- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Wounding Caenorhabditis elegans with Glass Wool
Published: Vol 11, Iss 2, Jan 20, 2021 DOI: 10.21769/BioProtoc.3885 Views: 3243
Reviewed by: Ayush RanawadeMatthias RieckherAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Infection of Caenorhabditis elegans with Vesicular Stomatitis Virus via Microinjection
Adam Martin [...] Don B. Gammon
Nov 20, 2017 9305 Views

Induction of Skeletal Muscle Injury by Intramuscular Injection of Cardiotoxin in Mouse
Xin Fu [...] Ping Hu
May 5, 2023 3298 Views

Quantifying Mechanical Strain–Induced Membrane Damage in Early Neuronal Cells Using an In Vitro Traumatic Brain Injury Model
Gia Kang [...] Andrew R. Harris
Feb 5, 2026 58 Views
Abstract
Research on wound healing majorly relies on rat, mice and other animal models. However, an alternative animal model ought to be brought in the field, pertaining to the stringent ethical issues owing to the use of animals in research. In this regard, Caenorhabdits elegans, a miniature model nematode gains the great attention of the researchers in wound healing. Though, the model is being explored in wound research for more than a decade, the existing protocols lack the acquisition of large wound population that in turn could enable the utility of global genomics (G), proteomics (P) and metabolomics (M) based approaches. In order to overcome the inadequacy of the existing protocols, the protocol described here affords the acquisition of voluminous wound population in C. elegans using truncated glasswool pieces to enable the utility of high throughput analytical techniques.
Graphic abstract
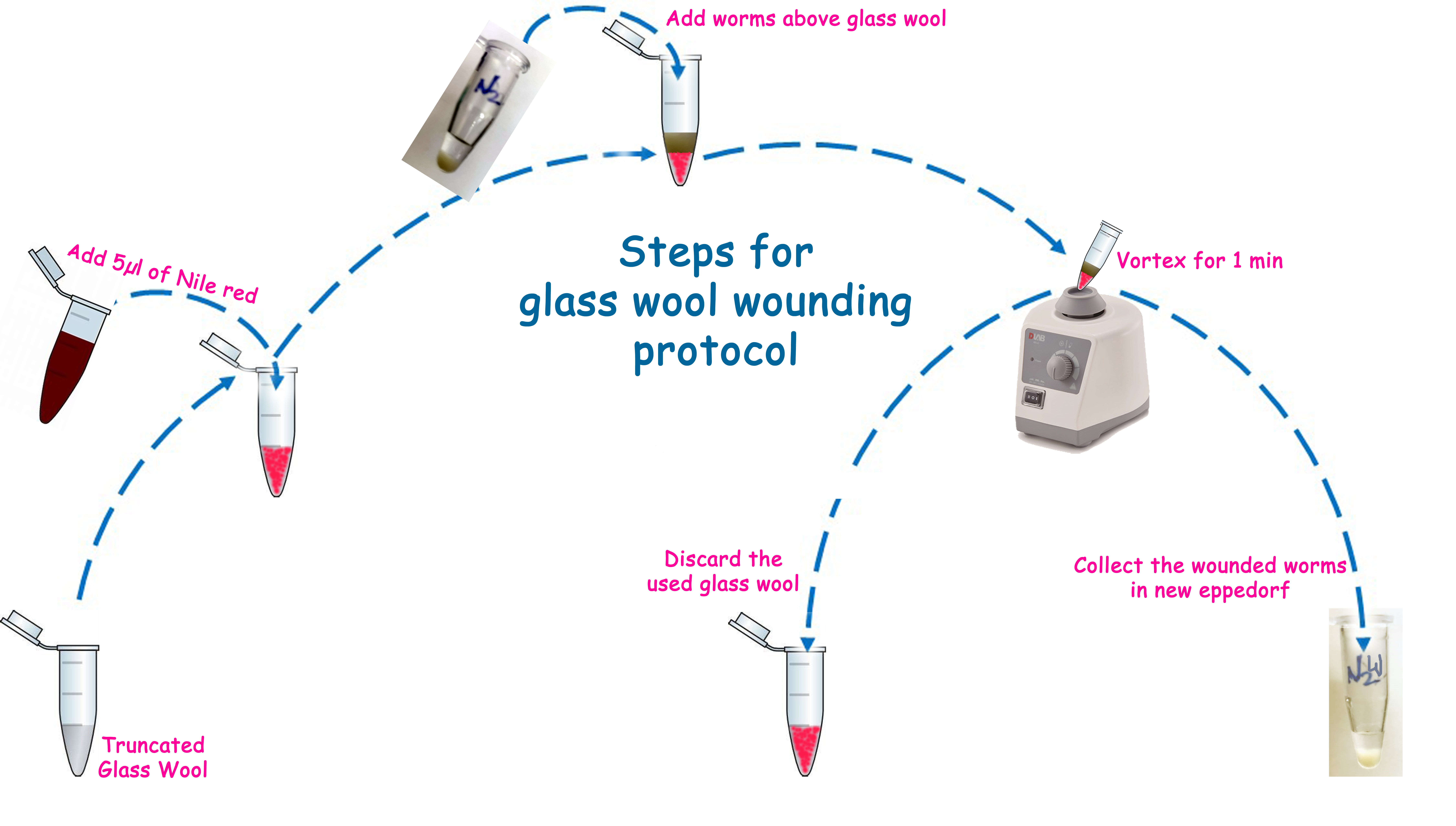
Steps involved in glass wool wounding protocol.
Background
Wounding of C. elegans is being done by fungal infection of Drechmeria coniospora (Pujol et al., 2008), micro-injection needle (Xu and Chisholm, 2014), femtosecond laser treatment (Chung et al., 2006) and micrometer-scale fine glass shards (Zhang et al., 2015). Most of these existing protocols involves individual wounding of worms in order to observe the roles of interested molecular players upon injury and during the process of healing. Existing methods have provided loads of new and useful insights from the mechanism of healing in C. elegans which was also proven to be extrapolatable to higher mammalian models. Nevertheless, the methods were inadequate to obtain large wound population in a shortframe of time and hence the further utility of high throughput alaytical techniques in G, P and M studies becomes tedious. In the era where high throughput analyses become the future of research, a new alternate protocol is required to obtain large wound population in single stretch at quick frame of time. Incidentally, glass wool wounding protocol described over here was established (Pooranachithra et al., 2019) inspired by the protocol described by Zhang et al., (2015). In addition to high throughput analytical techniques, the method was also found to be reliable for high throughput screening of active therapeutics for improved wound healing.
Materials and Reagent
- Glass wool (HIMEDIA, catalog number: RM1232)
- 50 ml Falcon tubes (TARSONS, catalog number: 546021 )
- 1.5 ml Eppendorf tubes (TARSONS, catalog number: 500010)
- 200 µl and 1 ml pipete tips (TARSONS, catalog numbers: 521010 and 521020)
- Microscope slides (LABTECH, catalog number: 217101) and Cover slips (BLUE STAR, 22 mm 10 g)
- 60 x 15 mm Petri plates
- C. elegans strain, N2 (Wild type) (CGC, Minnesota)
- E. coli OP50 (CGC, Minnesota)
- Nile red (Sigma, catalog number: 72485-100MG)
- Methanol (Fisher Scientific, catalog number: 43637G)
- KH2PO4
- K2HPO4
- MgSO4
- CaCl2
- Cholesterol
- Ethanol
- NaCl
- Peptone
- Agar
- Na2HPO4
- Tryptone
- Yeast extract
- House hold bleach solution
- KPO4 (1 M) (see Recipes)
- MgSO4 (1 M) (see Recipes)
- CaCl2 (1 M) (see Recipes)
- 5 mg/ml Cholesterol in 95% ethanol (see Recipes)
- NGM plates (see Recipes)
- M9 buffer (see Recipes)
- Nile red solution (see Recipes)
- LB broth (see Recipes)
Equipment
Scissors
T20, T200 and T1000 pipets (TARSONS, catalog numbers: 030020 ; 030040 ; 030050 )
Erlenmeyer flask (BOROSIL, catalog number: 4100029 )
Marter and Pestle (Thermo Scientific, catalog number: 533669 )
Dissecting stereo microscope (NIKON, model: SMZ 1000 )
20 °C C. elegans incubator (SANYO, Cooled incubator, model: MIR 554 )
37 °C bacterial incubator with shaker (SCIGENICS BIOTECH, ORBITEK, LETTD)
Vortex Mixer (DLAB, MX-S, 8031102000 )
Autoclave (EQUITRON, Vertical Autoclave)
Software
Microsoft Excel
Procedure
Preparation of truncated glass wool stock for wounding of C. elegans
Pour 30 ml of M9 buffer into a sterile 50 ml Falcon tube and add glass wool as much as it can hold.
Cut the glass wool into pieces using a scissors and make sure the glass wool is immerced in the buffer to avoid floating of fine glass shards in the air.
Continue to cut the glass wool until it’s volume reaches 5 ml as displayed in Figure 1 and transfer the course truncated glass wool pieces to a morter and pestle along with the M9 buffer.
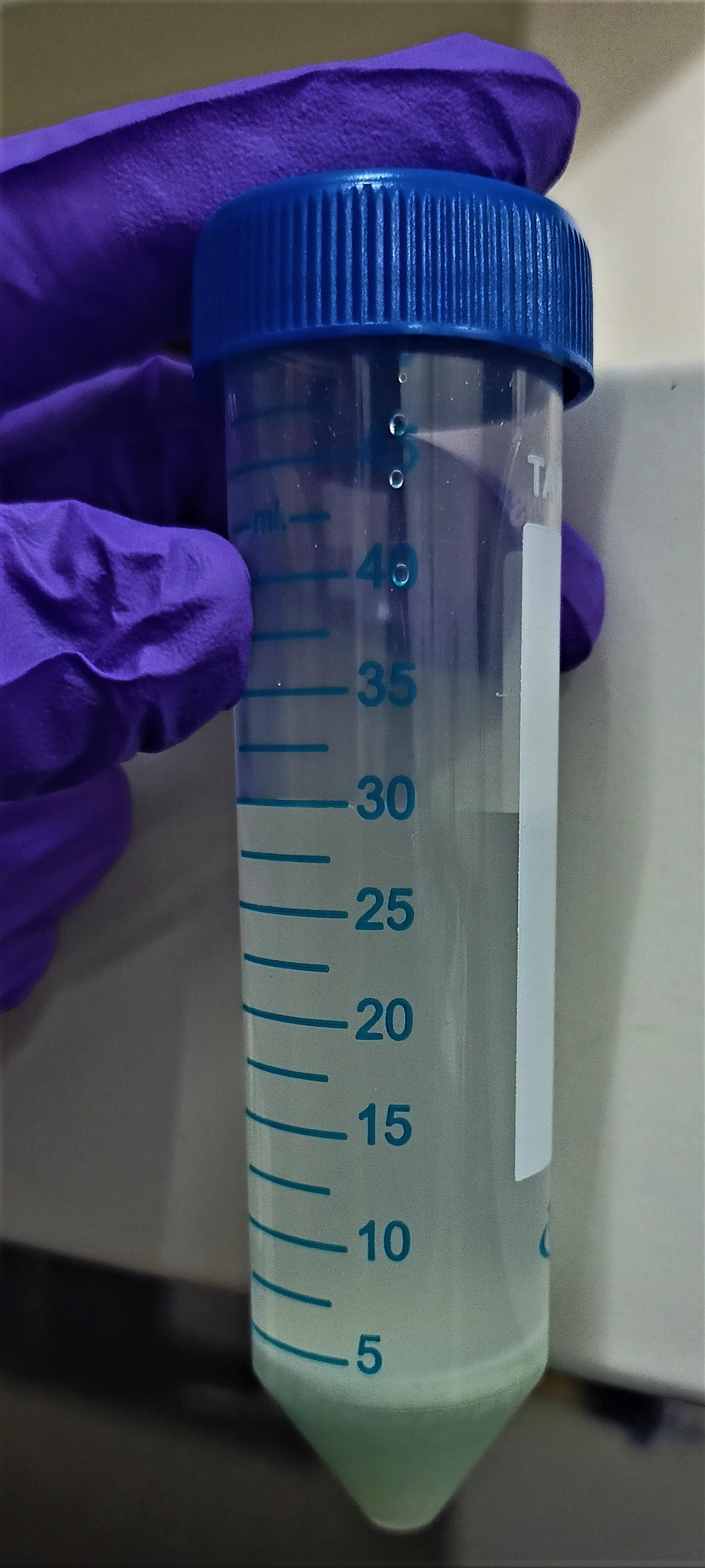
Figure 1. Truncated glass wool pieces transferred to a sterile 50 ml Falcon tube after cutting/grindingGrind it for 20 min to make it fine for the further purpose of injuring the miniature model, C. elegans.
After grinding, transfer the truncated glass wool pieces to a new 50 ml Falcon tube and autoclave the same at 121 °C for 20 min.
After autoclave, store it in room temperature and make sure that the glass wool is autoclaved before every wounding process to avoid contamination and ensure sterile wounding of C. elegans.
Preparation of worms for wounding
Bleach the gravid hermaphrodites by exposing to equal volume of 5 M KOH and house hold bleach solution (1:1 ratio) for 50 s and transfer the eggs to an Nematode Growth Medium (NGM) plate seeded with E. coli OP50 following two M9 wash by centrifuging at 3,500 x g for 1 min.
Incubate the plates at 20 °C until the worms reach young adult stage (~2.5 days). Before going for wounding of C. elegans, collect the worms in a 1.5 ml Eppendorf tube by washing from the plate using M9 buffer.
Glass wool wounding of C. elegans
Transfer 50 mg of autoclaved truncated glass wool pieces in a 1.5 ml Eppendorf tube by pipetting using 1 ml pipet tips and add 5 µl of nile red (1%) to it by gentle mixing for traking the site of injury after glass wool wounding.
Remove the M9 content as much as possible by pipetting and add equal volume of young adult stage worms above the glass pieces.
Vortex the tubes strictly for 1 min by directly keeping it on the vortex mixer. Rotate the tube periodically to cover all the worms during vortex.
Use another Eppendorf tube for wounding while handling higher volumes of worms to avoid the increase in propotion of uninjured worms in the resulted population.
Use maximum of 100 µl of worms containing more than 5,000 numbers of worms/tube and ensure the usage of 1.5 ml Eppendorf tubes for successful wounding of C. elegans.
Recovery of wounded worms after injury
Follow the below steps to recover the worms after injury based on the requirement.
Recovery of wounded worms for microscopic imaging
After vortex wounding, add 500 µl of M9 on the walls of the Eppendorf tube without disturbing the worms.
Later, transfer the worms rested above the glass pieces by pipetting (T20 pipet) to a glass slide and cover it with a cover slip.
Visualise the worms under stereo microscope and score the injured worms by differentiating the site of injury spoted in red by the application of nile red as shown in the Figure 2.
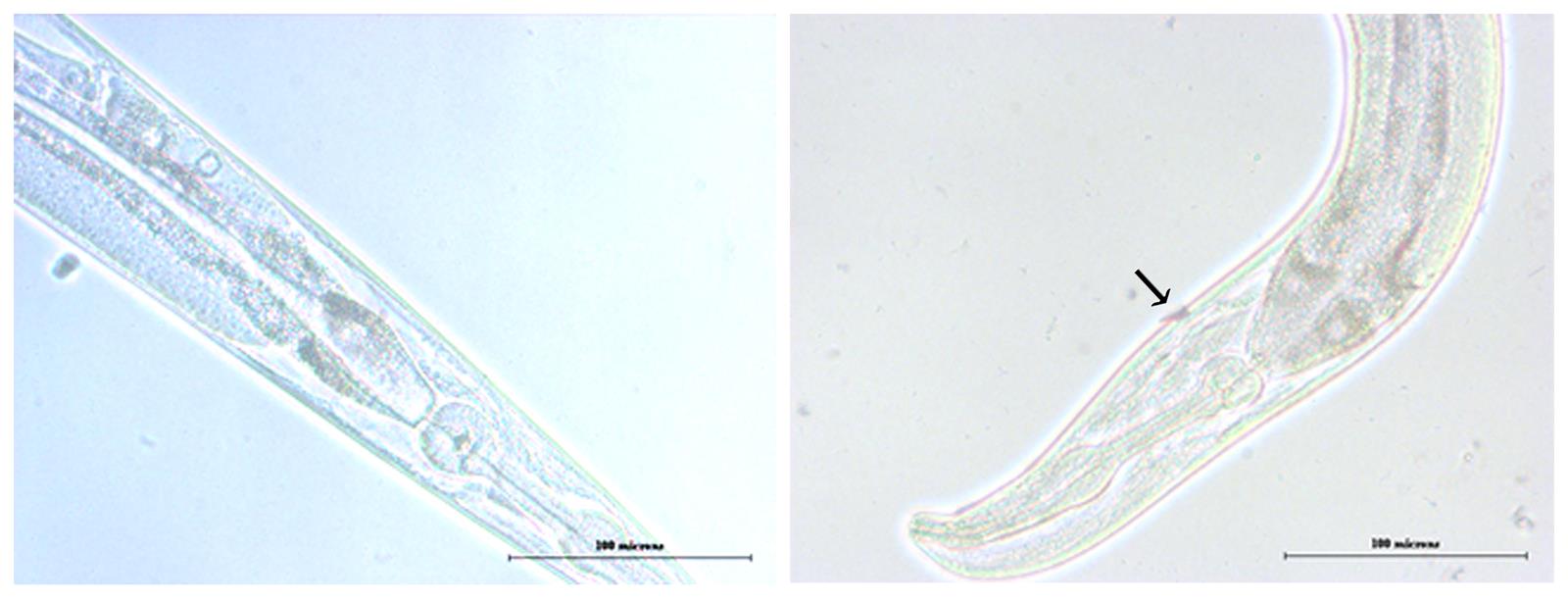
Figure 2. Glass wool wounded worm with unwounded control. Unwounded worm with intact skin layer (Left); Wounded worm with injury created by piercing with truncated glass wool indicated with an arrow mark (Right). Scale bars express 100 microns.Recovery of wounded worms for high throughput analysis
After vortex wounding, add 1 ml of M9 and gently mix the content by pipetting using T1000 pipet.
Let the worms settle above the glass piece and repeat the same for twice to clear out the Nile red and the inserted glass wool pieces.
After washing, let the worms settle and transfer them all to a new Eppendorf tube by pippeting.
Use edge cut tips with T1000 pipet for handling and washing of worms after glass wool wounding.
Data analysis
Score the worms for injured, uninjured and disintegrated under the microscope after the wounding process.
Collect the scores in triplicate to ensure for obtaining > 75% injured, < 20% uninjured and < 5% disintegrated worms as mean values with ± 5% standard errors.
Use the formula (Number of injured worms/Total Number of worms in the slide)*100 for calculating the % of injured worms.
Use excel for calculating standard One way ANOVA for statistical assessment.
Notes
Ensure wearing of gloves and masks while preparing and handling glass wool (Compulsory).
Make sure the truncated glass wool pieces are immersed in buffer to avoid floating of the same in air.
Make sure the glass wool stock is sterile by autoclaving it before every wounding process.
Ensure the stage of worms to be young adult or adult as the method is reliable only for the adults, with the size of worms (1 mm) to be maximum.
For wounding of larval animals, fine glass wool powders can be used.
Nile red in the study was used as a colored solution that aids in localization and depth of injury. Additionally, any dying agents could be used in the place. However, Nile red is recommended for improved traceability of injury by glass wool wounding in C. elegans. Application of Nile red is preferred in case of microscopic observations where it could be ignored for direct subjugation of wounded population for high throughput analyses.
Make sure that the time taken for vortexing with glass wool not to exceed 1 min, as the depth of injury increases with an increase in time taken for vortexing as shown in Figure 3.
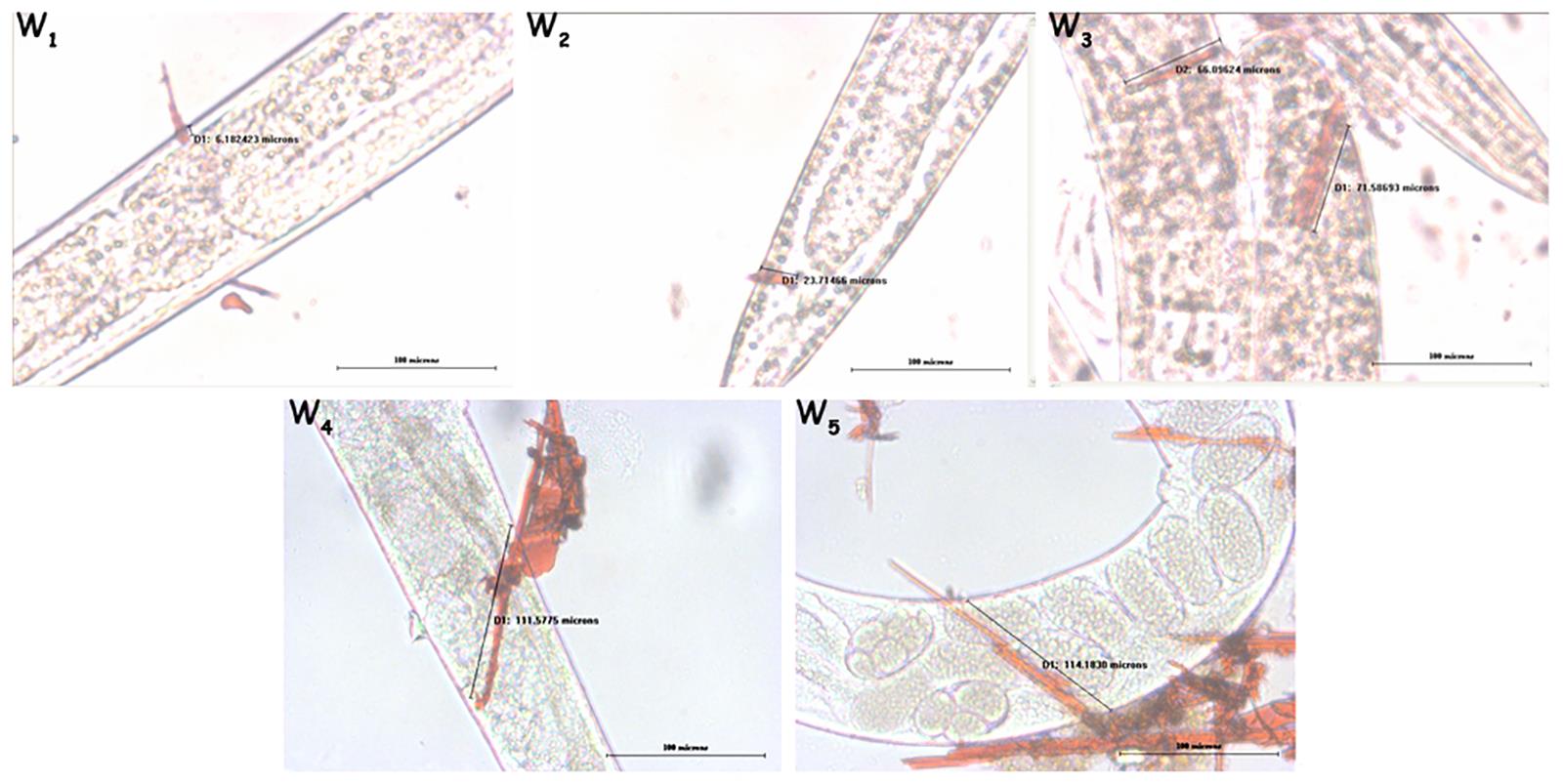
Figure 3. Worms wounded by vortex for various time points. Vortex for 1, 3, 5, 7 and 10 min are represented as W1, W2, W3, W4 and W5, respectively, in the figure.Ensure wiping the work space after completion of work with wet cotton to get rid of any spilled out fine pieces of glass.
Recipes
KPO4 (1 M)
For 100 ml, add 10.8 g of KH2PO4, 3.6 g K2HPO4 and make it up to 100 ml, autoclave at 121 °C for 20 min
MgSO4 (1 M)
For 100 ml, add 12 g of MgSO4 and make it up to 100 ml, autoclave at 121 °C for 20 min
CaCl2 (1 M)
For 100 ml, add 11 g of CaCl2 and make it up to 100 ml, autoclave at 121 °C for 20 min
5 mg/ml Cholesterol in 95% ethanol
Dissolve 5 mg of Cholesterol in 95% of ethanol
NGM plates
Prepare the plates by adding 3 g NaCl, 2.5 g peptone and 17 g agar in Erlenmeyer flask
Make up the volume to 1 L with deionized water
Autoclave the contents at 121 °C for 20 min and add 1 ml of 1 M CaCl2, 1 ml of 1 M MgSO4, 1 ml of 5 mg/ml cholesterol in ethanol, and 25 ml 1 M KPO4, once it reaches the bearable temperature
After mixing it thoroughly, dispense the NGM solution to 2/3 of the 60 x 15 mm Petri plates. Let the plates to solidify and seed them with E. coli OP50 food source (addition of Nystatin and streptomycin is recommended to avoid contamination of NGM during preparation)
For seeding purpose, use 200 µl of E. coli OP50 culture (OD = 0.5) grown in LB broth and incubate the plates for overnight at 37 °C (store the plates at 4 °C for later use)
M9 buffer
For 1,000 ml, add 3 g of KH2PO4, 6 g of Na2HPO4, 5 g of NaCl, 0.25 g of MgSO4·7H2O and make up the volume to 1,000 ml with deionized water, autoclave at 121 °C for 20 min
Nile red solution
Disolve 10 mg of Nile red in 1 ml of absolute methanol
LB broth
For 100 ml, add 1 g of Tryptone, 0.5 g of Yeast extract, 1 g of NaCl and make it up to 100 ml with deionized water, autoclave at 121 °C for 20 min
Acknowledgments
Authors thank the Caenorhabditis Genetics Center, which is funded by the National Institute of Health, National Centre for Research Resources for providing the nematode strains. MP acknowledges the ITC-LSTC, Bengaluru for the financial assistance in the form of ITC-Project Fellow. Instrumentation Facility provided by Department of Science and Technology (DST), Government of India through DST PURSE [Grant No. SR/S9Z-415 /2010/42(G)], DST FIST [Grant No. SR/FST/LSI-639/2015(C)], UGC through SAP-DRS1 [Grant No. F.5-1/2018/DRS-II (SAPII)] and RUSA 2.0 [Grant No. F. 24- 51/2014-U, Policy (TN Multi-Gen), Dept of Edn, GOI] are thankfully acknowledged.
Competing interests
Authors declare that there is no competing interest to disclose.
References
- Chung, S. H., Clark, D. A., Gabel, C. V., Mazur, E. and Samuel, A. D. (2006). The role of the AFD neuron in C. elegans thermotaxis analyzed using femtosecond laser ablation. BMC Neurosci 7: 30.
- Pooranachithra, M., Bhaskar, J. P., Murali, D., Das, S. S., JebaMercy, G., Krishnan, V. and Balamurugan, K. (2019). Unravelling the wound healing ability and mode of action of pyridine carboxamide oxime using Caenorhabditis elegans as potential prescreen wound model. Life Sci 235: 116859.
- Pujol, N., Cypowyj, S., Ziegler, K., Millet, A., Astrain, A., Goncharov, A., Jin, Y., Chisholm, A. D. and Ewbank, J. J. (2008). Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol 18(7): 481-489.
- Xu, S. and Chisholm, A. D. (2014). Methods for skin wounding and assays for wound responses in C. elegans. J Vis Exp(94): 51959.
- Zhang, Y., Li, W., Li, L., Li, Y., Fu, R., Zhu, Y., Li, J., Zhou, Y., Xiong, S. and Zhang, H. (2015). Structural damage in the C. elegans epidermis causes release of STA-2 and induction of an innate immune response. Immunity 42(2): 309-320.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Pooranachithra, M., Bhaskar, J. P. and Balamurugan, K. (2021). Wounding Caenorhabditis elegans with Glass Wool. Bio-protocol 11(2): e3885. DOI: 10.21769/BioProtoc.3885.
Category
Immunology > Animal model > Other
Cell Biology > Tissue analysis > Injury model
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












