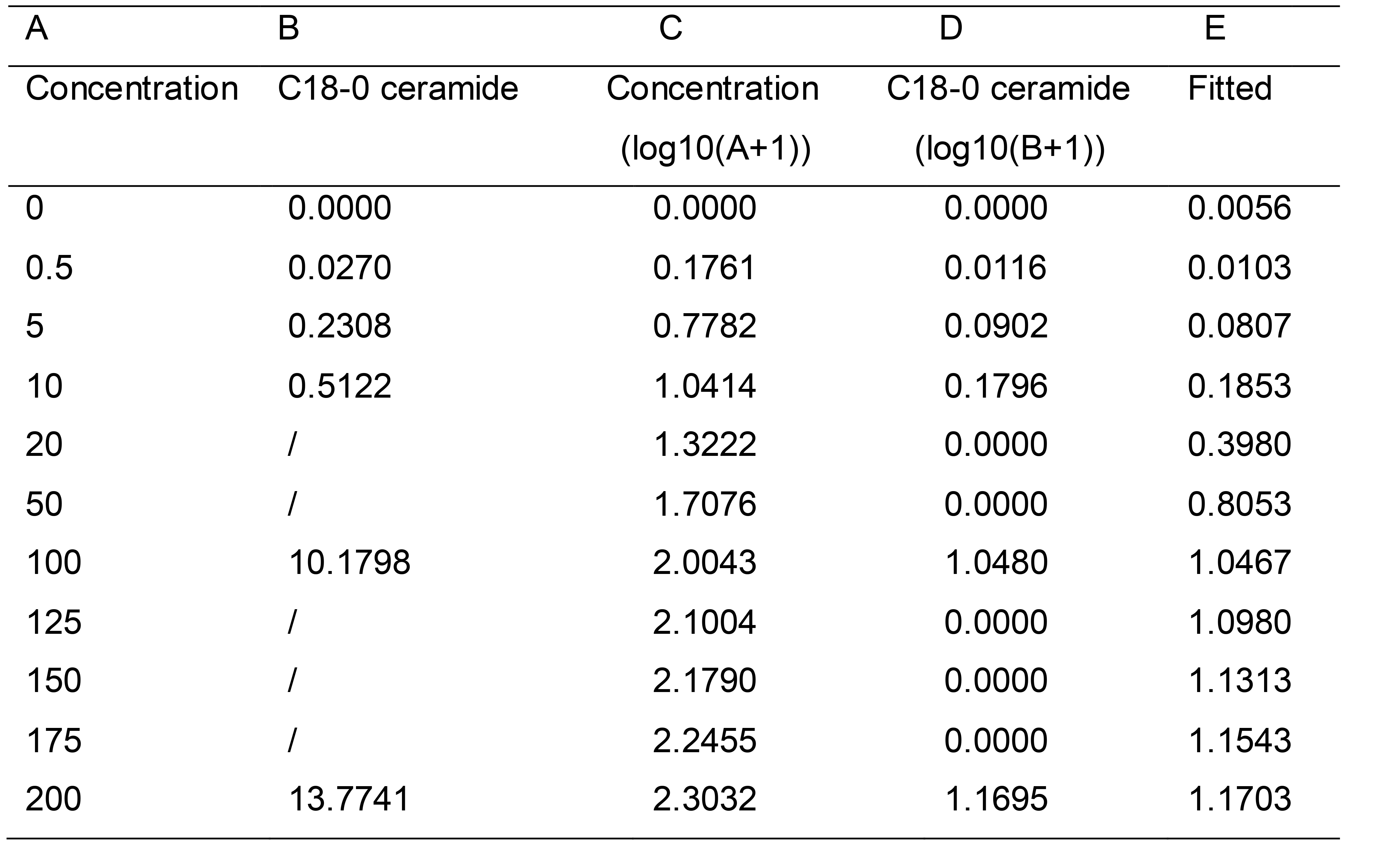
A standard curve with standards from 0.5 ng/ml to 200 ng/ml was generated for quantitative analysis. Curves consisted of triplicates of one blank sample and five calibration points at a concentration ranging from 0.5 ng/ml to 200 ng/ml (Table 4). The amount of each lipid species was calculated according to the sample peak area compared with the normalized internal standard peak area. We used the fitting curve to show the relationship between the concentration and peak area, followed by the fitting curve equation. Three or more biological repeats were recommended (Figure 2).
Table 4. The data points of ceramide standard curve
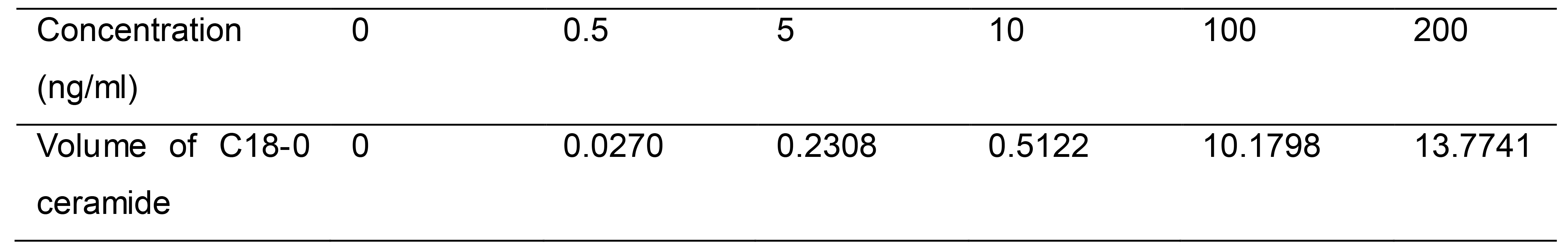
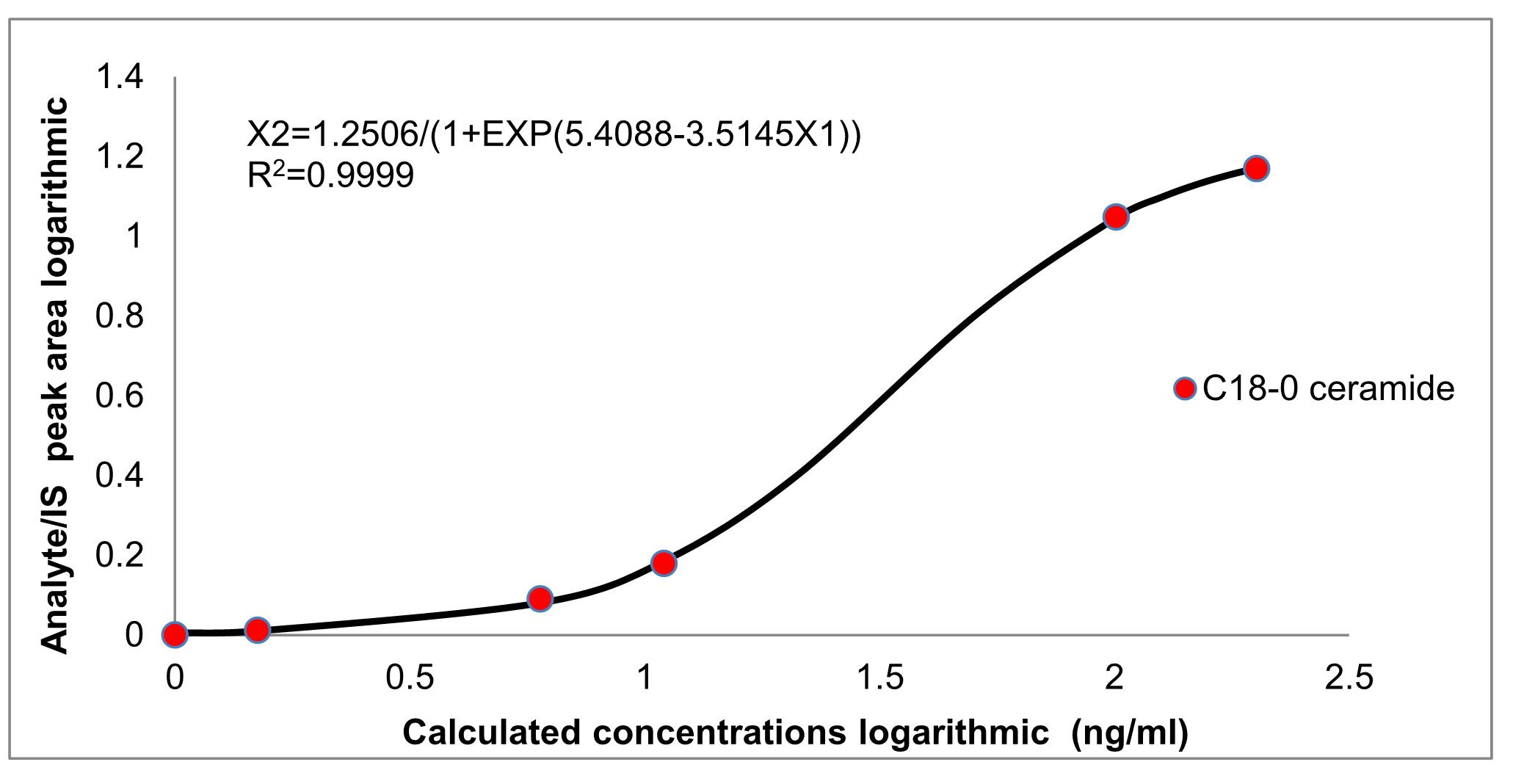
Figure 2. Example of ceramide standard curve. The following ceramides amounts were used: 0 ng/ml, 0.5 ng/ml, 5 ng/ml, 10 ng/ml, 100 ng/ml, 200 ng/ml. The x-coordinate is the concentration and the y-coordinate is the Analyte/IS peak area. For the generation of the standard curve, we converted the horizontal and vertical to logarithmic form.C18 sphingoid bases are the major backbone of most sphingolipids in mammals (Jarne et al., 2018). So we chose the C18-0 as a representative to describe the standard curve (Figure 2). For calibration line measurement, equal amounts and 100 ng Cer (d18:0/12:0) as internal standard were added into different calibration points. The calibration lines from 0.5 ng/ml to 200 ng/ml were converted to a fitted curve, with r values consistently greater than 0.9999 during validation (Table 5). The logistic curve in DPS (Data Processing) software was used to fit the equation.
Table 5. The fitting value of ceramide standard curve
For acquisition, the multiple reaction monitoring (MRM) mode and the software PeakView were used. The internal standard with the mass transition 482.00 m/z -> 264.40 m/z, the retention time of the internal standard is 13.19 min. The Cer (d18:1/16:0) standard with the mass transition 538.91 m/z -> 264.40 m/z, the retention time of the Cer (d18:1/16:0) standard is 16.45 min. Since the retention times and compound-specific ionization and fragmentation values are highly dependent on the used instrumentation, the given values list in Table 2 may be used as a reference but should be individually determined for different instrument setups (Figure 3; Figure 4).
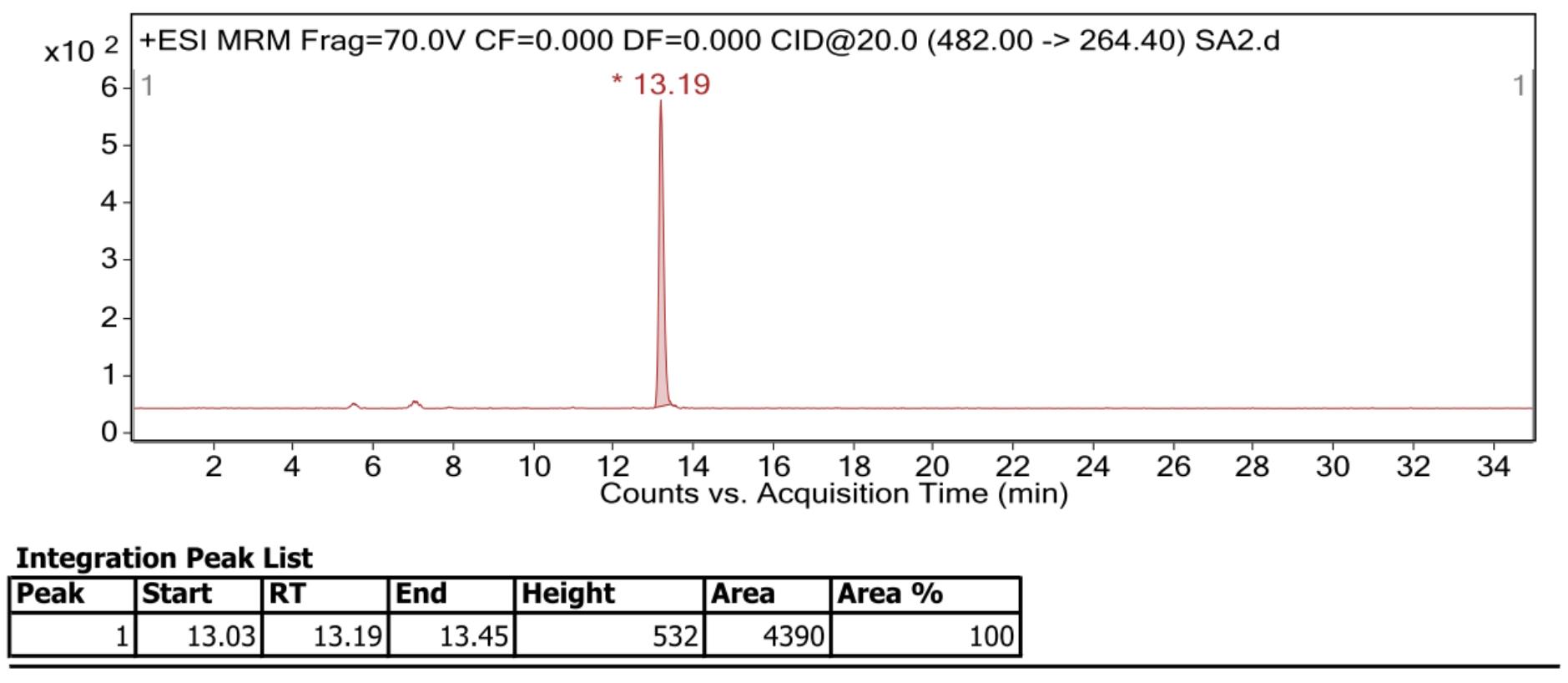
Figure 3. Example of internal standard mass spectrometry acquired with ESI ion source in positive mode from Nilaparvata lugens. Representative signal of internal standard Cer (d18:0/12:0) was plotted. Retention time of internal standard was slightly different from the table 2 list (13.19 min vs. 13.28 min).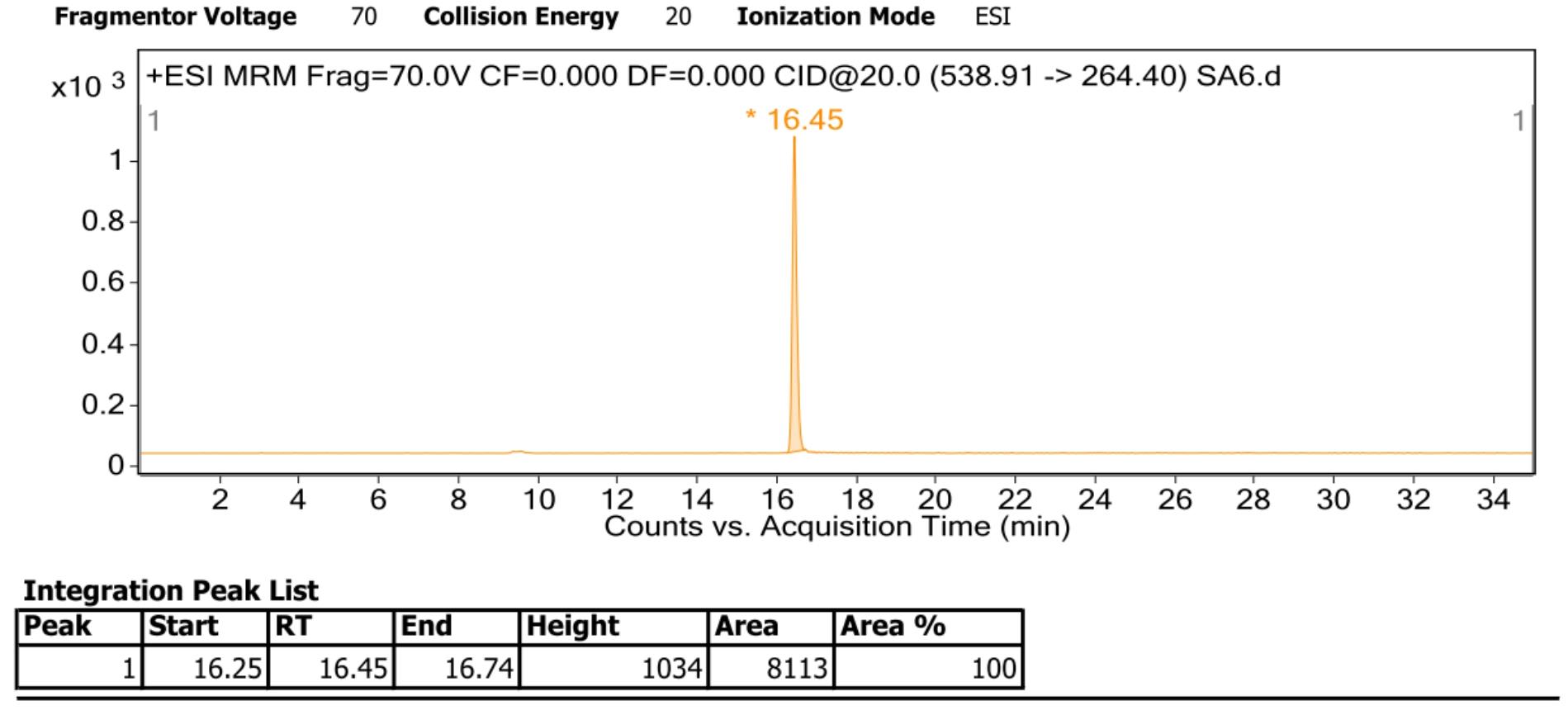
Figure 4. Example of Cer (d18:1/16:0) standard mass spectrometry acquired with ESI ion source in positive mode from Nilaparvata lugens. Representative signal of Cer (d18:1/16:0) was plotted. Retention time of internal standard was slightly different from the table 2 list (16.45 min vs. 16.55 min).Different ceramides were identified by comparing MS/MS ions of analysts with those of sphingolipid standards in ChemSpider base (http://www.chemspider.com/) through the software PeakView (Http://scie.com.cn/products/software/peakview-software). The maximum allowed error for the reliability was set to ± 10 ppm.
Recipes
Solvent extraction solution A
Ethyl acetate:isopropanol:water, 60:30:10 (vol/vol/vol)
Solvent extraction solution B
Methanol:0.1% formic acid, 9:1 (vol/vol)
Solution A
MQ water containing 0.1% formic acid
Solution B
100% Methanol
Internal standard
100 μg/ml Cer (d18:0/12:0) dissolved in HPLC-grade MeOH
Acknowledgments
This work was founded by the National Natural Science Foundation of China (31871962), and the Key R & D Plan of Zhejiang Province (2018C04G2011264). We thank Lijuan Mao from the Analysis Center of Agrobiology and Environmental Sciences Zhejiang University for the help in chemical analysis. The protocol was taken from the publication of Bielawski et al. 2010 with minor modified (Bielawski et al., 2010).
Competing interests
The authors declare that no competing financial interest.
References
- Bielawski, J., Pierce, J. S., Snider, J., Rembiesa, B., Szulc, Z. M. and Bielawska, A. (2010). Sphingolipid analysis by high performance liquid chromatography-tandem mass spectrometry(HPLC-MS/MS). Adv Exp Med Biol 688: 46-59.
- Bode, C. and Graler, M. H. (2012). Quantification of sphingosine-1-phosphate and related sphingolipids by liquid chromatography coupled to tandem mass spectrometry. Methods Mol Biol 874: 33-44.
- Cutignano, A., Chiuminatto, U., Petruzziello, F., Vella, F. M. and Fontana, A. (2010). UPLC-MS/MS method for analysis of sphingosine 1-phosphate in biological samples. Prostaglandins Other Lipid Mediat 93(1-2): 25-29.
- Dobrzyn, A. and Gorski, J. (2002). Ceramides and sphingomyelins in skeletal muscles of the rat: content and composition. Effect of prolonged exercise. Am J Physiol Endocrinol Metab 282(2): E277-285.
- Dobrzyn, A., Knapp, M. and Gorski, J. (2004). Effect of acute exercise and training on metabolism of ceramide in the heart muscle of the rat. Acta Physiol Scand 181(3): 313-319.
- Futerman, A. H. and Hannun, Y. A. (2004). The complex life of simple sphingolipids. EMBO Rep 5(8): 777-782.
- Gorska, M., Dobrzyn, A., Zendzian-Piotrowska, M. and Namiot, Z. (2002). Concentration and composition of free ceramides in human plasma. Horm Metab Res 34(8): 466-468.
- Hannun, Y. A. and Obeid, L. M. (2008). Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol 9(2): 139-150.
- Jarne, C., Saviron, M., Lapieza, M. P., Membrado, L., Orduna, J., Galban, J., Garriga, R., Morlock, G. E. and Cebolla, V. L. (2018). High-Performance Thin-Layer Chromatography Coupled with Electrospray Ionization Tandem Mass Spectrometry for Identifying Neutral Lipids and Sphingolipids in Complex Samples. J AOAC Int 101(6): 1993-2000.
- Lahiri, S. and Futerman, A. H. (2007). The metabolism and function of sphingolipids and glycosphingolipids. Cell Mol Life Sci 64(17): 2270-2284.
- Mao, C. and Obeid, L. M. (2008). Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim Biophys Acta 1781(9): 424-434.
- Preiss, J. E., Loomis, C. R., Bell, R. M. and Niedel, J. E. (1987). Quantitative measurement of sn-1,2-diacylglycerols. Methods Enzymol 141: 294-300.
- Shi, X.-X., Huang, Y.-J., Begum, M.-A., Zhu, M.-F., Li, F.-Q., Zhang, M.-J., Zhou, W.-W., Mao, C. and Zhu, Z.-R. (2018). A neutral ceramidase, NlnCDase, is involved in the stress responses of brown planthopper, Nilaparvata lugens (Stål). Sci Rep 8(1): 1130.
- Shi, X.-X., Zhang, H., Chen, M., Zhang, Y.-D., Zhu, M.-F., Zhang, M.-J., Li, F.-Q., Wratten, S., Zhou, W.-W., Mao, C. and Zhu, Z.-R. (2019). Two sphingomyelin synthase homologues regulate body weight and sphingomyelin synthesis in female brown planthopper, N. lugens (Stål). Insect Molecular Biology 28(2): 253-263.
- Tserng, K. Y. and Griffin, R. (2003). Quantitation and molecular species determination of diacylglycerols, phosphatidylcholines, ceramides, and sphingomyelins with gas chromatography. Anal Biochem 323(1): 84-93.
- Watts, J. D., Gu, M., Polverino, A. J., Patterson, S. D. and Aebersold, R. (1997). Fas-induced apoptosis of T cells occurs independently of ceramide generation. Proc Natl Acad Sci U S A 94(14): 7292-7296.
- Yano, M., Kishida, E., Muneyuki, Y. and Masuzawa, Y. (1998). Quantitative analysis of ceramide molecular species by high performance liquid chromatography. J Lipid Res 39(10): 2091-2098.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wang, N., Shi, X., Zhang, C., Zhou, W. and ZHU, Z. (2021). Extraction and Quantification of Sphingolipids from Hemiptera Insects by Ultra-Performance Liquid Chromatography Coupled to Tandem Mass Spectrometry. Bio-protocol 11(4): e3923. DOI: 10.21769/BioProtoc.3923.
Category
Biochemistry > Lipid > Membrane lipid
Developmental Biology > Cell growth and fate > Differentiation
Cell Biology > Cell metabolism > Lipid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








