- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ecdysone Quantification from Whole Body Samples of Drosophila melanogaster Larvae
Published: Vol 11, Iss 3, Feb 5, 2021 DOI: 10.21769/BioProtoc.3915 Views: 4256
Reviewed by: Pradeep Kumar BhaskarLijuan DuAnonymous reviewer(s)
Abstract
Steroid hormones strictly control the timing of sexual maturation and final body size both in vertebrates and invertebrates. In insects, the steroid hormone ecdysone controls the timing of the molts between larval instars as well as the transition to metamorphosis. Growth during the final instar accounts for over 80% of the increase in final mass in insects, and the duration of this growth period is driven by a sequence of small ecdysone pulses that ultimately induce metamorphosis. Historically the biologically active form of ecdysone, 20-hydroxyecdysone (20E), was quantified using radio-immunoassays, bioassays, or chromatography assays. However, these assays are methodologically complicated and often time consuming. Furthermore, collecting samples for precise measurements of ecdysone concentrations using these assays is limited in small insects like Drosophila melanogaster. Here, we describe an accurate and sensitive method to collect carefully-staged third instar larvae suitable for preparing samples for ecdysone quantification using a commercially-available 20E enzyme immunoassay (EIA). Because we resynchronize larval development at the molt to the final instar, collect large samples, and weigh each sample, we are able to detect a small ecdysone peak early in the final instar known as the critical weight ecdysone peak. This method detects peaks as low as 6 pg 20E/mg larval sample, allowing us to quantify other small ecdysone peaks in flies – the necessary prerequisite for eventually determining their regulation and function.
Keywords: 20-hydroxyecdysone (20E)Background
In a broad range of animals, development is tightly regulated by steroid hormones. In mammals, the onset of puberty is primarily regulated by sex steroid hormones (Koyama et al., 2020). In insects, the steroid hormone ecdysone regulates the timing of molting, including the timing of the metamorphic molts. Because small fluctuations in the concentrations of ecdysone are predicted to trigger key developmental events, developing methods to detect changes in ecdysone concentrations is crucial for scientists aiming to evaluate steroid hormone function. In this protocol, we describe how to collect high-quality samples from developmentally synchronized larvae, and how to prepare these samples for ecdysone quantifications using a commercially-available enzyme immunoassay kit.
Materials and Reagents
1.5 ml microcentrifuge tubes
Paper towels
Parafilm (Bemis Company, PM996 )
Aluminum foil
Disposable pestles
Crystal cuvette
200 parental virgin female Drosophila melanogaster and 150 parental males
Precoated (Mouse Anti-Rabbit IgG) ELISA 96-Well Strip Plate (Cayman Chemicals, catalog number: 400005 ), store at 4 °C
We have successfully used these plates up to one year after purchase.
Absolute methanol (VWR, Methanol absolute ≥ 99.8% ACS, catalog number: M1240-4LTGL ), store at room temperature
Absolute ethanol (VWR, Ethanol absolute ≥ 99.8%, Electran Molecular biology grade, catalog number: 437435L ), store at room temperature
20-Hydroxyecdysone EIA Kit (Cayman Chemicals, catalog number: 501390 )
20-Hydroxyecdysone (SciTech Chemicals, catalog number: S3314-001 )
Sucrose (50 g sucrose in 250 ml ddH2O. Preferably freshly prepared. Otherwise store at 4 °C for up to a week. Warm up to be about 20-25 °C before use)
Materials need for fly husbandry (dry yeast, plastic disposable vials, plugs, 60 mm tissue culture plates, embryo collection chambers)
Double-distilled water
Dry ice
70% ethanol
Molasses
Sugar
Cornmeal
Yeast Extract
Agar
Cornmeal/molasses medium (fly medium, see Recipes)
Equipment
Dumont #3 Forceps (Fine Science Tools, catalog number: 11231-30 ) or entomology forceps (Fisher Scientific, catalog number: S72110 )
Ultra-microbalance (Sartorius, Sartorius Cubis Series, SE2)
Cordless pestle motor (VWR International, VWR Cordless Pestle Motor, catalog number: 47747-370 )
Microcentrifuge with refrigeration
SpeedVac (Savant, SpeedVac Plus SC10A, catalog number: SC110A-8E210146-4K )
Microplate reader, Victor3 (PerkinElmer, Victor3, catalog number: 1420-050 )
Equipment needs for fly husbandry (fly station including a CO2 pad, CO2 supply system, humidity-controlled 25 °C fly incubator)
Spectrophotometer
-80 °C freezer
Rocker for agitation
Procedure
Collecting developmentally synchronized larvae
Cross approximately 200 parental virgin female Drosophila melanogaster and 150 parental males and maintain them on a fly medium sprinkled with instant dry yeast for 2 d at 25 °C.
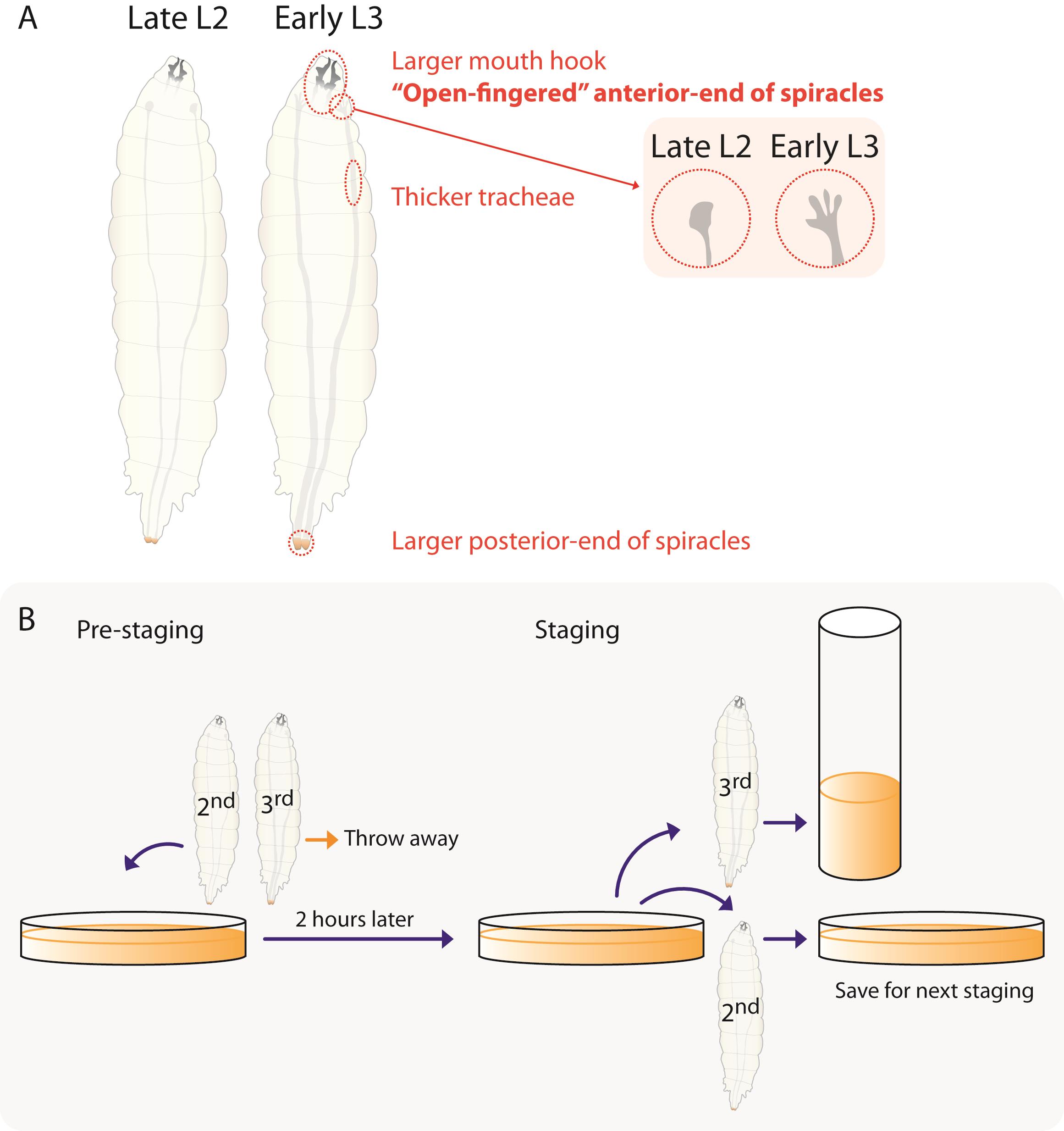
Figure 1. Schemes for precise staging. A. Morphological differences between late L2 and early L3 larvae. Comparing to L2 larvae, L3 larvae show 1) larger mouth hooks, 2) branched “finger-like” anterior spiracles, 3) thicker tracheae, and 4) larger orange-color posterior spiracles. In contrast, L2 larvae show 1) smaller mouth hooks, 2) “closed” anterior spiracles, 3) thinner tracheae, and yellow-color posterior spiracles. We mainly rely on the anterior spiracle morphology. B. Scheme of staging. At pre-staging, we collect only L2 larvae on new fly medium plates (pre-staging). Two hours later, we collect third instar larvae in food vials and put the rest of the L2 larvae in a new food plate for the next staging. If required, the larvae can be separated by sex, using either sex-specific genetic markers, such as paternally-inherited X or Y chromosome with a ubiquitous GFP marker that will only be expressed in males (for example see Testa et al., 2013), or using a transgenic sex lethal promoter to drive the expression of GFP in females only, or by assessing the presence or absence of visible gonad (the developing testes, Perry et al., 2014).To establish the larval cultures, allow the females to lay eggs for 2-4 h on 60 mm tissue culture plates filled with a cornmeal/molasses standard fly medium without any additional yeast at 25 °C. Standardize embryo density to 200 larvae/plate. Excess embryos can be either removed or transferred onto new larval culture plates with a small piece of fly medium to avoid damaging embryos. Due to the high numbers of larvae required for these assays, we recommend establishing a minimum of 4 larval culture plates (approximately 800 larvae) for every time point to be sampled.
Incubate the larval cultures in an incubator at 25 °C and 60% relative humidity under constant light. Constant light conditions are preferred because ecdysone secretion is circadian gated.
Once larvae develop to the end of the second instar (L2; in wildtype, generally 70-72 h after embryo collection), pour 20% sucrose solution (weight/volume) into the larval culture plates. This causes the larvae to float to the surface. We used the morphology of the anterior spiracles to distinguish between second and third instar larvae (Bodenstein, 1950, see also Figure 1A). Remove all third instar larvae and transfer second instar larvae onto a new larval culture plate using either Dumont #3 forceps or entomology forceps. We usually remove all L3 larvae from sucrose solution and transfer L2 larvae by double-checking one by one. This is necessary for pre-staging. (Figure 1B, see also Park et al., 2002, for details of morphological changes during ecdysis in D. melanogaster).
Two hours after transferring second instar larvae, collect all newly-molted L3 larvae into vials and transfer L2 larvae onto another new plate. Raise these newly-molted L3 larvae in a standard plastic fly vial with a normal cornmeal/molasses medium (usually up to 30 larvae/vial) without any additional yeast until the desired developmental time. The time transferred on a new fly medium is set as 0 h after L3 ecdysis (AL3E, see also Figure 1B). If required, newly molted larvae can be separated by sex either by examining the larvae for the presence of visible gonad (the developing testes) or by using genetic markers like paternally-inherited X or Y chromosome that express GFP (only in males, as in Testa et al., 2013) or a transgenic line that uses the sex lethal promoter to drive GFP expression (only in females, Perry et al., 2014).
Repeat this process every two hours until enough larvae have been collected for your experiment. We flood each larval culture plate a maximum of five times, to avoid excess handling of the larvae.
Standard preparation for ecdysone quantification (see also Figure 2)
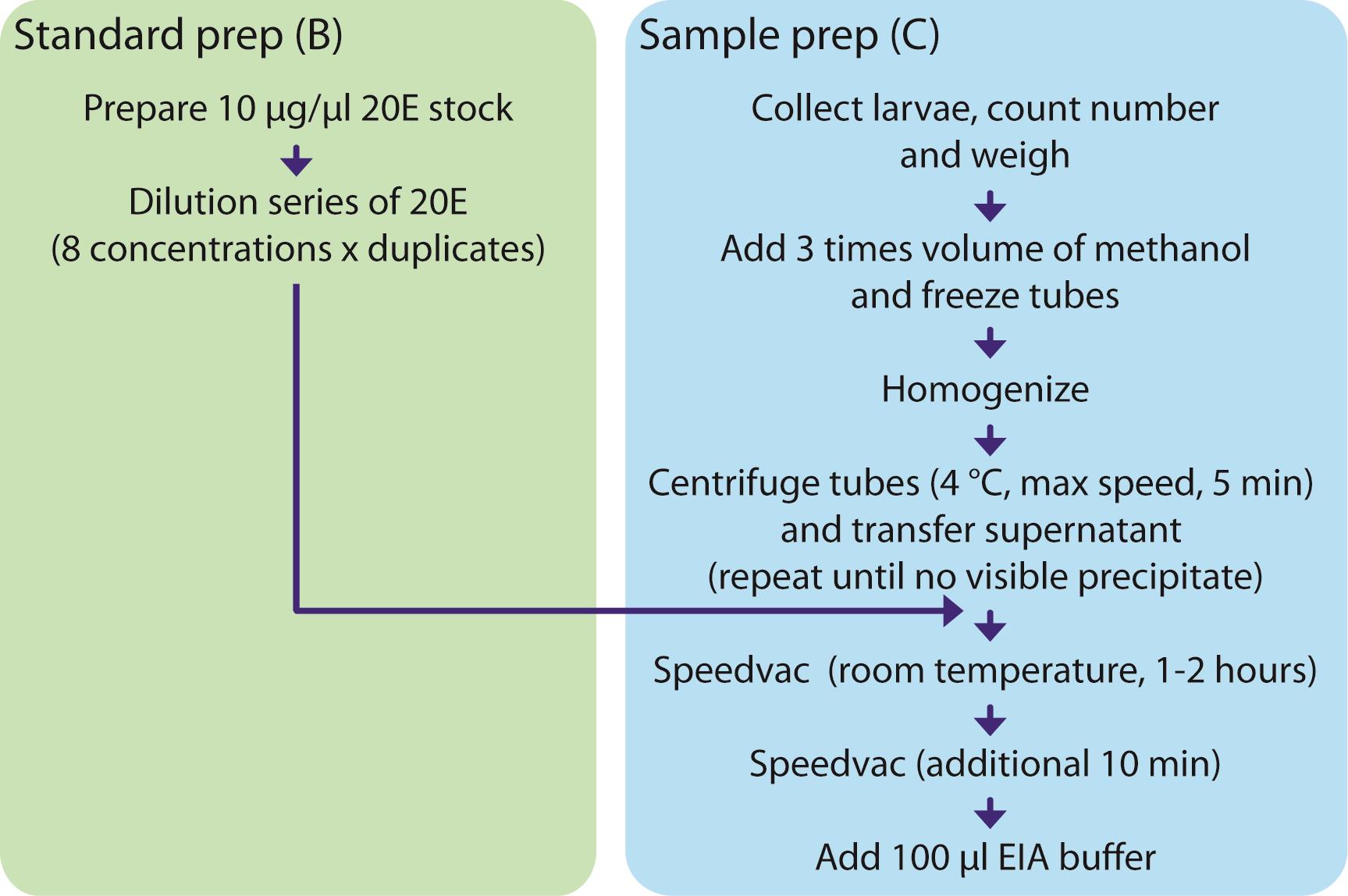
Figure 2. A schematic procedure of standard (Procedure B) and sample (Procedure C) preparationsPrepare a 10 µg/µl 20E stock solution in molecular biology or HPLC grade absolute ethanol.
To check the accuracy of the stock solution concentration, dilute the stock solution to 1:200 in absolute ethanol, and measure optical density at 240 nm in a crystal cuvette. The extinction coefficient of 20E in absolute ethanol is 12,656.52 and the molecular weight of 20E is 480.6. Therefore, the formula will be:
20E concentration (μg/μl) = (OD240) × (Dilution ratio; it will be 200 in this case) × 480.6/12,676.52
Dilute the stock solution to be 1,000 pg/μl, and make a dilution series: 1,000, 500, 250, 125, 62.5, 31.25, 15.625, and 7.8125 pg/μl for the standard curve using absolute ethanol.
Prepare standard solutions by taking 80 µl of the dilution series from Step B3, such that you obtain 80,000, 40,000, 20,000, 10,000, 5,000, 2,500, 1,250, 625 pg/tube respectively. We prepare two replicate tubes for each concentration (see Figure 3).
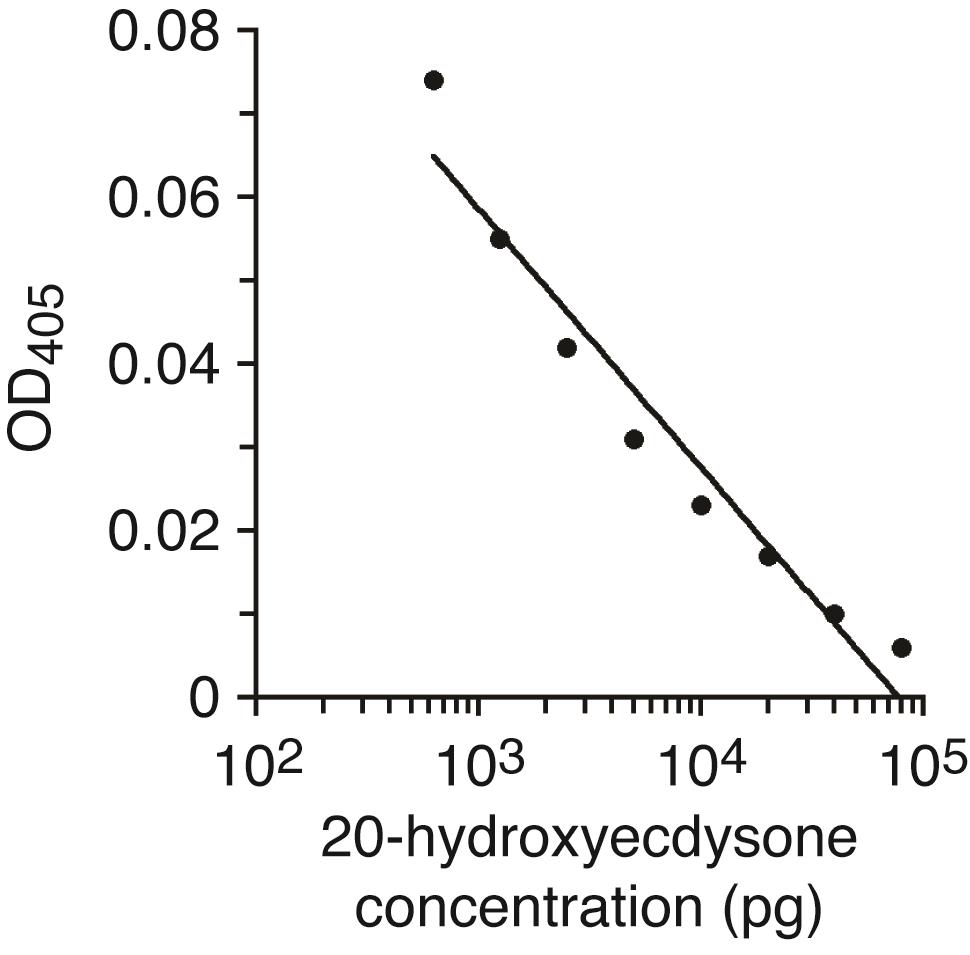
Figure 3. Standard curve for ecdysone quantification. Using this curve, we calculated ecdysone concentration as: Ecdysone (pg) = 68,473.88209 × e^(-70.40127 × OD405). R2 = 0.92.Evaporate ethanol completely using a Speedvac at room temperature simultaneously with Step C9. This typically takes 1-2 h. An additional 10 min may be necessary to completely remove all of the ethanol from the tubes. Pellets are usually invisible.
Sample preparation for ecdysone quantification (see also Figure 2)
At the desired time points, recover staged larvae from the fly medium using a 20% sucrose solution.
Wash collected larvae twice in double-distilled water to remove any fly medium residuals and wash out sucrose solution. Exclude larvae that appear noticeably smaller or larger for their cohort at this step, as it is likely that these larvae have not been correctly staged.
After drying larvae briefly on a small piece of paper towel for about a minute, weigh a group of larvae for one biological replicate together on an ultra-micro balance.
Usually, 20-30 mg of larvae (50-60 larvae for the onset of the L3 and 35-40 larvae for 24 h after the molt to the L3) is sufficient for accurate ecdysone quantification.
Place all weighed larvae in a 1.5 ml microcentrifuge tube, add three times the volume of absolute methanol (30 μl methanol for 10 mg larvae), and freeze them immediately on dry ice.
Keep all samples at -80 °C until you are ready for ecdysone quantification (Parvy et al., 2005).
When you are ready to begin ecdysone quantification, place all tubes on dry ice and homogenize frozen larvae carefully using disposable pestles and a cordless hand-pestle motor (VWR International).
Centrifuge tubes at 4 °C at maximum speed for 5 min.
Carefully transfer supernatant into new 1.5 ml microcentrifuge tubes. Repeat Steps C7 and C8 1-2 times until there is no visible precipitate.
Evaporate methanol completely using a Speedvac at room temperature for 1-2 h. An additional 10 min of evaporation may be necessary for complete removal of methanol from all tubes. A small precipitate with lipid from the larval fat body should be visible.
Ecdysone quantification by an enzyme immunoassay (EIA)
Redissolve the pellets in 100 µl of EIA buffer. Use only 50 µl of these sample solutions for the assay, saving the remaining 50 µl of this solution. In case the ecdysone concentration of the sample is too high, this sample can be diluted between 2- and 10-fold to be in a range of standard curve.
Set up a plate as outlined in the ecdysone EIA manufacturer’s instructions (Porcheron et al., 1989; 501390 .pdf">https://www.caymanchem.com/pdfs/ 501390 .pdf).
Cover the plate with a piece of parafilm and incubate overnight at 4 °C with gentle agitation.
Reconstitute Ellman’s reagent shortly before use.
Empty the plate by turning it over. Rinse each well with 300 μl wash buffer. Repeat four more times.
After the 5th wash, blot the plate on a few layers of clean paper towel to ensure any liquid will be removed completely.
Add 200 μl of Ellman’s reagent to each 96 well. Make sure the plate is sealed with a piece of parafilm and covered by a piece of aluminum foil, as the Ellman’s reagent is light sensitive. Incubate the plate at room temperature with gentle agitation.
Clean the bottom of the plate with 70% ethanol using paper towels.
Read the plate at a wavelength of 405 nm. Read the plate several times (approximately every 10 min until 60 min have passed post-incubation) to ensure that you capture ideal color development, meaning all standard samples from Step B4 show yellowish color but the color is not yet fully saturated. Multiple readings are necessary because color development continues with time until eventually the color of all samples becomes saturated.
Data analysis
Draw the standard curve as explained in the ecdysone EIA manufacturer’s instructions. Our standard curve for Figure 1A in our publication (Koyama et al., 2014) is shown in Figure 3. Based on this curve, we calculated ecdysone concentration using the following formula:
Ecdysone (pg) = 68,473.88209 × e^(-70.40127 × OD405).
We then corrected the ecdysone concentration in the sample for sample weights obtained in Step C3 to obtain pg ecdysone/mg larval weight.
Notes
Detecting the smaller pulses of ecdysone in third instar larvae requires both accurately staging the larvae from the molt to the third instar and weighing the samples (Warren et al., 2006).
Recipes
Cornmeal/molasses medium (fly medium)/1 L
Weigh and mix all ingredients as below except for Niapagin in a plastic beaker
Molasses 45 g
Sugar 75 g
Cornmeal 70 g
Yeast Extract 20 g
Agar 10 g
Boiling Water 1,100 ml
10% Niapagin 25 ml
Add boiling water gradually
Transfer the medium into 2 L flasks with screw caps (up to 1,800 ml)
Autoclave at 121 °C for 30 min
When the medium temperature reaches 45-50 °C, add Niapagin
Acknowledgments
We thank for our funding sources, Fundação Calouste Gulbenkian (Portugal) and Australian Research Council (FT170100259) to CKM and the Fundação para a Ciênica e a Tecnologia (Portugal) to TK (SFRH/BPD/74313/2010).
Competing interests
Both TK and CKM do not have any competing interests to declare.
References
- Bodenstein, D. (1950). The postembryonic development of Drosophila. In: Biology of Drosophila. Demerec, M. (Ed.). New York: Cold Spring Harbor Laboratory Press, pp. 275-367.
- Koyama, T., Rodrigues, M. A., Athanasiadis, A., Shingleton, A. W. and Mirth, C. K. (2014). Nutritional control of body size through FoxO-Ultraspiracle mediated ecdysone biosynthesis. Elife 3: e03091.
- Koyama, T., Texada, M. J., Halberg, K. A. and Rewitz, K. (2020). Metabolism and growth adaptation to environmental conditions in Drosophila. Cell Mol Life Sci 77: 4523-4551.
- Park, Y., Filippov, V., Gill, S. S. and Adams, M. E. (2002). Deletion of the ecdysis-triggering hormone gene leads to lethal ecdysis deficiency. Development 129(2): 493-503.
- Parvy, J. P., Blais, C., Bernard, F., Warren, J. T., Petryk, A., Gilbert, L. I., O’Connor, M. B. and Dauphin-Villemantaet, C. (2005). A role for betaFTZ-F1 in regulating ecdysteroid titers during post-embryonic development in Drosophila melanogaster. Dev Biol 282: 84-94.
- Perry, J. C., Harrison, P. W. and Mank, J. E. (2014). The ontogeny and evolution of sex-biased gene expression in Drosophila melanogaster. Mol Biol Evol 31: 1206-1219.
- Porcheron, P., Moriniere, M., Grassi, J. and Pradelles, P. (1989). Development of an enzyme immunoassay for ecdysteroids using acetylcholinesterase as label. Insect Biochem 19: 117-122.
- Testa, N. D., Ghosh, S. M. and Shingleton, A. W. (2013). Sex-specific weight loss mediates sexual size dimorphism in Drosophila melanogaster. PLoS One 8: e58936.
- Warren, J. T., Yerushalmi, Y., Shimell, M. J., O'Connor, M. B., Restifo, L. L. and Gilbert, L. I. (2006). Discrete pulses of molting hormone, 20-hydroxyecdysone, during late larval development of Drosophila melanogaster: correlations with changes in gene activity. Dev Dyn 235(2): 315-26.
Article Information
Copyright
Koyama and Mirth. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Koyama, T. and Mirth, C. K. (2021). Ecdysone Quantification from Whole Body Samples of Drosophila melanogaster Larvae. Bio-protocol 11(3): e3915. DOI: 10.21769/BioProtoc.3915.
- Koyama, T., Rodrigues, M. A., Athanasiadis, A., Shingleton, A. W. and Mirth, C. K. (2014). Nutritional control of body size through FoxO-Ultraspiracle mediated ecdysone biosynthesis. Elife 3: e03091.
Category
Developmental Biology
Biochemistry > Other compound > Steroid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









