- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Histochemical Staining of Suberin in Plant Roots
Published: Vol 11, Iss 3, Feb 5, 2021 DOI: 10.21769/BioProtoc.3904 Views: 7801
Reviewed by: Samik BhattacharyaGanesh Chandrakant NikaljeMather A Khan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Plate Growth Assay to Quantify Embryonic Root Development of Zea mays
Jason T. Roberts [...] David M. Braun
Oct 20, 2023 2241 Views

Live Imaging of the Shoot Apical Meristem of Intact, Soil-Grown, Flowering Arabidopsis Plants
Gabriele Bradamante
Jun 20, 2024 2273 Views

Micrografting Technique of Hevea brasiliensis In Vitro Plantlets
Florence Dessailly [...] Julie Leclercq
Feb 20, 2025 1480 Views
Abstract
Histological stains are useful tools for characterizing cell shape, arrangement and the material they are made from. Stains can be used individually or simultaneously to mark different cell structures or polymers within the same cells, and to visualize them in different colors. Histological stains can be combined with genetically-encoded fluorescent proteins, which are useful for understanding of plant development. To visualize suberin lamellae by fluorescent microscopy, we improved a histological staining procedure with the dyes Fluorol Yellow 088 and aniline blue. In the complex plant organs such as roots, suberin lamellae are deposited deep within the root on the endodermal cell wall. Our procedure yields reliable and detailed images that can be used to determine the suberin pattern in root cells. The main advantage of this protocol is its efficiency, the detailed visualization of suberin localization it generates in the root, and the possibility of returning to the confocal images to analyze and re-evaluate data if necessary.
Keywords: SuberinBackground
Suberin is a complex polyester that, together with cutin and lignin, forms a physical barrier in land plants. In Arabidopsis (Arabidopsis thaliana), suberin primarily accumulates in the cell walls of epidermal leaf cells, in the seed coat, in the periderm and in the cell wall of root endodermal cells. In the root, suberin deposition is dynamic process (Barberon et al., 2016; Andersen et al., 2018; Ursache et al., 2020) that is modulated by various nutritional stresses, drought stress, and phytohormone signaling pathways (Baxter et al., 2009; Kosma et al., 2014; Barberon et al., 2016). Suberin in root endodermal cells acts as a barrier that controls the uptake of water and nutrients (Baxter et al., 2009; Krishnamurthy et al., 2009 and 2011; Wang et al., 2019), but also provides a protective layer against various pathogens (Thomas et al., 2007; Ranathunge et al., 2008; Holbein et al., 2019; Zhou et al., 2020; Emonet et al., 2020). Moreover, suberin is also involved in periderm formation, the tissue that envelops secondary stems as part of the bark. Given its position deep within the root, it is technically challenging to study suberin. Developing a reliable tool to study the multifunctional activities of suberin and its precise localization in cells would contribute to new discoveries in many areas of plant research. A protocol for suberin staining was described previously by Lux et al. (2005) and subsequently modified over the years (Naseer et al., 2012; Fujita et al., 2020; Ursache et al., 2020). Here, we provide a standardized protocol to stain suberin and to acquire data from 10-15 roots at a single tile-scan image by confocal microscopy in a short time. This protocol will enhance the efficiency of data generation with possibility to re-analyze data if necessary.
Materials and Reagents
Square dishes 120 × 120 × 17 mm (Greiner Bio-One GmbH, catalog number: 688102 )
Flat-bottom 6-, 12-, and 24-well plates with lid (Stemcell Technology, catalog number: 100-0096 )
Adhesive or any available tape
2 ml Eppendorf tubes
Falcon tubes 15 or 50 ml (Stemcell Technology, catalog numbers: 38010, 100-0090; 38009, 100-0092 )
Aluminum foil
Parafilm “M” (Merck, catalog number: P7543-1EA )
SuperFrost microscope slides (Thermo Scientific, catalog number: 12362098 )
Coverslips (Thermo Scientific, catalog number: 12393138)
Note: Material needed for the protocol is illustrated in Figure 1.
Chambered cover glass (VWR, chamber slides-one chamber, NuncTM Lab-TekTM, catalog number: 734-2056 )
Arabidopsis seeds
GROWTH CONDITIONs: Sow surface-sterilized Arabidopsis seeds on half-strength MS agar square plates (45.5 ml). Stratify plates for 2 days at 4 °C in the dark and then transfer to growth chambers under a 16-h-light/8-h-dark photoperiod at 21 °C. Allow seedlings to grow on vertically-oriented plates for 5 days.
Note: 5-day old seedlings are the standard age of seedlings in our experimental setups. The age of seedlings can be adapted according to experiments.
Milli-Q Water (H2O)
Sucrose (VWR International, catalog number: 27483.294 )
Murashige and Skoog basal salt mixture (MS salts) (Duchefa Biochemie, catalog number: M0221.0050 )
2-[N-morpholino] ethanesulfonic acid (MES) (Duchefa Biochemie, catalog number: M1503.0100 )
Agar (Lab M, catalog number: MC029 )
Potassium hydroxide (KOH) (Merck KGaA, catalog number: 1.05021.1000 )
Ethanol (EtOH) (Sigma-Aldrich, catalog number: 32221-2.5L )
Lactic acid (Honeywell, FlukaTM, catalog number: 15636730 )
Fluorol Yellow 088 (Santa Cruz Biotechnology, catalog number: 81-37-8 )
Aniline blue (Sigma-Aldrich, catalog number: 66687-07-8), 0.5% in MilliQ Water
Seed sterilization with ethanol (see Recipes)
Half-strength MS medium (see Recipes)
0.03% Fluorol Yellow 088 solution (see Recipes)
0.5% Aniline blue solution (see Recipes)

Figure 1. Required material and equipment. A. Five-day-old Arabidopsis seedlings. Scale bar: 1 cm. B. Materials needed to stain samples. C. Dissolving Fluorol Yellow 088 in lactic acid in a 50 ml Falcon tube attached to a vortex mixer with adhesive tape. D. Dissolved Fluorol Yellow (left) and aniline blue (right) solutions.
Equipment
Growth chamber to grow plant materia (Percival, CLF, Plant Climatics, AR75-L3 )
Vortex mixer (Instrumentfirman Labora, model: Vortex-Genie 2 )
Water bath (Fisher Scientific)
Soft stainless steel tweezers (3B DISSECTION SOFT TWEEZERS, LABWORLD)
Inverted confocal microscope (Zeiss, model: LSM880 )
Note: A fully motorized X, Y, Z scanning stage is required to perform tile scan image acquisition experiments.
Objectives: 10× or 20× (suitable for monitoring the whole specimen or root, e.g., Figure 3C). For higher resolution images, a higher magnification objective, such as 40× or 63×, is required (Figure 3D)
Fluorescence signal detection system for GFP and other fluorescent reporters (Shaner et al., 2007)
Software
Software operating the confocal microscope
ImageJ (Abràmoff et al., 2004)
Microsoft Excel
Prism (GraphPad)
Procedure
Sample staining
Add 15-20 seedlings into each well containing 2 ml of Fluorol Yellow 088 (FY088) (e.g., 12-well plate, depending on the number of samples) (Figure 2A).
Close the lid and seal the plate with parafilm. Place the plate in a water bath on an aluminum foil raft at 70 °C for 20 min (Figures 2B-2D).
Note: Bend the edges of the aluminum foil to prevent sinking. During the incubation time in the FY088 solution, prepare the 0.5% aniline blue solution and keep in the dark at room temperature until use.
Transfer the seedlings from the FY088 solution to a new 12-well plate containing 2 ml water and incubate for 1 min at room temperature (Figure 2E).
Remove water by pipetting and add 2 ml of 0.5% aniline blue solution to each sample (Figure 2F). Incubate for 20 min in the dark at room temperature.
After incubation in aniline blue, move the seedlings to a new -, 12-, and 24-well plates containing 2-3 ml water (Figure 2G) and incubate for 10 min in the dark.
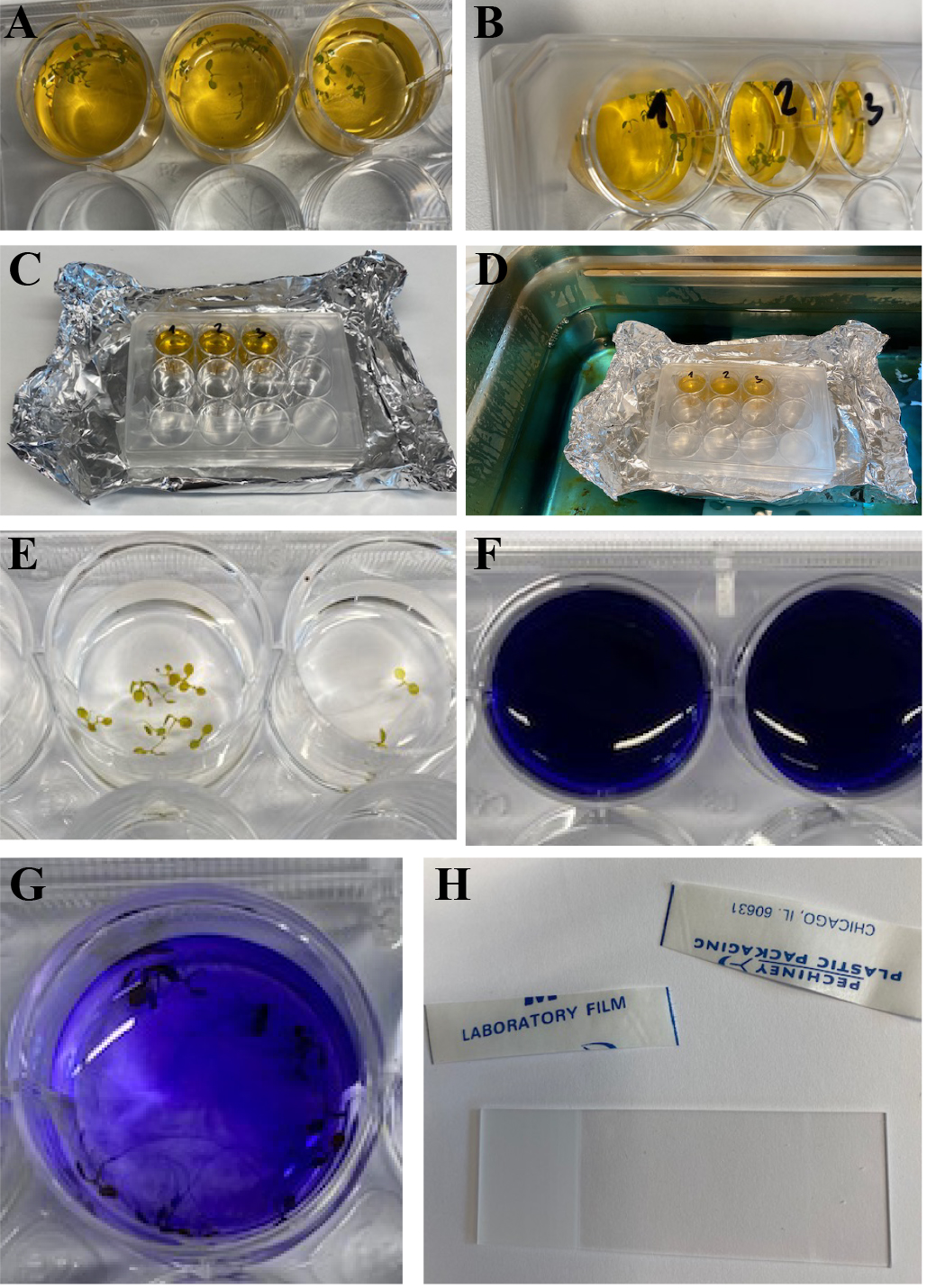
Figure 2. Sample Staining. A. Twelve-well plate containing Arabidopsis seedlings in 2 ml FY088 solution. B. Plate sealed with parafilm. C. Sealed well plate placed on an aluminum foil raft. D. Samples placed into a water bath at 70 °C for 20 min. E. Seedlings washed in 2 ml of water. F. Well plate containing 2 ml aniline blue solution with seedlings. G. Wash aniline blue in 2 ml water. H. Preparation of microscope slide with parafilm strips to make a “spacer”.
Sample preparation/mounting
Cut two strips of parafilm and wrap 2-3 times around both ends of the microscope slide (Figures 2H and 3A).
Add a few drops of water (30-50 μl) in the center of the slide between two parafilm strips. Position the seedlings onto this liquid using tweezers and cover with a cover glass slide (Figures 3A and 3B).
For instructions on using a one-well chambered cover glass and positioning the seedlings, please follow the protocol described previously: Marhavý and Benková, (2015).
Sample imaging
Prepare an inverted confocal microscope for use; set lasers (for GFP: 488 nm excitation, emission 500-550 nm) and objectives.
Note: For a broad overview of suberin localization in the root, we recommend performing tile scan imaging using either a 10× or 20× objective (in dry or water-immersion mode). For higher resolution images, we recommend using a 40× or 63× objective immersed in water or oil.
Parameters for image capture on a Zeiss LSM 880 shown in Figure 3C: Image size – x: 21793.72 µm, y: 18702.12 µm; Scaling X – 1510 µm, Scaling Y – 1510 µm; Tile scan – tiles1, overlap in percent: 20.0, tiling mode: rectangular grid (stitched image); scan mode – plane, zoom 1.1; Pixel dwell – 0.85 µs; master gain – 482; digital gain – 1.5; digital offset – 0.00; filters – SP 555; beam splitters – MBS: MBS 488 nm, MBS_InVis: plate, FW1: Rear.
Note: Parameters should be adjusted according to the specimen used.
Mount a glass slide or chamber with seedlings and place on the stage of the confocal microscope.
Activate the position list and tile scan (25 × 34 tiles) with 20% overlap between images.
Notes:
Adjust the number of tiles according to the approximate scanning area. Alternatively, activate automatic stitching (this can also be done later using the confocal software).
Parameters should be adjusted according to the size of the samples.
Find the root closest to the central point of the scanning area and mark the position.
Deactivate the position and start scanning.
Notes:
With the above-mentioned parameters, it will take approximately 15 min to obtain the final image. This provides enough time to prepare the next sample, as described above.
If the laboratory does not have suitable confocal microscope, images can be taken by a fully motorized Fluorescence stereo microscope with a tile scan option.
Export confocal images in TIFF or JPEG format. Open images in ImageJ or other software and analyze your images (Figures 3C, 3D).
Note: Be exact in terms of both the timing and procedure for each step to ensure reproducibility of your results.

Figure 3. Sample preparation and imaging. A. A 30 μl drop of water placed on the microscope slide between two parafilm strips. B. Place 10-15 seedlings onto each slide and cover with a cover slip. C. Tile scan of stained Arabidopsis seedlings. Yellow fluorescence indicates suberin deposition in the root. The white frame surrounds the area that is expanded in D for a detailed image of suberin lamellae. Scale bar: 1 mm. D. Detailed image of suberin lamellae in endodermal root cells. Scale bar: 100 μm.
Data analysis
Open image in ImageJ Set scale; Known distance; Unit of length (mm or cm); click/mark Global and press OK. Go to ImageJ, select drawing line (segmented or freehand line) and draw a line along the root, from base to tip, press “M” to measure root length. Repeat the procedure to measure the length of the suberized region of the root. Copy the results to Excel or Prisma GraphPad for further processing (for example, to calculate suberin occupancy in the root).
Note: We do not recommend analyzing FY fluorescence intensity, since this can be influenced by discrepancies in staining time, washing, and/or imaging.
Recipes
Seed sterilization with ethanol
Transfer seeds to 2 ml Eppendorf tubes (maximum volume of seeds to be sterilized per tube should not exceed 3 mm from the bottom of the tube)
Add 1 ml 70% EtOH, shake for 5 s and allow seeds to settle for 10 min
Dry seeds under the clean bench
Half-strength MS medium (1 L)
Combine 10 g sucrose, 2.3 g MS salts, and 0.5 g MES, adjust the pH to 5.9 with KOH, add 8 g agar and fill with ddH2O to 1 L
Autoclave medium for 20-30 min, and allow to cool to 55 °C before pouring plates
0.03% Fluorol Yellow 088 solution
Fluorol Yellow 088 in lactic acid at room temperature (Figures 1C, 1D) must be freshly prepared before each experiment
Note: To speed up dissolving FY 088 in lactic acid, attach a 50 ml Falcon tube to a vortex mixer with adhesive tape and vortex for 10-15 min (Figure 1C).
0.5% Aniline blue solution
Aniline blue in Milli-Q Water
Store in a dark place at room temperature
Acknowledgments
We are thankful to Niko Geldner for all the discussion and feedback to develop this protocol. This work was supported by the Swedish research Council a Starting grant to Peter Marhavý. Shahid Siddique was supported by a grant from the United States Department of Agriculture (project no. CA-D-ENM-2562-RR).
The protocol was improved based on previously published work of Lux et al. (2005).
Competing interests
There are no conflicts of interest or competing interest.
References
- Abràmoff, M. D., Magalhães, P. J. and Ram, S. J. (2004). Image processing with ImageJ. Biophotonics Int 11(7): 36-43.
- Andersen, T. G., Naseer, S., Ursache, R., Wybouw, B., Smet, W., De Rybel, B., Vermeer, J. E. M. and Geldner, N. (2018). Author Correction: Diffusible repression of cytokinin signalling produces endodermal symmetry and passage cells. Nature 559(7714): E9.
- Barberon, M., Vermeer, J. E., De Bellis, D., Wang, P., Naseer, S., Andersen, T. G., Humbel, B. M., Nawrath, C., Takano, J., Salt, D. E. and Geldner, N. (2016). Adaptation of Root Function by Nutrient-Induced Plasticity of Endodermal Differentiation. Cell 164(3): 447-459.
- Baxter, I., Hosmani, P. S., Rus, A., Lahner, B., Borevitz, J. O., Muthukumar, B., Mickelbart, M. V., Schreiber, L., Franke, R. B. and Salt, D. E. (2009). Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PLoS Genet 5(5): e1000492.
- Emonet, A., Zhou, F., Vacheron, J., Heiman, C. M., Tendon, V. D., Ma, K. W., Schulze-Lefert, P., Keel, C. and Geldner, N. (2020). Spatially Restricted Immune Responses Allow for Root Meristematic Activity During Bacterial Colonisation. bioRxiv. doi: https://doi.org/10.1101/2020.08.03.233817.
- Fujita, S., De Bellis, D., Edel, K. H., Koster, P., Andersen, T. G., Schmid-Siegert, E., Denervaud Tendon, V., Pfister, A., Marhavy, P., Ursache, R., Doblas, V. G., Barberon, M., Daraspe, J., Creff, A., Ingram, G., Kudla, J. and Geldner, N. (2020). SCHENGEN receptor module drives localized ROS production and lignification in plant roots. EMBO J 39(9): e103894.
- Holbein, J., Franke, R. B., Marhavy, P., Fujita, S., Gorecka, M., Sobczak, M., Geldner, N., Schreiber, L., Grundler, F. M. W. and Siddique, S. (2019). Root endodermal barrier system contributes to defence against plant-parasitic cyst and root-knot nematodes. Plant J 100(2): 221-236.
- Kosma, D. K., Murmu, J., Razeq, F. M., Santos, P., Bourgault, R., Molina, I. and Rowland, O. (2014). AtMYB41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types. Plant J 80(2): 216-229.
- Krishnamurthy, P., Ranathunge, K., Franke, R., Prakash, H. S., Schreiber, L. and Mathew, M. K. (2009). The role of root apoplastic transport barriers in salt tolerance of rice(Oryza sativa L.). Planta 230(1): 119-134.
- Krishnamurthy, P., Ranathunge, K., Nayak, S., Schreiber, L. and Mathew, M. K. (2011). Root apoplastic barriers block Na+ transport to shoots in rice(Oryza sativa L.). J Exp Bot 62(12): 4215-4228.
- Lux, A., Morita, S., Abe, J. and Ito, K. (2005). An improved method for clearing and staining free-hand sections and whole-mount samples. Ann Bot 96(6): 989-996.
- Marhavý, P. and Benková, E. (2015). Real-time Analysis of Lateral Root Organogenesis in Arabidopsis. Bio-protocol 5(8): e1446.
- Naseer, S., Lee, Y., Lapierre, C., Franke, R., Nawrath, C. and Geldner, N. (2012). Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci U S A 109: 10101-10106.
- Ranathunge, K., Thomas, R. H., Fang, X., Peterson, C. A., Gijzen, M. and Bernards, M. A. (2008). Soybean root suberin and partial resistance to root rot caused by Phytophthora sojae. Phytopathology 98(11): 1179-1189.
- Shaner, N. C., Patterson, G. H. and Davidson, M. W. (2007). Advances in fluorescent protein technology. J Cell Sci (24): 4247-4260.
- Thomas, R., Fang, X., Ranathunge, K., Anderson, T. R., Peterson, C. A. and Bernards, M. A. (2007). Soybean root suberin: anatomical distribution, chemical composition, and relationship to partial resistance to Phytophthora sojae. Plant Physiol 144(1): 299-311.
- Ursache, R., De Jesus Vieira-Teixeira, C., Tendon, V. D., Gully, K., De Bellis, D., Schmid-Siegert, E., Andersen, T, G., Shekhar, V., Calderon, S., Pradervand, S., Nawrath, C., Geldner, N. and Vermeer, J. E. M. (2020). GDSL-domain containing proteins mediate suberin biosynthesis and degradation, enabling developmental plasticity of the endodermis during lateral root emergence. bioRxiv. doi: https://doi.org/10.1101/2020.06.25.171389.
- Wang, P., Calvo-Polanco, M., Reyt, G., Barberon, M., Champeyroux, C., Santoni, V., Maurel, C., Franke, R. B., Ljung, K., Novak, O., Geldner, N., Boursiac, Y. and Salt, D. E. (2019). Surveillance of cell wall diffusion barrier integrity modulates water and solute transport in plants. Sci Rep 9(1): 4227.
- Zhou, F., Emonet, A., Denervaud Tendon, V., Marhavy, P., Wu, D., Lahaye, T. and Geldner, N. (2020). Co-incidence of Damage and Microbial Patterns Controls Localized Immune Responses in Roots. Cell 180(3): 440-453 e418.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Marhavy, P. and Siddique, S. (2021). Histochemical Staining of Suberin in Plant Roots. Bio-protocol 11(3): e3904. DOI: 10.21769/BioProtoc.3904.
Category
Plant Science > Plant developmental biology > General
Molecular Biology > DNA > DNA cloning
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









