- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Using 14C-acetate Pulse-chase Labeling to Study Fatty Acid and Glycerolipid Metabolism in Plant Leaves
Published: Vol 11, Iss 3, Feb 5, 2021 DOI: 10.21769/BioProtoc.3900 Views: 4069
Reviewed by: Agnieszka ZienkiewiczAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Fluorescent Biosensor Imaging of Nitrate in Arabidopsis thaliana
Yen-Ning Chen and Cheng-Hsun Ho
Aug 20, 2023 3635 Views

Vegetative Propagation of Cannabis sativa and Resin Obtained From its Female Inflorescences
Sebastián D´Ippolito [...] Silvana L. Colman
Feb 20, 2025 1735 Views

A New Approach to Detect and Semi-quantify All Molecular Species and Classes of Anionic Phospholipids Simultaneously in Plant Samples
Manon Genva [...] Laetitia Fouillen
Apr 20, 2025 1718 Views
Abstract
Lipids metabolism is comprised of networks of reactions occurred in different subcellular compartments. Isotopic labeling is a good way to track the transformations and movements of metabolites without perturbing overall cellular metabolism. Fatty acids, the building blocks of membrane lipids and storage triacylglycerols, are synthesized in plastids. The immediate precursor for fatty acid synthesis is acetyl-CoA. Exogenous acetate is rapidly incorporated into fatty acids in leaves and isolated plastids because it can diffuse freely through cellular membranes, enter the plastid where it is rapidly metabolized to acetyl-CoA. Therefore, isotope-labeled acetate is often used as a tracer for the investigation of fatty acid synthesis and complex lipid metabolism in plants and other organisms. The basic principle of isotope labeling and its recent technological advances have been reviewed (Allen et al., 2015). The present protocol describes the use of 14C-labeled acetate to determine rates of fatty acid synthesis and degradation and to track the metabolism of glycerolipids in leaves. This method, which is often referred to as acetate pulse-chase labeling, has been widely used to probe various aspects of lipid metabolism (Allen et al., 2015), including the role of autophagy in membrane lipid turnover (Fan et al., 2019) and the interplay between lipid and starch metabolism pathways (Yu et al., 2018).
Keywords: 14C-acetate pulse-chaseMaterials and Reagents
Pipette tips
90 mm Petri dish (Thermo Fisher Scientific, catalog number: 101R20 )
1.5 ml and 0.5 ml Eppendorf safe-lock microcentrifuge tube
TLC Silica gel 60 plate (Merck)
Filter paper
Pencil
3-4-week-old Arabidopsis plants
14C-acetate (American Radiolabeled Chemicals, catalog number: ARC 0101D-1 mCi )
Double-distilled water (ddH2O)
MES (Sigma-Aldrich, catalog number: M3671 )
MS salt (Gentaur, catalog number: LSP03-50LT )
Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9333 )
Chloroform (Thermo Fisher Scientific, catalog number: C607 )
Methanol (VWR Chemicals, catalog number: BDH1135-4LG )
Acetic acid (Macron, catalog number: MK-8817-500 )
Acetone (Macron, catalog number: MK244310 )
Toluene (VWR Chemicals, catalog number: BDH1151-4LG )
Hexane (VWR Chemicals, catalog number: BDH1129-4LG )
Diethyl ether (Macron, catalog number: MK085010 )
(NH4)2SO4 (Mallinckrodt Chemicals, catalog number: 3512-12 )
Nitrogen gas
Scintillation liquid (PerkinElmer, catalog number: 6013199 )
Tween-20 (Sigma-Aldrich, CAS number: 9005-64-5 )
H3PO4 (Sigma-Aldrich, CAS number: 7664-38-2 )
Labeling medium (see Recipes)
Lipid extraction solution (see Recipes)
KCl solution (see Recipes)
Developing solvent 1 (see Recipes)
Developing solvent 2 (see Recipes)
Equipment
Pipettes
Analytical balance (Mettler Toledo, model: AE100 )
Shaker (Bellco Biotechnology, model: 7644-10115 )
Vortexer (Denville Scientific, model: S7030 )
Fume hood
Nitrogen evaporator (Thermo Fisher Scientific, model: TS-18825 )
Oven (Thermo Fisher Scientific, model: 13-247-650G )
Personal safety equipment: lab coat, nitrile gloves, and eye protection
Cork borer
TLC developing tank
Scissors
Survey Meter (Ludlum Measurements, model: Model 3 )
Autoradiography cassette (FUJIFILM, model: BAS 2040 )
Phosphor screen (FUJIFILM, model: BAS-SR-2040 )
Scintillation analyzer (PerkinElmer, model: Tri-Carb 2900TR )
Typhoon FLA 7000 biomolecular imager (GE Healthcare)
-80 °C and -20 °C freezer
Centrifuge (Eppendorf, model: 5425 )
Software
ImageQuant5.2 (GE Healthcare)
Procedure
Radioactive labeling (pulse)
Pour 10 ml labeling medium into a Petri dish. Cut the fully expanded rosette leaves and use cork borer (~8 mm diameter, or smaller, depending on the size of the source leaves) to prepare leaf disks. Float 40-50 leaf disks with the abaxial side down on the labeling medium in Petri dishes.
Place the Petri dishes on a rotator (50 rpm/s), add 0.1 mCi of 14C-acetate to the labeling medium to start the labeling at room temperature (22-25 °C) under a light intensity of 80 µmol m-2 s-1.
After 1 h of incubation with the radiotracer (pulse), remove the labeling medium and add 15 ml ddH2O to the Petri dish to wash the leaf disks on the rotator for 3-4 times, 5 min each.
Weigh a 1.5 ml Eppendorf safe-lock microcentrifuge tube. Transfer leaf disks (6-8 disks per sample) to clear filter paper and blot them dry. Transfer the leaf disks in the weighted Eppendorf tube and weigh it again. Calculate the fresh weight (FW) of the leaf disks. The remaining leaf disks are incubated in the labeling medium without isotope under the same conditions as described for the pulse and then used for the chase (Procedure B).
Extract lipids from 1 h-pulsed leaf disks:
Add lipid extraction buffer with a pipette (700 μl per 100 mg FW) to the Eppendorf tubes with leaf disks mentioned above, and mix by vortexing for 30 min.
Add 1 M KCl buffer (0.5 volume of extraction buffer) to the extract, vortex and centrifuge at 12,000 × g at room temperature for 1 min. Lipids stay in the lower chloroform phase. Transfer the chloroform phase to a new Eppendorf tube and store it at -20 °C for further use.
Note: The labeling medium with 14C-acetate can be reused, if desired. We recommend freezing promptly at -20 °C, with appropriate labels and storage for isotopes, if it is to be reused.
Chase
Liquid scintillation counting
Aliquot 10 μl lipid extract to 0.5 ml Eppendorf tubes, dry them in the fume hood. Add 20 μl hexane to dissolve the lipids, then add 200 μl scintillation liquid and mix by vortexing.
Radioactivity associated with total lipids was determined by liquid scintillation counting using a Tri-carb (Perkin-Elmer).
Thin layer chromatography (TLC) and phosphor imaging
Preparation of TLC plates: submerge a 20 cm × 20 cm Silica Gel 60 (Merck) TLC plate into 0.15 M (NH4)2SO4 solution for 30 s, air dry the plate for at least 2 days.
On the day of experiments, bake the TLC plates in an oven at 120 °C for 3 h.
Draw a straight line (1 cm from the edge of the plate) across the plate using a pencil. Slowly spot 60-80 μl of lipid extract on the line using a 100 μl pipette under a slow stream of N2 in a fume hood. Keep the lipid spots smaller than 1 cm in diameter.
Polar lipids separation
When the lipid spots completely dried in the fume hood, add 50 ml of the developing solvent 1 into a sealable TLC developing tank and put the TLC plate into the tank with the sample end facing down. Seal the tank and let the solvent ascend to half of the plate.
Take out the plate, completely dry it in the fume hood. Pure out the developing solvent 1 from the tank, and let the tank completely dry in the fume hood.
Neutral lipids separation
Pour 50 ml of the developing solvent 2 into the tank, place the TLC plate back into the tank with the sample end facing down.
When the developing solvent 2 front has reached 1 cm from the top of the plate, take out the plate and dry completely in the fume hood.
Note: Here we use two-phase TLC to separate polar lipids and neutral lipids. However, polar lipids and neutral lipids can be separated by two individual TLC plates using the developing solvent 1 and the developing solvent 2, respectively.
Phosphor imaging
Wrap the TLC plate with Saran wrap. Place it in the autoradiography cassette with the silica side upward, cover it with phosphor screen and close the cassette.
Place the cassette in -80 °C freezer for 1-2 days before taking out the phosphor screen for phosphor imaging captured by a Typhoon FLA 7000 imager (GE Healthcare).
Quantify the relative radioactivity associated with individual bands by the ImageQuant software.
Harvest leaf disks at different time points (we use 1 day, 2 day, 3 day) and extract lipids as described before.
Data analysis
Phosphor imaging captured by a Typhoon FLA 7000 imager is shown in Figure 1A. The label associated with monogalactosyldiacylglycerol (MGDG) and triacylglycerol (TAG) during different time points is shown in Figures 1B and 1C, respectively. The relative radioactivity associated with individual lipid bands was quantified by the ImageQuant software. Radioactivity associated with total fatty acids after 1 h of labeling and 3 d of chase were determined by liquid scintillation counting. Rates of fatty acid synthesis and degradation are shown in Figures 1D and 1E, respectively.
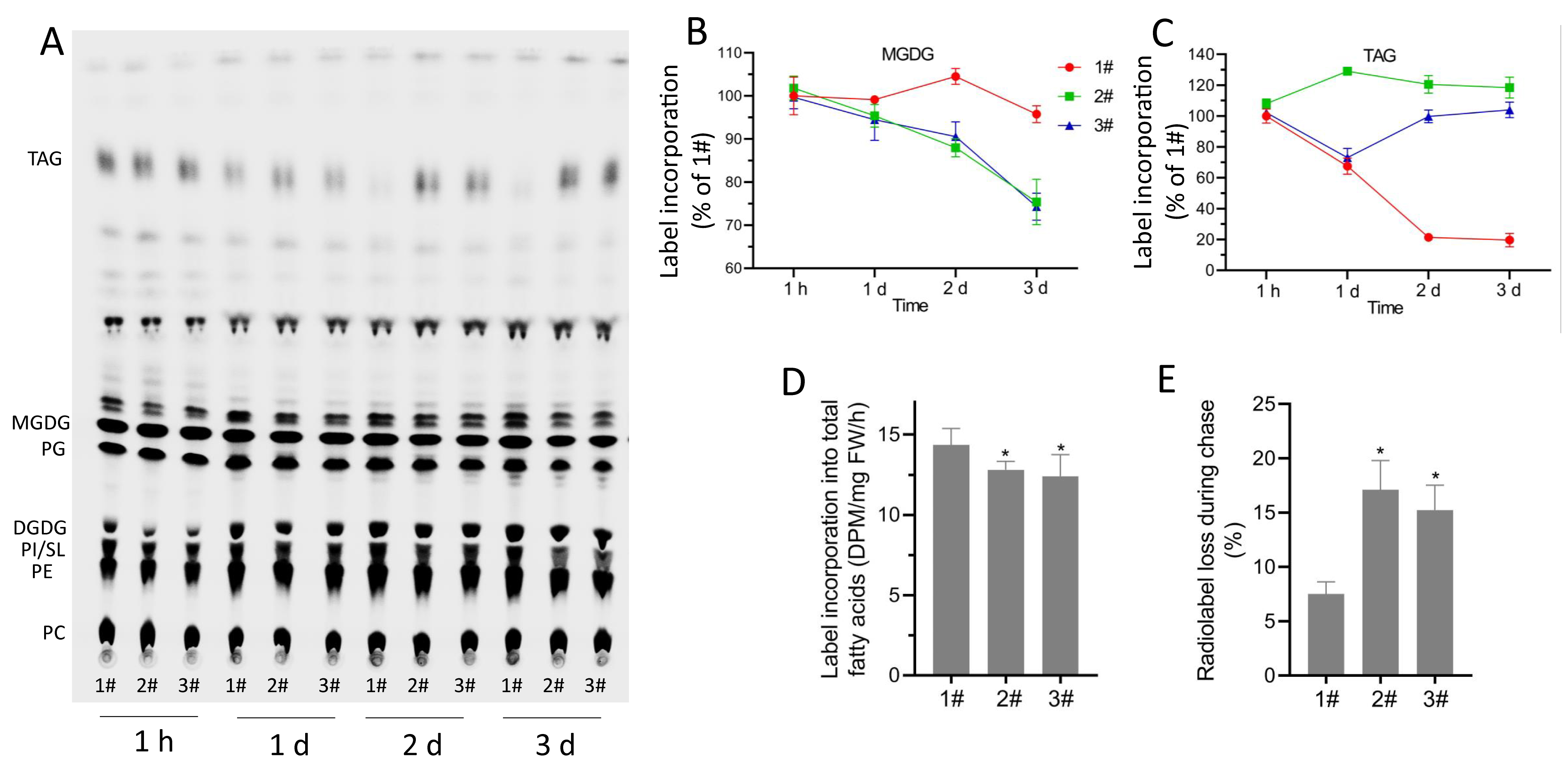
Figure 1. Assays for synthesis and degradation by pulse-chase 14C-acetate labeling. A. Phosphor imaging of lipids. B-C. Changes in radioactivity associated with MGDG and TAG during 3-d chase. D. Rate of 14C-acetate incorporation into total fatty acids in leaves of different lines. E. Radiolabel loss during the 3-d chase following 1 h of incubation with 14C-acetate. Data are means of three independent experiments ± SD. TAG: triacylglycerol; MGDG: monogalactosyldiacylglycerol; PG: phosphatidylglycerol; DGDG: digalactosyldiacylglycerol; PE: phosphatidylethanolamine; PI/SL: phosphatidylinositol/sulfoquinovosyldiacylglycerol; PC: phosphatidylcholine.
Recipes
Labeling medium
Lipid extraction solution
KCl solution
Developing solvent 1
Developing solvent 2
20 mM MES, pH 5.5
One-tenth strength of MS salts
0.01% Tween20
Methanol/chloroform/formic acid = 2/1/0.1
1 M KCl
0.2 M H3PO4
Acetone/toluene/H2O = 91/30/8
Hexane/diethyl ether/acetic acid = 70/30/1
Acknowledgments
We acknowledge the financial support from the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences (DE-SC0012704).
Competing interests
The authors declare no conflicts of interest or competing interests.
References
- Allen, D. K., Bates, P. D. and Tjellstrom, H. (2015). Tracking the metabolic pulse of plant lipid production with isotopic labeling and flux analyses: Past, present and future. Prog Lipid Res 5897-120.
- Fan, J., Yu, L. and Xu, C. (2019). Dual role for autophagy in lipid metabolism in Arabidopsis. Plant Cell 31(7): 1598-1613.
- Yu, L., Fan, J., Yan, C. and Xu, C. (2018). Starch Deficiency Enhances Lipid Biosynthesis and Turnover in Leaves. Plant Physiol 178(1): 118-129.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yu, L., Zhou, C., Fan, J. and Xu, C. (2021). Using 14C-acetate Pulse-chase Labeling to Study Fatty Acid and Glycerolipid Metabolism in Plant Leaves. Bio-protocol 11(3): e3900. DOI: 10.21769/BioProtoc.3900.
Category
Biochemistry > Lipid
Plant Science > Plant biochemistry > Metabolite
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








