- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Measurement of Membrane Attack Complex in RPE Cells
Published: Vol 11, Iss 4, Feb 20, 2021 DOI: 10.21769/BioProtoc.3916 Views: 4493
Reviewed by: Lokesh KalekarMartin V KolevVemika Chandra

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Sensitive and Adaptable Turn-On Maturation (ATOM) Fluorescent Biosensors for Detecting Subcellular Localization of Protein Targets in Cells
Harsimranjit Sekhon [...] Stewart N. Loh
Mar 20, 2025 2263 Views

Assay for Site-Specific Homologous Recombination Activity in Adherent Cells, Suspension Cells, and Tumor Tissues
Yuki Yoshino [...] Natsuko Chiba
Apr 5, 2025 2433 Views

Preparation of Testicular Cells for Immunofluorescence Analysis of Manchette in Elongating Spermatids
Changmin Niu [...] Zhibing Zhang
Jun 20, 2025 2374 Views
Abstract
Initiation of the complement system results in the formation of a multiprotein pore termed the membrane attack complex (MAC, C5b-C9). MAC pores accumulate on a cell surface and can result in cell lysis. The retinal pigment epithelium (RPE) is a single monolayer of pigmented epithelial cells located at the posterior poll of the eye that forms the outer blood retinal barrier. RPE cells are highly polarized with apical microvilli and basolateral contact with Bruch’s membrane. In order to obtain biologically relevant polarized RPE cultures in vitro, RPE cells are seeded onto the apical side of a transwell filter and cultured for 4 weeks in low serum media. MAC formation on RPE cells has been reported to be sub-lytic. MAC formation can be achieved in vitro by introduction of normal human serum (NHS) to media following serum starvation for 24 h. NHS contains all serum complement proteins required to initiate complement activation and MAC formation. We combined in vitro RPE polarization and complement activation to visualize MAC formation in vitro utilizing confocal microscopy allowing for high resolution MAC imaging.
Keywords: Membrane attack complex (MAC)Background
The Complement system is an evolutionary conserved innate immune pathway. There are three major independent yet overlapping pathways for complement activation that converge at the C3 convertase, the classical, the lectin and the alternative pathways. In the classical pathway, immune complexes (Antigen-Antibody complexes) bind C1 via the C1q subcomponent, then the C1s protease subunit cleaves complement factors C4 and C2. Fragments of these (C4bC2b) forms an enzyme complex ‘C3 convertase’, that cleaves C3 to C3b and releases the anaphylatoxin, C3a. The binding of C3b to the C3 convertase generates the C5 convertase (C4bC2bC3b). The lectin pathway is an analogous system, except that the initiating step is the binding by lectins to repetitive sugars on microbial surfaces. Mannose-associated serine proteases (MASPs) take the place of the C1 proteases. The alternative pathway (AP) continuously self-activates at a low level to generate C3b that deposits on pathogens or debris. C3b or C3(H2O) engages the rate limiting alternative pathway components, factors B (FB) and D (FD), to form the alternative C3 convertase (C3bBb), which in turn cleaves more C3 into C3a and C3b. The binding by another C3b to the C3 convertase generates the C5 convertase (C3bBbC3b). Properdin (P) is a positive regulator that stabilizes both the AP C3 and C5 convertases. The C5c convertase subsequently cleaves C5 to release the potent anaphylatoxin C5a, while C5b engages the terminal pathway and initiates the formation of the membrane attack complex (MAC). In this way activation of the complement system results in the assembly of transmembrane pores; MAC (Figure 1). MAC formation on gram-negative bacteria causes cell lysis. In nucleated cells MAC deposition can result in cell death by apoptosis (Nauta et al., 2002), direct lysis (Koski et al., 1983) or in some cases MAC can be sub-lytic and promote an inflammatory response (Niculescu and Rus, 2001). The number of MAC pores which form on a cells surface is directly related to cell lysis and as such MAC formation is tightly regulated by complement inhibitors. CD59 is a direct inhibitor of complement and is expressed on the cell surface of various host tissues (Meri et al., 1991). In addition to expression of complement inhibitors some nucleated cells can endocytose or exocytose MAC pores to prevent cell lysis (Morgan et al., 1987).
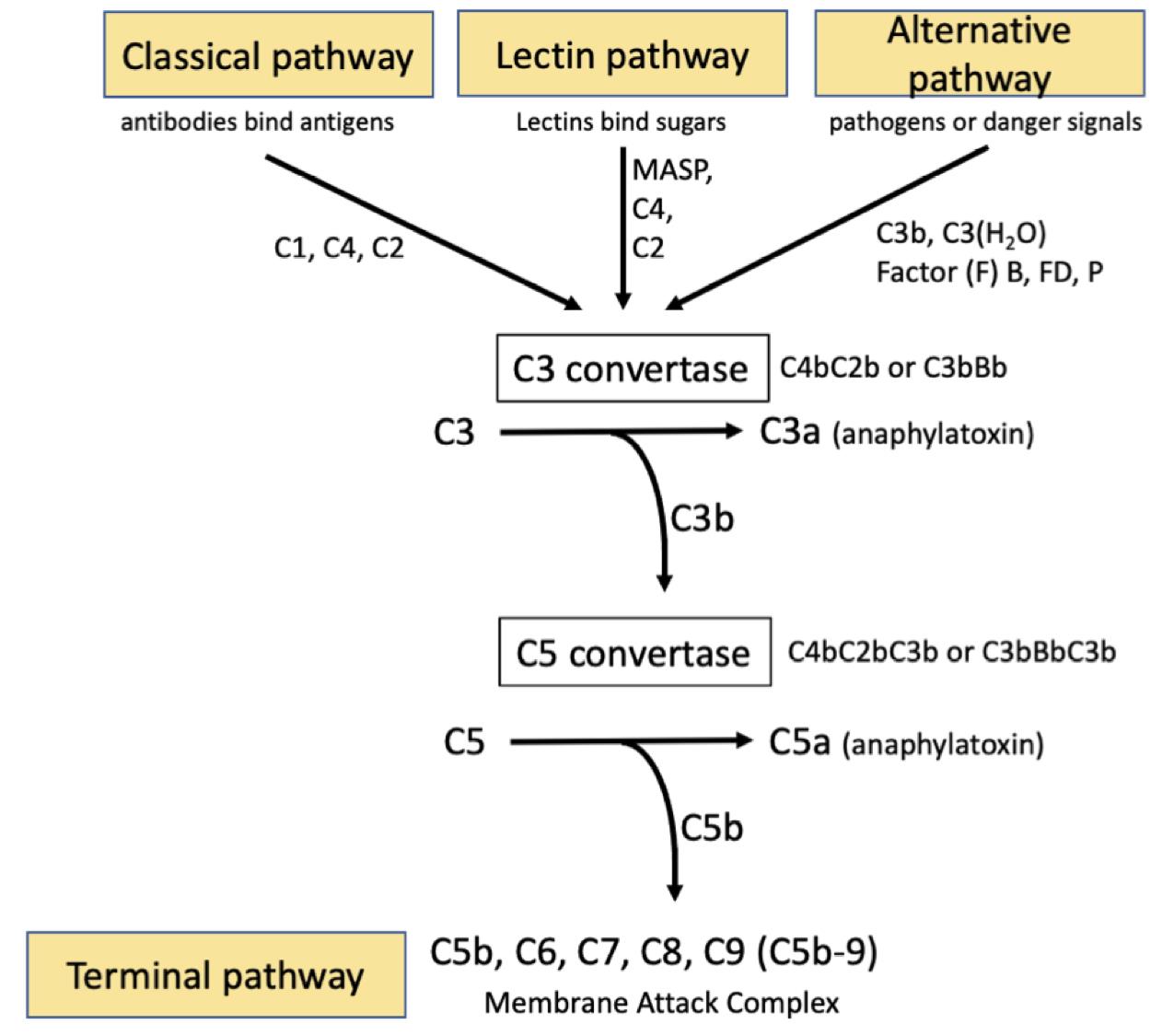
Figure 1. The three independent complement activating pathways leading to C3 convertase activation
Aberrant activation of complement and increased MAC formation is involved in the pathogenesis of age-related macular degeneration. MAC accumulation has been observed in donor eye tissue and is most abundant in the choroid in choriocapillaris endothelial cells (Mullins et al., 2014). MAC deposition is also observed although less readily on the retinal pigment epithelium (RPE). This may be explained by the finding that the RPE can rapidly endocytose and clear MAC pores (Georgiannakis et al., 2015). Sub-lytic MAC does not appear to be responsible for RPE cell death, however, it may act as a pro-inflammatory signaling mechanism contributing to overactivation of the innate immune system and leading ultimately to tissue degeneration (Mulfaul et al., 2020).
The RPE is a highly specialized monolayer of pigmented epithelial cells located between the neural retina and the choriocapillaris. In vivo, the RPE forms polarized monolayers with apical microvilli which interconnect with the overlying photoreceptor outer segments. The basolateral RPE has basal infoldings in close contact with Bruch’s membrane. The RPE is therefore an essential component of the outer blood retinal barrier and is necessary for maintenance of healthy vision. Monolayer polarization is essential to RPE function. In order to achieve a polarized monolayer of RPE cells in vitro, cells can be seeded on transwell filters and cultured for at least 4 weeks (Figure 2). This results in the formation of apical microvilli and polarized expression of tight junction proteins (Kannan et al., 2006; Sonoda et al., 2009). Polarizing RPE in vitro allows us to closer mimic RPE behavior in the mammalian eye.
In order to study the contribution of MAC formation on the RPE in AMD we first polarized both the immortalized RPE cell line ARPE-19 or primary human fetal RPE (hfRPE). We then incubated cells with normal human serum (NHS) which contains all of the necessary components to drive complement activation and MAC deposition. NHS alone resulted in no visible MAC formation on ARPE-19 cells and only a small number of MAC were observed on hfRPE cells (Mulfaul et al., 2020). The apparent lack of MAC on RPE cells in spite of the presence of NHS is likely due to RPE expression of complement regulators including CD59 which directly inhibits MAC complex formation (Yang et al., 2009). Furthermore, if MAC complex’s do form the RPE can efficiently remove MAC by internalization of the MAC pore by endocytosis (Georgiannakis et al., 2015). We found that stimulation of RPE with an inflammatory insult prior to supplementation with NHS allowed MAC pores to persist on RPE cells that could be imaged by confocal microscopy. In a previous report, we demonstrated that the oxidative protein modification 2-(ω-carboxyethyl)pyrrole CEP, which is found in abundance in AMD donors (Gu et al., 2003) and is known to activate TLR2 and CD36, increased complement activation and MAC deposition on the RPE (Mulfaul et al., 2020). RPE cells activated with CEP in the presence of complement rich serum accumulate immunofluorescent measurable MAC pores. This protocol can be used as an in vitro model to study whether therapeutic targets can reduce MAC deposition on the RPE as we have previously demonstrated using an anti-TLR2 inhibitor (Mulfaul et al., 2020).
Materials and Reagents
T75 cell culture flasks (Corning, catalog number: CLS3275 )
15 ml conical tube (Corning, catalog number: CLS430791 )
0.4 μm polyester transwell inserts (VWR, catalog number: 734-1581 )
Polylysine coated slide (Thermo Fisher Scientific, catalog number: 10219280 ).
ARPE-19 cells (ATCC, catalog number: CRL-2302 )
Alexa Fluor 488 Goat Anti-Rabbit (Thermo Fisher Scientific, catalog number: A11034 )
Normal Goat Serum (Sigma-Aldrich, catalog number: G9023-10ml )
Collagen IV (Sigma-Aldrich, catalog number: C5533 )
DMEM/F-12 Ham (Sigma-Aldrich, catalog number: D8437-500ML )
Fetal Bovine Serum (Sigma-Aldrich, catalog number: F9665-500ML )
Pennicillin Streptomycin (Sigma-Aldrich, catalog number: P4333-100ML )
Phosphate Buffered Saline (Sigma-Aldrich, catalog number: D8662-500ML )
Trypsin-EDTA (Biosciences, catalog number: 25200-056 )
Normal human serum (Sigma-Aldrich, catalog number: H4522-20ml )
ZO-1 (Thermo Fisher Scientific, catalog number: 40-2200 )
Anti-C5b-9 (Santa Cruz Biotech, clone ae11, catalog number: sc58935 )
Hoechst (Sigma-Aldrich, catalog number: B2261-25MG )
Mowiol® 4-88 (Sigma-Aldrich, catalog number: 81381-50G )
Glycerol (Sigma-Aldrich, catalog number: G5516 )
Tris-HCl (Thermo Fisher Scientific, catalog number: 10724344 )
Paraformaldehyde (Sigma-Aldrich, catalog number: 158127 )
NaOH (Sigma-Aldrich, catalog number 567530 )
Triton X-100 (Sigma-Aldrich, catalog number: x100-500ML )
L-glutamine (Thermo Fisher Scientific, catalog number: 10378-016 )
Mowiol 4-88 (see Recipes)
4% PFA (see Recipes)
Equipment
Fume hood
Hemocytometer
-80 °C freezer
4 °C fridge
Tweezers
Biological Safety Cabinet Class ll
Heat Block
Confocal laser scanning microscope Axio Observer Z1 Inverted Microscope equipped with a Zeiss LSM 700 T-PMT Scanning unit and a 40x plan.
Procedure
Transwell preparation
Coat 0.4 μm polyester transwell inserts with 100 μg/ml of collagen IV in PBS by pipetting the appropriate volume of collagen onto the apical side of the transwell filter.
Leave the transwell filters to air dry with the lid off in a cell culture grade biosafety cabinet.
Gently aspirate off any additional collagen from each transwell filter.
Wash each filter twice in PBS prior to cell seeding.
Cell seeding ARPE-19
ARPE-19 cells are maintained in DMEM/F-12 Ham with 10% FBS and 1% PS in T75 cell culture flasks. Cells are passaged at 80% confluency (we have used them up to passage 30).
Aspirate medium from T75 flask and wash cell monolayer twice with 5 ml of sterile PBS.
Add 3 ml of pre-warmed trypsin and incubate at 37 °C for 5 min until cells begin to detach.
Add 3 ml of DMEM F-12.
Use a 10 ml stripette to aspirate 6 ml of trypsin/media and flush the bottom wall of the T75 to remove all attached cells.
Transfer cell suspension to a 15 ml conical tube.
Pellet cells by centrifugation at 205 × g for 5 min.
Gently aspirate media.
Resuspend cell pellet in 1 ml of fresh media.
Count cells using a hemocytometer.
Pipette 600 μl of DMEM/F-12 Ham containing 10% FBS into the basolateral side of the transwell filter.
Seed cells at a density of 1.7 × 105 cells per cm2 in the upper transwell chamber in 200 μl of DMEM/F-12 Ham containing 10% FBS, (transwell surface area 0.33 cm2 per well).
Two days later replace media with DMEM/F-12 containing 1% FBS.
Replace media every Monday, Wednesday and Friday for 4 weeks (Figure 2).

Figure 2. Polarization of RPE cells grown on transwell filters. A. Schematic representation of the RPE. Polarised RPE cells display apical microvilli and ZO-1 tight junction protein expression. B. RPE cells seeded on the apical side of a transmembrane filter and maintained for at least 4 weeks form polarised monolayers as indicated by defined ZO-1 immunohistochemistry in both (C) ARPE-19 cells and (D) primary hfRPE cells. Scale bars = 20 µm.
Cell seeding primary human fetal RPE (hfRPE) cells
Cells were kindly provided by Dr. Arvydas Maminishkis from the National Eye Institute (NEI), Bethesda, USA, and were received as a confluent monolayer of P-0 (Maminishkis et al., 2006).
hfRPE cells are received in a T25 with filled with media.
After receiving gently aspirate all media from the cell monolayer.
Add 5 ml of fresh media Alpha MEM (w/o L-glutamine Lonza BE12-169F), plus Pen/strep + L-glutamine, 5% FBS.
Leave cells to rest for at least 1 day prior to plating.
Aspirate cell culture media.
Wash cells with 50 ml of PBS.
Note: This will fill the entire flask.
Remove 50 ml of PBS and repeat for a second wash step.
Pipette 5 ml of warm trypsin-EDTA into the flask.
Place flask in the incubator for 15 min.
Use a spinal needle with a 3 ml syringe to aspirate the trypsin from the RPE monolayer.
Direct the flow of the needle against the wall of the flask which has adherent cells.
Press down on the syringe to stream wash the cells from the wall of the flask.
Use a 5 ml pipette to collect the cell suspension into a sterile conical tube.
Immediately add RPE media containing 15% FBS to the cells in the conical tube.
Repeat steps 9-15 a second time to ensure all cells are dislodged.
Pellet cells by centrifugation at 205 × g for 5 min.
Gently aspirate media.
Resuspend cell pellet in 1 ml of fresh media (no need for FCS).
Count cells using a hemocytometer.
Add 600 μl of RPE media containing 15% FCS to the basolateral well of each transwell.
Seed cells at a density of 100 × 103 cells per cm2 into the apical transwell chamber in 200 μl of RPE media containing 15% FCS, transwell surface area 0.33 cm2 per well.
48 h later aspirate media from both apical and basolateral transwell chambers.
Pipette 200 μl of RPE media containing 5% FCS into the apical and 600 μl into the basolateral chamber.
Continue to change RPE media every Monday, Wednesday and Friday for 4 weeks.
Validation of tight junction formation
Note: Validation of tight junction formation can be conducted in parallel with the MAC formation assay.
After 4 weeks in culture RPE cells on transwell inserts are fixed by adding 200 μl of 4% paraformaldehyde (PFA) to the apical membrane and 600 μl of 4% PFA to the basolateral chamber.
Cells are incubated in PFA for 10 min at room temperate.
Gently aspirate PFA taking care not to poke a hole in the transwell filter or disturb the cells.
Wash cells on filter 3 times for 5 min with 200 μl of PBS in the apical compartment and 600 μl on the basolateral side.
Pipette 200 μl or 600 μl of 0.05% Triton X-100 into the apical and basolateral chambers respectively.
Cells are incubated in 0.05% Triton X-100 for 10 min at room temperature to permeabilize the cell membrane.
Aspirate Triton X-100.
Pipette 200 μl or 600 μl of 5% normal goat serum (NGS) into the apical and basolateral chambers respectively.
Block cells in NGS for 1 h at room temperature.
Aspirate 5% NGS.
Pipette 100 μl of ZO-1 antibody (1:100 in 5% NGS) into the apical side of the transwell filter.
100 μl of antibody was pipetted into the bottom of an empty 6-well plate and the transwell inserts were placed to sit directly on the antibody.
Inserts were incubated overnight at 4 °C.
Aspirate antibody from apical chamber.
Place transwell into a new plate with 600 μl of PBS in the basolateral chamber and pipette 100 μl of PBS into the apical side.
Repeat this wash step 3 times.
Pipette 100 μl or 600 μl of PBS with goat anti-rabbit 488 (1:500) into the apical and basolateral chambers respectively.
Incubate transwells in secondary antibody for 2 h at room temperature.
Aspirate off secondary antibody.
Pipette 200 μl or 600 μl of PBS into the apical and basolateral chambers respectively.
Repeat PBS wash 3 times.
Aspirate off last PBS
Add 200 μl of Hoechst (1:10,000) to the apical transwell.
Incubate transwell for 2 min.
Wash transwells 3 times in PBS.
Gently remove all liquid from the transwell filter.
Turn the transwell upside down on a clean surface.
Gently use a sterile scalpel to cut around the circumference of the membrane.
Use a sterile tweezer to grip one side of the filter after the first incision.
Gently use the tweezers to loosen the membrane as you continue to cut slowly with the blade. Take care not to scratch the cell monolayer with the tweezers.
Once the membrane is cut free use the tweezers to mount the membrane basolateral side down on to a polylysine coated slide. i.e., cells should be facing up on top of the slide.
Pipette 50-100 μl of Mowiol® 4-88 mounting media onto the membrane and cover with cover slip.
MAC formation Assay
48 h prior to MAC formation assay remove all RPE media from both apical and basolateral transwell chambers.
Pipette 600 μl of serum free DMEM/F-12 Ham into the basolateral transwell chamber.
Pipette 200 μl of serum free DMEM/F-12 Ham into the apical chamber.
Thaw stock bottle of normal human serum (NHS) rapidly at 37 °C.
Aliquot NHS into single use aliquots (300-500 μl).
Store aliquoted NHS at -80 °C.
On the day of the MAC assay remove two aliquots of NHS from -80 °C.
Thaw aliquots on ice.
Heat inactivate one aliquot of NHS by placing in either a heat block or water bath set to 56 °C.
Incubate serum for 30 min at 56 °C inverting the tube every 10 min.
Allow the heat inactivated serum to cool to room temperature.
Designate which transwells will be used for apical or basolateral MAC formation.
Aspirate media from transwells.
Add 600 μl of serum free media to the basolateral chamber and 180 μl of serum free media to the apical chamber of wells designated for apical MAC assessment.
Add 540 μl of serum free media to the basolateral chamber and 200 μl of serum free media to the apical chamber of wells designated for basolateral MAC assessment.
Pipette 20 μl of either NHS or heat-inactivated (Hi) HiNHS to the apical chamber of wells containing 180 μl of media.
Pipette 60 μl of either NHS or HiNHS to the basolateral chamber of wells containing 540 μl of media.
Incubate RPE with NHS or HiNHS for 2-24 h.
After 24 h fix cells by adding 200 μl of 4% PFA to the apical membrane and 600 μl of 4% PFA basolateral chamber.
Incubate cells in PFA for 10 min at room temperature.
Gently aspirate PFA taking care not to poke a hole in the transwell filter or disturb the cells.
Wash cells on filter 3 times for 5 min with 200 μl of PBS in the apical compartment and 600 μl on the basolateral side.
Pipette 200 μl or 600 μl of 5% bovine serum albumin (BSA) into the apical and basolateral chambers respectively.
Block cells in BSA for 1 h at room temperature.
Aspirate 5% BSA.
Pipette 100 μl of C5b-9 MAC antibody (1:25 in 5% BSA) into the apical side of the transwell filter.
100 μl of antibody was pipetted into the bottom of an empty 6-well plate and the transwell inserts were placed to sit directly on the antibody, the plate is lidded and sealed with parafilm.
Inserts were incubated overnight at 4 °C in a fridge.
Aspirate antibody from apical chamber.
Place transwell into a new plate with 600 μl of PBS in the basolateral chamber and pipette 100 μl of PBS into the apical side.
Repeat this wash step 3 times.
Pipette 100 μl or 600 μl of PBS with goat anti-mouse 647 (1:500) and phalloidin (1:500) into the apical and basolateral chambers respectively.
Incubate transwells in secondary antibody for 2 h at room temperature.
Aspirate off secondary antibody.
Pipette 200 μl or 600 μl of PBS into the apical and basolateral chambers respectively.
Repeat PBS wash 3 times.
Aspirate off last PBS
Add 200 μl of Hoechst (1:10,000) to the apical transwell.
Incubate transwell for 2 min.
Wash transwells 3 times in PBS.
Gently remove all liquid from the transwell filter.
Turn the transwell upside down on a clean surface.
Gently use a sterile scalpel to cut around the circumference of the membrane.
Use a sterile tweezer to grip one side of the filter after the first incision.
Gently use the tweezers to loosen the membrane as you continue to cut slowly with the blade. Take care not to scratch the cell monolayer with the tweezers.
Once the membrane is cut free use the tweezers to mount the membrane on to a polylysine coated slide (Thermo Scientific).
Pipette 50-100 μl of Mowiol® 4-88 mounting media onto the membrane and cover with cover slip.
MAC formation on ARPE-19 cells (Figure 3) and hfRPE cells (Figure 4) was imaged using a confocal laser scanning microscope.
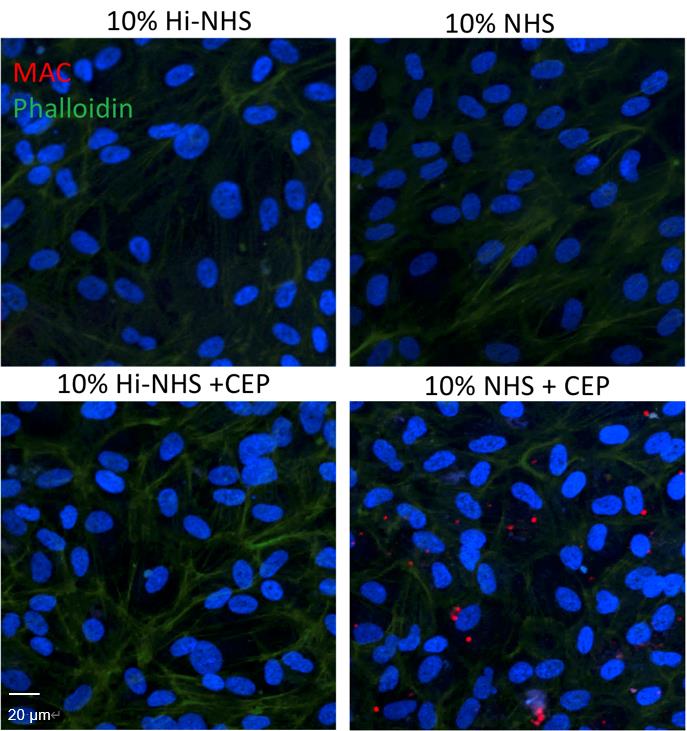
Figure 3. MAC assay on ARPE-19 cells. MAC formation (red) was observed in ARPE-19 cells stimulated with CEP (that induces the alternative complement pathway) and 10% NHS as a source of complement for 24 h. No staining was observed in the NHS alone or HiNHS control. Scale bar = 20 μm.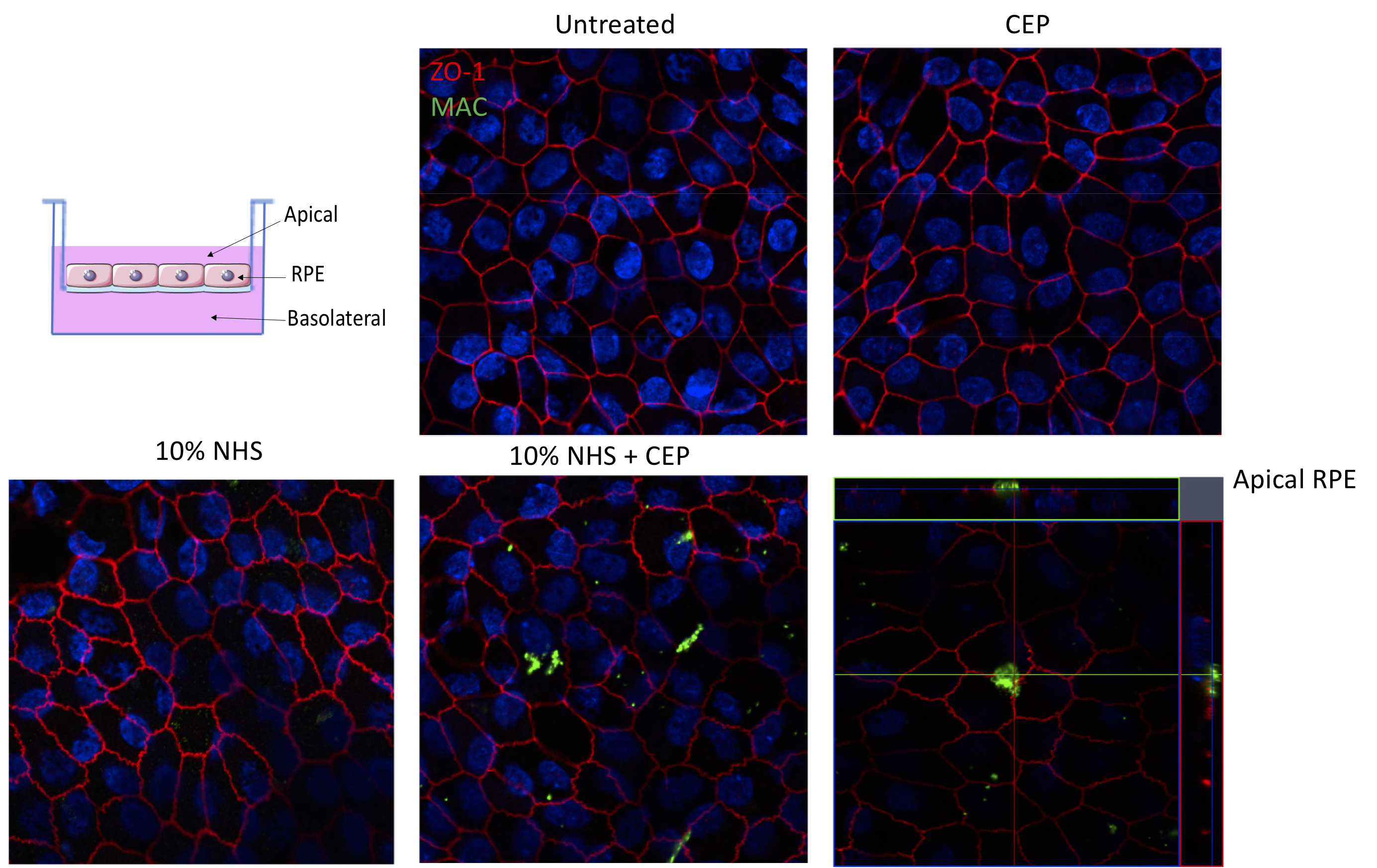
Figure 4. MAC Assay on hfRPE. MAC formation (green) was observed in hfRPE cells stimulated with CEP and 10% NHS as a source of complement for 2 h. No staining was observed in the NHS alone or HiNHS controls. Scale bar = 20 μm.
Data analysis
Staining was analysed using a confocal laser scanning microscope Axio Observer Z1 inverted microscope equipped with a Zeiss LSM 700 T-PMT scanning unit and a 40× plan.
Recipes
Mowiol 4-88
2.4 g Mowiol
6 g glycerol
6 ml H2O
Stir for several hours at room temperature
Add 12 ml 0.2 M Tris-HCl (pH 8.5) and heat to 50 °C for 10 min
Centrifuge Mowiol at 5,000 × g for 15 min
Aliquot and store at -20 °C
4% PFA
Add 4 g PFA to 50 ml of H2O
Add 1 ml of 1 M NaOH, stir gently on a heating block at ~60 °C until PFA is dissolved
Add 10 ml of 10× PBS and allow the mixture to cool to room temperature
Adjust pH to 7.4
Adjust the final volume to 100 ml with H2O
Filter the solution through a 0.45-μm membrane filter to remove any particulate matter
Make the PFA solution fresh prior to use, or store in aliquots at −20 °C for several months
Notes:
Avoid repeated freeze/thawing.
Preparation of PFA can give off toxic fumes, use a fume hood as a precautionary measure.
Acknowledgments
This research was funded by the BrightFocus Foundation (M2016030), United States, Health Research Board Ireland (HRB-HRA/2013.290) and Science Foundation Ireland (SFI 15/CDA/3497), Royal Victoria Eye and Ear Hospital (RVEEH), Ireland, National Children's Research Centre (NCRC) , Ireland, Irish Research Council (IRCLA/2017/295), Ireland. The original research paper that displayed this protocol was published in cell reports (Mulfaul et al., 2020).
Competing interests
Authors have no financial or non-financial competing interests.
References
- Georgiannakis, A., Burgoyne, T., Lueck, K., Futter, C., Greenwood, J. and Moss, S. E. (2015). Retinal pigment epithelial cells mitigate the effects of complement attack by endocytosis of C5b. J Immunol 195(7): 3382-3389.
- Gu, X., Meer, S. G., Miyagi, M., Rayborn, M. E., Hollyfield, J. G., Crabb, J. W. and Salomon, R. G. (2003). Carboxyethylpyrrole protein adducts and autoantibodies, biomarkers for age-related macular degeneration. J Biol Chem 278(43): 42027-42035.
- Kannan, R., Zhang, N., Sreekumar, P. G., Spee, C. K., Rodriguez, A., Barron, E. and Hinton, D. R. (2006). Stimulation of apical and basolateral VEGF-A and VEGF-C secretion by oxidative stress in polarized retinal pigment epithelial cells. Mol Vis 12: 1649-1659.
- Koski, C. L., Ramm, L. E., Hammer, C. H., Mayer, M. M. and Shin, M. L. (1983). Cytolysis of nucleated cells by complement: cell death displays multi-hit characteristics. Proc Natl Acad Sci U S A 80(12): 3816-3820.
- Maminishkis, A., Chen, S., Jalickee, S., Banzon, T., Shi, G., Wang, F. E., Ehalt, T., Hammer, J. A. and Miller, S. S. (2006). Confluent monolayers of cultured human fetal retinal pigment epithelium exhibit morphology and physiology of native tissue. Invest Ophthalmol Vis Sci 47(8): 3612-3624.
- Meri, S., Waldmann, H. and Lachmann, P. J. (1991). Distribution of protectin(CD59), a complement membrane attack inhibitor, in normal human tissues. Lab Invest 65(5): 532-537.
- Morgan, B. P., Dankert, J. R. and Esser, A. F. (1987). Recovery of human neutrophils from complement attack: removal of the membrane attack complex by endocytosis and exocytosis. J Immunol 138(1): 246-253.
- Mulfaul, K., Ozaki, E., Fernando, N., Brennan, K., Chirco, K. R., Connolly, E., Greene, C., Maminishkis, A., Salomon, R. G., Linetsky, M., Natoli, R., Mullins, R. F., Campbell, M. and Doyle, S. L. (2020). Toll-like receptor 2 facilitates oxidative damage-induced retinal degeneration. Cell Rep 30(7): 2209-2224 e2205.
- Mullins, R. F., Schoo, D. P., Sohn, E. H., Flamme-Wiese, M. J., Workamelahu, G., Johnston, R. M., Wang, K., Tucker, B. A. and Stone, E. M. (2014). The membrane attack complex in aging human choriocapillaris: relationship to macular degeneration and choroidal thinning. Am J Pathol 184(11): 3142-3153.
- Nauta, A. J., Daha, M. R., Tijsma, O., van de Water, B., Tedesco, F. and Roos, A. (2002). The membrane attack complex of complement induces caspase activation and apoptosis. Eur J Immunol 32(3): 783-792.
- Niculescu, F. and Rus, H. (2001). Mechanisms of signal transduction activated by sublytic assembly of terminal complement complexes on nucleated cells. Immunol Res 24(2): 191-199.
- Sonoda, S., Spee, C., Barron, E., Ryan, S. J., Kannan, R. and Hinton, D. R. (2009). A protocol for the culture and differentiation of highly polarized human retinal pigment epithelial cells. Nat Protoc 4(5): 662-673.
- Yang, P., Tyrrell, J., Han, I. and Jaffe, G. J. (2009). Expression and modulation of RPE cell membrane complement regulatory proteins. Invest Ophthalmol Vis Sci 50(7): 3473-3481.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mulfaul, K. and Doyle, S. (2021). In vitro Measurement of Membrane Attack Complex in RPE Cells. Bio-protocol 11(4): e3916. DOI: 10.21769/BioProtoc.3916.
Category
Immunology > Complement analysis
Neuroscience > Cellular mechanisms
Molecular Biology > Protein > Detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








