- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Low- and High-resolution Dynamic Analyses for Magnetic Resonance Spectroscopy Data
Published: Vol 11, Iss 2, Jan 20, 2021 DOI: 10.21769/BioProtoc.3892 Views: 3626
Reviewed by: Zijian ZhangHélène LégerMasashi AsaiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Precise Targeting of Single Microelectrodes to Orientation Pinwheel Centers
Xue Mei Song [...] Anna Wang Roe
Jun 5, 2020 3958 Views

Visual-stimuli Four-arm Maze test to Assess Cognition and Vision in Mice
Jean-Philippe Vit [...] Maya Koronyo-Hamaoui
Nov 20, 2021 5290 Views

Mouse Vestibulo-Ocular Reflex Testing for Otolith Organs and Horizontal Semicircular Canal
Tong Zhao [...] Fangyi Chen
Nov 20, 2025 1236 Views
Abstract
Magnetic resonance spectroscopy (MRS) can be used to measure in vivo concentrations of neurometabolites. This information can be used to identify neurotransmitter involvement in healthy (e.g., perceptual and cognitive processes) and unhealthy brain function (e.g., neurological and psychiatric illnesses). The standard approach for analyzing MRS data is to combine spectral transients acquired over a ~10 min scan to yield a single estimate that reflects the average metabolite concentration during that period. The temporal resolution of metabolite measurements is sacrificed in this manner to achieve a sufficient signal-to-noise ratio to produce a reliable estimate. Here we introduce two analyses that can be used to increase the temporal resolution of neurometabolite estimates produced from MRS measurements. The first analysis uses a sliding window approach to create a smoothed trace of neurometabolite concentration for each MRS scan. The second analysis combines transients across participants, rather than time, producing a single “group trace” with the highest possible temporal resolution achievable with the data. These analyses advance MRS beyond the current “static” application by allowing researchers to measure dynamic changes in neurometabolite concentration and expanding the types of questions that the technique can be used to address.
Keywords: Magnetic resonance spectroscopyBackground
Magnetic resonance spectroscopy (MRS) can be used in vivo to measure the concentration of neurometabolites within the brain. The blood-oxygen-level-dependent signal measured using functional magnetic resonance imaging (fMRI) can provide evidence of neural activity. However, MRS can provide evidence that can be used to distinguish between different types of activity, e.g., excitatory and inhibitory. For the purpose of understanding neural mechanisms, identifying the involvement of neurotransmitter systems that support sensory/cognitive processes can be more informative than locating regions of neural representation.
The methodological approaches used to investigate sensory/cognitive processing with MRS can be separated into two categories: correlational and functional. The common correlational approach is to measure the concentration of metabolites in a group of participants, within a brain region of interest, and test whether individual variance of a particular metabolite is related to performance on a task supported by the sensory/cognitive process (Edden et al., 2009; Rideaux and Welchman, 2018).
A common functional approach used to identify metabolite involvement is to measure baseline metabolite concentration while a participant is “at rest” and compare this with another measurement taken while the participant performs a task. A difference in metabolite concentration observed between the two measurements may be taken as evidence of its involvement in the sensory/cognitive processes supporting the task (Kurcyus et al., 2018; Rideaux et al., 2019).
While MRS can be used to differentiate excitatory and inhibitory transmission within the brain, compared to fMRI, it is severely limited in terms of its temporal resolution, which is constrained by the signal-to-noise ratio of measurements acquired using the technique. Although the duration that restricts the temporal resolution of MRS (i.e., the relaxation time) can be similar to that of fMRI (i.e., ~2 s), in order to yield a reliable estimate of metabolite concentration from within the brain, multiple transients must be combined to reach a sufficient signal-to-noise ratio (Mikkelsen et al., 2018). For example, it is common to combine between 200-300 transients (~10 min) to produce a single measure of metabolite concentration (Puts and Edden, 2012).
The information provided using this approach is severely limited. In particular, by reducing the measure of neurometabolite concentration to a single estimate averaged across an experimental condition, dynamic changes in concentration during this period are obscured. By contrast, establishing the temporal dynamics in neurometabolite concentration reveals the timing and magnitude of change in different neurotransmitters and novel relationships between neurotransmitters. This information is essential for a comprehensive understanding of the role of neurotransmitters in sensory and cognitive processes, and may inform our understanding of the balance between excitation and inhibition, which is thought to be a key factor in multiple neurological and psychiatric illnesses (Bradford, 1995; Rubenstein and Merzenich, 2003; Kehrer et al., 2008).
Here we describe two analyses that can be used to estimate dynamic neurometabolite concentration from MRS spectra. The first analysis uses a sliding window approach to create a smoothed trace of neurometabolite concentration for each MRS scan. The second analysis combines transients across participants, rather than time, producing a single “group trace” with the highest possible temporal resolution achievable with the data.
Equipment
3-tesla MRI scanner unit (Siemens PRISMA, Siemens Healthcare)
32-channel head coil for MRI signal reception (Siemens PRISMA, Siemens Healthcare)
Software
MATLAB R2018B (The MathWorks, www.mathworks.com)
Procedure
Set up the subject in the MRI scanner using a 32-channel head coil.
Perform magnetic resonance imaging (MRI) anatomical scan with the following parameters:
Whole-brain T1-Magnetization Prepared Rapid Acquisition of Gradient Echoes (MPRAGE, sagittal 3D turbo-fast low angle shot sequence, acquisition matrix 256 × 256, isotropic resolution 1 mm, 176 slices, repetition time (TR) = 2250 ms, echo time (TE) = 3.02 ms, inversion time (TI) = 900 ms, flip angle 9°, acquisition time 5 min 38 s). Parameters were reproduced from Rideaux and Welchman (2019).
Perform magnetic resonance spectroscopy (MRS) scan, using the MRI anatomical scan to position the voxel, with the following parameters:
MEGA-PRESS sequence (Mescher et al., 1996 and 1998): TE = 68 ms, TR = 3000 ms; 256 transients of 2048 data points, acquisition time 12 min 48 s; 14.28 ms Gaussian editing pulse at 1.9 (ON) and 7.5 (OFF) ppm. Apply water suppression using variable power with optimized relaxation delays (VAPOR; Tkáč and Gruetter, 2005) and outer volume suppression. Position voxel (3 × 3 × 2 cm) medially in the occipital lobe; with the lower face aligned with the cerebellar tentorium and positioned so to avoid including the sagittal sinus and to ensure it remains within the occipital lobe (Figure 1). Perform automated shimming followed by manual shimming to achieve approximately 12 Hz water linewidth.
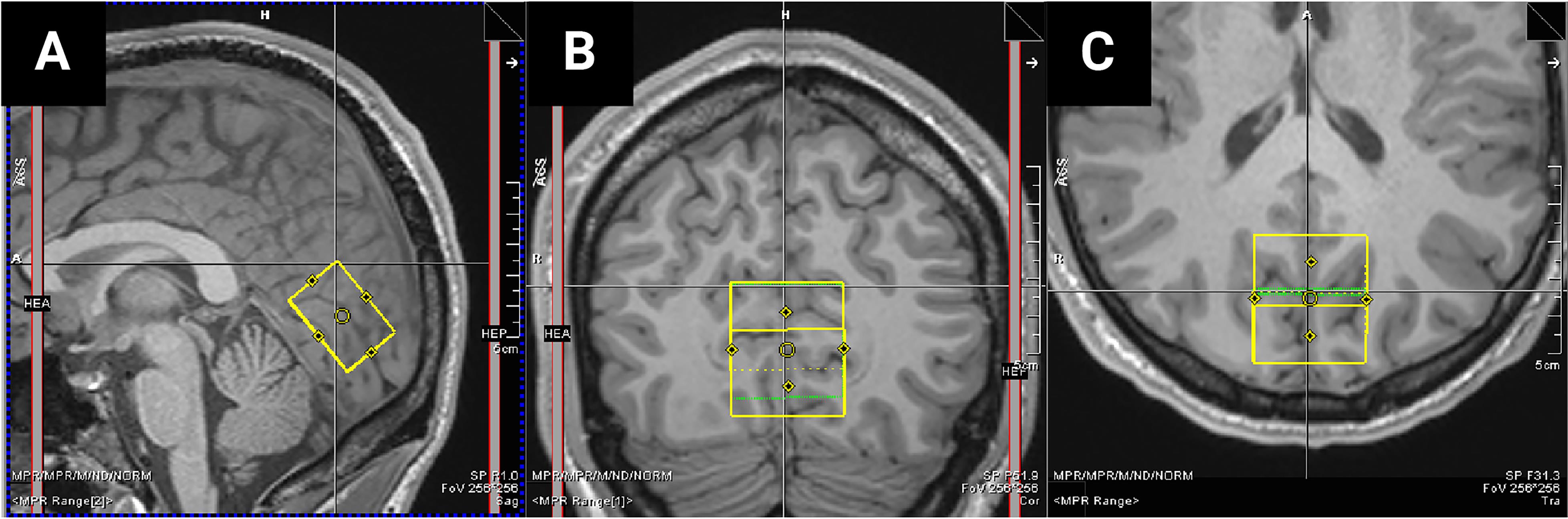
Figure 1. MRS voxel placement. Screenshot taken from Siemens MRI scanner terminal showing an example of voxel placement (yellow bounding box) in the (A) sagittal, (B) coronal, and (C) axial plane.
Data analysis
To preprocess the data, for all transients, fit a Lorentzian model to the creatine (Cr) signal at 3.0 ppm to obtain frequency, phase, area, and full width at half maximum (FWHM) parameters (Figure 2A). Remove outlier transients with any parameter estimates > 3 standard deviations from the mean within the scan. Align the frequency of the all transients using the “circshift” MATLAB function, shifting each spectrum by the difference between the estimated frequency of the Cr signal and the target frequency at 3.0 ppm (Figure 2B). Align the phase of each spectra with the following:
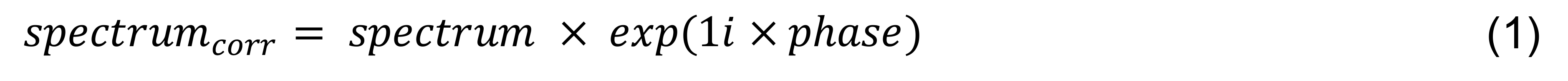
where spectrum and spectrumcorr denote the uncorrected and phase corrected spectra, respectively, and phase denotes the phase parameter estimate in radians of the Lorentzian model.
Follow the following steps to perform the sliding window analysis. For each scan, calculate the average of all i) OFF transients and ii) OFF – ON (i.e., difference spectrum) transients between the first acquisition and the size of the sliding window (i.e., 128; Figure 2C). Quantify the total creatine (tCr) and total N-acetylaspartate (tNAA) signal intensity by fitting a single Lorentzian model to the average OFF spectra at 3.0 ppm and 2.0 ppm, respectively. Quantify GABA+ (γ-aminobutyric acid and co-edited macromolecules; 3 ppm) and Glx (a complex comprising Glu and Glutamine (Gln); 3.8 ppm) signal intensity by fitting single- and double- Gaussian models, respectively (Figure 2D-2E). Calculate neurometabolite signal intensities as the area of the fitted peak/s. Repeat this process, starting at the second ON/OFF transient, rather than the first, until the last transient in the scan is reached. That is, slide the window across from the point where one edge is aligned to the first transient in the scan to the point where the other edge is aligned to the last transient in the scan. Omit data from further analysis for which the FWHM of any of the quantified neurometabolites (tCr, tNAA, GABA+, or Glx) is > 3 standard deviations from the mean across the scan. This analysis provides a temporal trace of metabolite concentration for each metabolite, for each scan (Figure 2F). Compare traces between subjects, or conditions, to test for differences in neurometabolite temporal dynamics. Additionally, calculate difference of each estimate from first measurement to assess significant changes over time. A cluster correction, with time as the clustered dimension, can be applied to avoid possible spurious differences.
Follow the following steps to perform the intersubject combination analysis. Calculate the average of the first i) OFF transients and ii) OFF-ON transients across all subjects. Quantify tCR, tNAA, GABA+, and Glx signal intensity in the same manner as described above. Move to next ON/OFF transient pair in the scan and repeat these steps. Continue to repeat for the entire scan. This analysis provides a temporal a single trace for each metabolite concentration with the highest possible temporal resolution achievable with the data (i.e., 6 s). Compare high temporal resolution trace between conditions to test for differences in neurometabolite temporal dynamics. Additionally, use cross correlation between dynamic neurometabolite estimates to assess high frequency interdependencies (Rideaux, 2020).
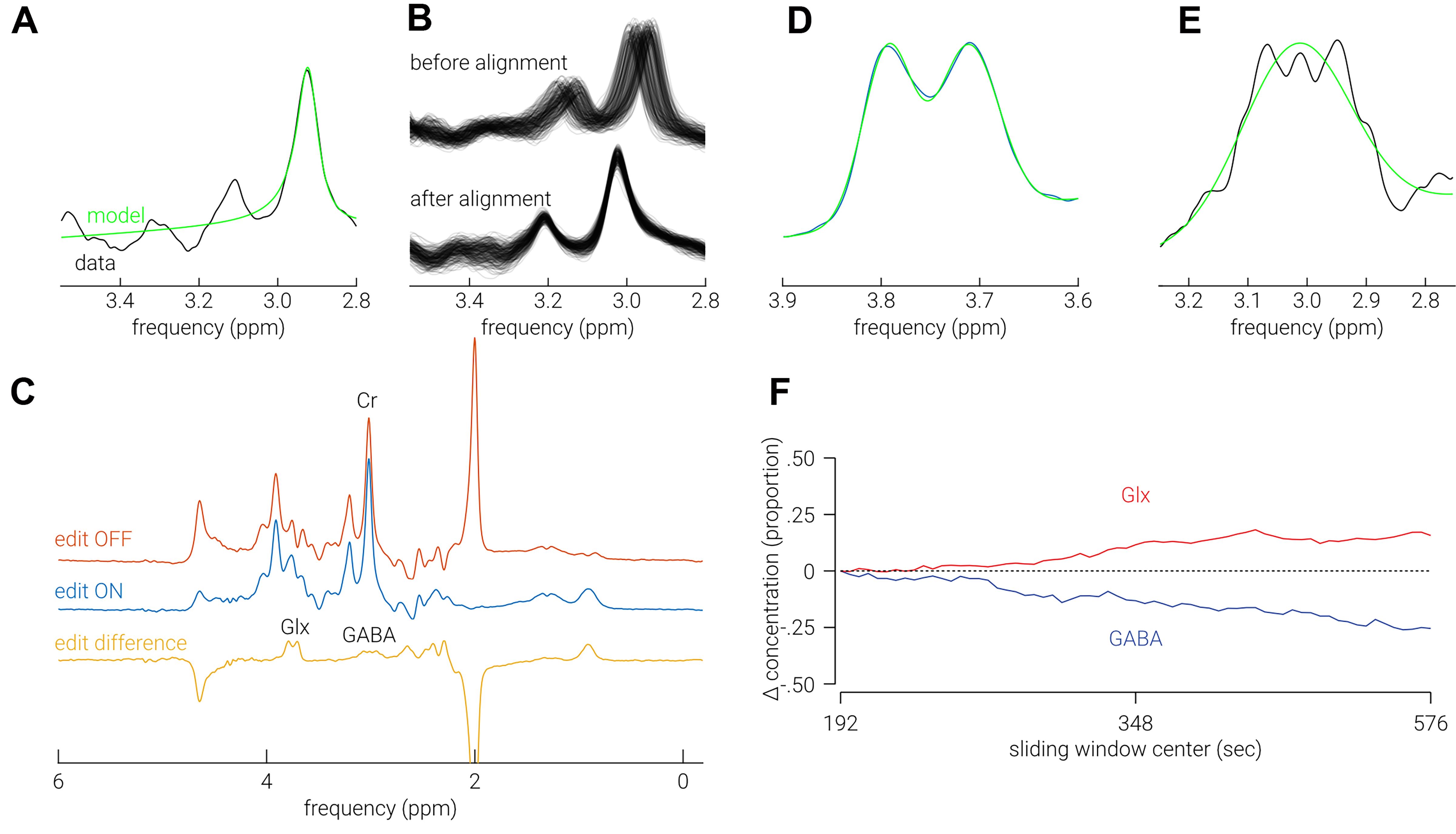
Figure 2. Analysis. A. Example Lorentzian model fit (green) to Cr signal (black). B. Spectra (n = 256) from representative scan before and after alignment. C. Average edit OFF (red), ON (blue), and difference (yellow) spectra. D. Example double-Gaussian model fit (green) to Glx signal (black). E. Example Gaussian model fit (green) to GABA signal (black). F. Example change in Glx/GABA concentration as a function of time using the sliding window method.
Notes
We aligned to the Cr signal, but other alignment methods can alternatively be used.
We used a window size of 128 transients, equal to half the length of the scan, but different window sizes may be used, with the constraint that the window must be suitably large to yield sufficient signal-to-noise ratio.
Acknowledgments
This research was supported by Leverhulme Trust Grant ECF-2017-573 and Isaac Newton Trust Grant 17.08(o). The protocol was derived from Rideaux (2020).
Competing interests
The authors declare no financial or non-financial competing interests.
Ethics
All experiments were conducted in accordance with the ethical guidelines of the Declaration of Helsinki and were approved by the University of Cambridge ethics committee, and all participants provided informed consent.
References
- Bradford, H. F. (1995). Glutamate, GABA and epilepsy. Prog Neurobiol 47(6): 477-511.
- Edden, R. A., Muthukumaraswamy, S. D., Freeman, T. C. and Singh, K. D. (2009). Orientation discrimination performance is predicted by GABA concentration and gamma oscillation frequency in human primary visual cortex. J Neurosci 29(50): 15721-15726.
- Kehrer, C., Maziashvili, N., Dugladze, T. and Gloveli, T. (2008). Altered Excitatory-Inhibitory Balance in the NMDA-Hypofunction Model of Schizophrenia. Front Mol Neurosci 1: 6.
- Kurcyus, K., Annac, E., Hanning, N. M., Harris, A. D., Oeltzschner, G., Edden, R. and Riedl, V. (2018). Opposite dynamics of GABA and glutamate levels in the occipital cortex during visual processing. J Neurosci 38(46): 9967-9976.
- Mescher, M., Merkle, H., Kirsch, J., Garwood, M. and Gruetter, R. (1998). Simultaneous in vivo spectral editing and water suppression. NMR in Biomedicine 11(6): 266-272.
- Mescher, M., Tannus, A., M., O’Neil Johnson and Garwood, M. (1996). Solvent suppression using selective echo dephasing. J Magn Reson- Series A 123(2): 226-229.
- Mikkelsen, M., Loo, R., Puts, N., Edden, R. and Harris, A. (2018). Designing GABA-edited magnetic resonance spectroscopy studies: considerations of scan duration, signal-to-noise ratio and sample size. J Neurosci Methods 303.
- Puts, N. A. and Edden, R. A. (2012). In vivo magnetic resonance spectroscopy of GABA: a methodological review. Prog Nucl Magn Reson Spectrosc 60: 29-41.
- Rideaux, R. (2020). Temporal dynamics of GABA and Glx in the visual cortex. eNeuro 7(4): 1-11.
- Rideaux, R., Goncalves, N. R. and Welchman, A. E. (2019). Mixed-polarity random-dot stereograms alter GABA and Glx concentration in the early visual cortex. J Neurophysiol 122(2): 888-896.
- Rideaux, R. and Welchman, A. E. (2018). Proscription supports robust perceptual integration by suppression in human visual cortex. Nat Commun 9(1): 1502.
- Rideaux, R. and Welchman, A. E. (2019). Contextual effects on binocular matching are evident in primary visual cortex. J Vision 159: 76-85.
- Rubenstein, J. L. and Merzenich, M. M. (2003). Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav 2(5): 255-267.
- Tkáč, I. and Gruetter, R. (2005). Methodology of H NMR spectroscopy of the human brain at very high magnetic fields. Appl Magn Reson 29(1): 139-157.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Rideaux, R. (2021). Low- and High-resolution Dynamic Analyses for Magnetic Resonance Spectroscopy Data. Bio-protocol 11(2): e3892. DOI: 10.21769/BioProtoc.3892.
Category
Neuroscience > Sensory and motor systems > Visual system
Neuroscience > Basic technology > Neurometabolite
Systems Biology > Metabolomics > Neurometabolite
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








