- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Antimicrobial Sensitivity Assay for Bdellovibrio bacteriovorus
Published: Vol 10, Iss 24, Dec 20, 2020 DOI: 10.21769/BioProtoc.3865 Views: 4985
Reviewed by: Alessandro DidonnaAmberley D. StephensAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

SIMBA Method—Simultaneous Detection of Antimicrobial and Anti-biofilm Activity of New Compounds Using Salmonella Infantis
Meta Sterniša [...] Anja Klančnik
Aug 5, 2023 2010 Views

Functional Assay for Measuring Bacterial Degradation of Gemcitabine Chemotherapy
Serkan Sayin and Amir Mitchell
Sep 5, 2023 1741 Views

Identification of Mycobacterium tuberculosis and its Drug Resistance by Targeted Nanopore Sequencing Technology
Chen Tang [...] Guangxin Xiang
Feb 5, 2025 1985 Views
Abstract
Bdellovibrio bacteriovorus, an obligate predatory bacterium [i.e., bacteria that kill and feed on other bacteria (prey)], has the potential to be used as a probiotic for the disinfection of surfaces or for the treatment of bacterial infections. One option is to use this organism in combination with antimicrobials to potentiate the effectiveness of treatments. In order to make this approach feasible more has to be known about the ability of B. bacteriovorus to resist antibiotics itself. Standard assays to determine the minimum inhibitory concentration (MIC) are not suitable for B. bacteriovorus, since the small size of this bacterium (0.25-0.35 by 0.5-2 μm) prevents scattering at OD600. Since these predatory bacteria require larger prey bacteria for growth (e.g., E. coli dimensions are 1 by 1-2 μm), the basis for the antimicrobial sensitivity assay described here is the reduction of the OD600 caused by prey lysis during growth. Previous studies on predatory bacteria resistance to antimicrobials employed methods that did not allow a direct comparison of antimicrobial resistance levels to those of other bacterial species. Here, we describe a procedure to determine B. bacteriovorus sensitivity to antimicrobials which can be compared to a reference organism tested as close as possible to the same experimental conditions. Briefly, minimal inhibitory concentration (MIC) values of B. bacteriovorus are determined by measuring the reduction in absorbance at 600 nm of mixed predator/prey cultures in presence and absence of different antimicrobial concentrations. Of note, this method can be modified to obtain antimicrobial MIC values of other predatory bacteria, using different conditions, prey bacteria and/or antimicrobials.
Background
Bdellovibrio bacteriovorus is a Gram-negative predatory bacterium that "predates" upon other Gram-negative bacteria species, growing at their expense and eventually killing them. B. bacteriovorus as well as other predatory bacteria have the potential to be used as probiotics for disinfection of surfaces or for the treatment of bacterial infections (Tyson and Elizabeth Sockett, 2017; Madhusoodanan et al., 2019). The combined use of B. bacteriovorus with antimicrobials may enhance the effectiveness of these treatments. This necessarily entails B. bacteriovorus to resist the action of antimicrobials at their working concentration. Little is known on the antimicrobial susceptibility of B. bacteriovorus. However, the sequenced genomic DNA encodes membrane drug transporters (efflux pumps) and other antimicrobial resistance proteins (Rendulic et al., 2004). The sensitivity of B. bacteriovorus to antimicrobials cannot be assessed by a standard broth dilution assay, as it requires the presence of prey bacteria to grow.
In a 2014 study, Pasternak et al. made a first attempt to determine the antimicrobial sensitivity of predatory bacteria (Micavibrio aeruginosavorus EPB and Bdellovibrio exovorus JSS) by developing a modified antimicrobial disc diffusion method. However, this method provides a relative measure of antimicrobial resistance levels for the species analyzed, but does not allow to compare these values with those of non-predatory bacteria species as it does not use the same conditions of standardized assays (predatory bacteria require the presence of prey bacteria to grow).
In a study by Im et al. (2017), B. bacteriovorus was used in combination with violacein (an antimicrobial active against Gram-positive bacteria) to treat mixed populations of multi-drug resistant (MDR) pathogens. The aim of the study was to demonstrate that this combined treatment was more effective than treatments with individual agents (B. bacteriovorus or violacein). The efficacy of the treatments was evaluated in terms of reduction in the titer of MDR bacterial cells and, therefore, did not provide an indication of the degree of general antimicrobial resistance in B. bacteriovorus.
The method presented in this study allows to determine the MIC values of antimicrobials for B. bacteriovorus. Standard broth dilution assays (performed on non-predatory bacteria) are performed in liquid broth containing antimicrobial dilution series (typically two-fold dilutions) and are inoculated with a defined number of bacterial cells (5 x 105 colony-forming units (CFU)/ml). In order to test antimicrobials on B. bacteriovorus, the initial numbers of both B. bacteriovorus and prey bacterial cells have to be considered (initial predator to prey ratio, PPR). The numbers of B. bacteriovorus cells can be estimated reliably only after conducting the test (by plating in double layer agar plates), as their viability is strongly affected by culture conditions. Higher initial PPRs (comparable to a higher inoculum in standard broth dilution assays) can lead to an increased estimate of the antimicrobial minimal inhibitory concentration (MIC; i.e., the lowest concentration (in mg/L) of the antimicrobial agent that prevents visible growth of a microorganism under defined conditions) (Wiegand et al., 2007). The presented method requires the determination of different MIC values of a single antimicrobial at different initial PPR values. For this, serial two-fold dilutions of B. bacteriovorus cells and a fixed number of prey bacteria are added to a master plate with serial two-fold antimicrobial dilutions. In order to quantify the degree of resistance to a specific antimicrobial, the MICs determined at different initial PPRs must be reduced to a single value. This MIC value is obtained by curve fitting the MIC values at each PPR for each antimicrobial and definition of the MIC at the arbitrary initial PPR of 0.02 (which constitutes a low ratio that allows B. bacteriovorus to grow well and lyse almost all prey cells within 24 h under the experimental conditions). In order to compare the MIC values of B. bacteriovorus with that of other bacterial species, such as the prey bacteria, it is necessary to determine the MIC values under conditions resembling experimental conditions employed for B. bacteriovorus. In this way, the ratio between predatory and prey bacteria MIC values provides a comparative measure of the sensitivity to the antimicrobials tested on both species.
This method may not allow the determination of absolute B. bacteriovorus MIC values for bactericidal antimicrobials at concentrations equal or higher than the minimal bactericidal concentration (MBC) for the prey bacteria (Marine et al., 2020). This is because B. bacteriovorus may not be able to grow on inactivated prey bacteria at concentrations equal or above their MBC, since this would result in a lower estimate of the MIC (this is true especially for lysed prey bacteria or those with a compromised structural integrity) (Marine et al., 2020). To overcome this limitation, prey bacteria may be transformed with resistance genes for the specific antimicrobial tested and used in the B. bacteriovorus MIC assay. However, these resistance genes should not encode for a resistance mechanism that inactivates the antimicrobial. One possibility might be the use of resistance genes encoding multidrug resistance pumps (e.g., AcrAB-TolC, MacAB-TolC, MdfA, etc.) that efflux antimicrobials out of the cell.
The protocol presented below may be adapted to test other predatory bacteria, or use different experimental conditions (i.e., culture broth, temperature, aeration, prey bacteria species), or can be used on different antimicrobials. The only limitation to our knowledge is the use of predatory bacteria that grow well within 24 h.
Materials and Reagents
Breathable sealing membrane (Breathe-EasyTM sealing membrane, Diversified Biotech, catalog number: Z380059 )
Centrifuge tubes, 15 ml (Sarstedt, catalog number: 62.554.502 )
Centrifuge tubes, 50 ml (Sarstedt, catalog number: 62.547.254 )
Clear 96-well plates (BRAND, catalog number: 781662 )
Deep 96-well plates, 2.2 ml wells (BRAND, catalog number: 701354 )
Microcentrifuge tubes, 1.5 ml (VWR, catalog number: 20170-038 )
Parafilm M (Bemis, catalog number: HS234526A )
Petri dishes (Sarstedt, catalog number: 82.1472.001 )
Bdellovibrio bacteriovorus HD100 (source: Sockett RE, Queen's Medical Centre Nottingham)
Escherichia coli BW25113 (source: laboratory collection)
Agar (AppliChem, catalog number: A0949 )
Anhydrous sodium acetate (Roth, catalog number: 6773 )
CaCl2·2H2O (Emsure, Merk, catalog number: 102382 )
HEPES free acid (Roth, catalog number: 9105 )
MgSO4·7H2O (Roth, catalog number: P027 )
Peptone (BD, BactoTM, catalog number: 211830 )
Tryptone (BD, BactoTM, catalog number: 211699 )
Yeast extract (BD, BactoTM, catalog number: 212720 )
NaCl (Applichem, catalog number: A4661.5000 )
CaCl2 stock solution (see Recipes)
MgSO4 stock solution (see Recipes)
YT broth (see Recipes)
YPSC broth (see Recipes)
Ca/Mg/HEPES buffer (see Recipes)
Bottom agar (see Recipes)
Top agar (see Recipes)
LB agar (see Recipes)
Equipment
Benchtop centrifuge (from any qualified supplier)
Bottles, 250 ml (VWR, catalog number: 215-1593 )
Bottles, 500 ml (VWR, catalog number: 215-1595 )
Bunsen burner (from any qualified supplier)
Cuvettes (Sarstedt, catalog number: 67.742 )
Flasks, 250 ml (VWR, catalog number: 214-0423 )
Freezer, -80 °C (from any qualified supplier)
Plate incubators (from any qualified supplier)
Laminar flow hood (from any qualified supplier)
Inoculation loop (from any qualified supplier)
Microcentrifuge (Thermo Fisher Scientific, catalog number: 75002471 )
Multichannel pipette (8x) 100-1,200 μl (Gilson, catalog number: FA10039 )
Multichannel pipette (8x) 30-300 μl (Sartorius, catalog number: LH-729140 )
Phase contrast microscope, magnification equal or above 1,000x (from any qualified supplier)
Pipette 0.5-10 μl (Eppendorf, catalog number: 3120000020)
Pipette 10-100 μl (Eppendorf, catalog number: 3120000046)
Pipette 100-1,000 μl (Eppendorf, catalog number: 3120000062)
Plate reader (TECAN, model: Infinite M200 PRO)
Plaque visualization chamber ( Marine et al., 2020 : Figure S9)
Refrigerator, 4 °C (from any qualified supplier)
Shakers (Infors HT, Multitron Standard)
Spectrophotometer (Eppendorf, Biospectrometer basic, catalog number: 6135000017)
Water bath (from any qualified supplier)
Microwave (from any qualified supplier)
Software
Prism 5 (GraphPad, https://www.graphpad.com/scientific-software/prism/)
However, any other statistics software can be used by setting the modified Gompertz equation (Lambert and Pearson, 2000).
WPS Office (Kingsoft, https://www.wps.com/)
Procedure
This protocol details every step of antimicrobial testing on B. bacteriovorus from culturing cells to setting up MIC experiments and determining the MIC point. A general outline of the workflow is summarized below:
Revival of B. bacteriovorus from glycerol stocks
Growth of B. bacteriovorus in liquid culture
Antimicrobial susceptibility testing
Enumeration on double layer agar plates
Antimicrobial susceptibility testing of prey bacteria
Note: All steps that require working in a laminar flow under sterile conditions are indicated with the symbol "*".
Revival of B. bacteriovorus from glycerol stocks
Day 1:
Open the glycerol stock tube containing E. coli and scrape the top of the frozen culture with a sterile inoculation loop*.
Streak the E. coli cells onto a section at the border of a LB 1% agar plate, by gently touching the LB agar with the inoculation loop containing the E. coli cells and streaking the loop back and forth. To produce single colonies, sterilize the loop and gently drag the bacteria across the surface of the agar from the first section to another section of the plate by streaking through the first section. Repeat this step one or two more times to spread the bacteria from the second or third section to a new section of the plate* ( Slonczewski et al., 2020 : Figure 4.11, p. 131).
Incubate plates upside down overnight in an incubator at 37 °C. These plates can be stored at 4 °C for 2 weeks.
Day 2:
Pour 5 ml YT broth inside a 50 ml tube. Inoculate the broth with 4 separately picked E. coli colonies using an inoculation loop* and incubate the culture at 37 °C with shaking at ~170 rpm overnight. This step has to be done late in the afternoon, so the E. coli cells do not overgrow.
Day 3:
Unscrew the lid of bottles containing bottom and top agar and melt the agar in a microwave (500 ml of bottom agar allow to cast around 20-25 plates; the amount of top agar required for 25 plates is 125 ml). Melting the agar may require several minutes, depending on the power setting and the amount of agar to be melted. Make sure that the cap of the bottle is partly unscrewed as to allow for boiling vapor to escape. We suggest to use a minimal power setting and to stop the microwave as soon as the liquified agar begins to boil. Take out the bottle and gently mix it by swirling (careful, when agitated the liquid agar can boil). Repeat heating in the microwave and mixing until all the agar is melted and there are no lumps visible. Then, briefly heat the agar one last time at a higher power setting to melt all remaining lumps not visible by eye.
Subsequently, place the bottles in a 55 °C water bath until they reach this temperature (Note 1).
Supplement both bottom and top agar with CaCl2 and MgSO4 to a final concentration of 2 mM and 1 mM, respectively*.
Pour a layer (20-25 ml) of bottom agar in a Petri dish (diameter of 10 cm) and distribute it equally by gently swirling the Petri dish*.
Centrifuge 5 ml of an E. coli overnight culture and resuspend it in the exhausted broth of culture (which was used to grow cells) to a final OD600 of ~10*.
Note: The relationship between OD600 and cell biomass approximates linearity only at low absorbance values (usually up to 0.3-0.4, depending on the instrument and cuvettes). Therefore, serial dilutions (e.g., 1:2) of the E. coli overnight culture should be prepared for measuring the OD600. The construction of a graph of absorbance against dilution factors allows to determine the linear range of values. Linear regression can be used to calculate the OD600 value of the undiluted overnight culture.
Add 500 μl of the concentrated overnight E. coli culture and 5 ml of the top agar in a 10 ml test tube*.
After capping the tube with a sterile lid, gently mix by inversion of the tube for 2 times (avoiding the formation of bubbles)*.
Quickly pour the agar onto the Petri dish containing the solidified bottom agar and distribute it evenly by gently swirling the Petri dish*. Leave the dish under the laminar flow hood for the agar to solidify with the lid semi-open.
Pipet 50 μl of a thawed B. bacteriovorus glycerol stock (containing 20% (v/v) glycerol) in the center of the double layer agar plate (i.e., a 2-layer plate consisting of bottom and top agar) with E. coli cells within the top agar*.
Leave this plate under the laminar flow hood with the lid semi-open to dry the B. bacteriovorus suspension*.
After the B. bacteriovorus suspension has dried (after approx. 30 min), seal the plate-lid and plate with parafilm to prevent it from drying out.
Incubate the plate for 2 to 3 days at 29 °C until a clear zone (plaque) on the agar is visible (due to E. coli lysis) at the center of the plate where the B. bacteriovorus suspension has been applied (Figure 1).
Note: Ca/Mg/HEPES double layer agar plates can be stored at room temperature for 2 weeks without significantly affecting B. bacteriovorus viability (Lambert et al., 2008).

Figure 1. B. bacteriovorus plaque on a double-layer agar plate. B. bacteriovorus revived from a glycerol stock form a large plaque on a double-layer plate (containing E. coli in the top agar) at the point of application. Some of the agar has been taken with a pipette from the center of the plaque (holes in the agar).
Grow B. bacteriovorus in liquid culture
Note: B. bacteriovorus predation efficiency is higher in presence of slow growing or stationary phase bacteria preys. Broth of cultures such as DNB (diluted nutrient broth), YT (yeast extract, trypton) and YPSC (yeast extract, peptone, sodium, calcium) have a low concentration of nutrients and result in slow bacterial growth rates and were originally used for the concomitant culturing of B. bacteriovorus and its preys (inoculation of fresh broth with both predatory and prey bacteria, at the same time). Although, most research groups grow B. bacteriovorus on stationary phase prey bacteria from overnight cultures, they still use DNB or YT broth (YPSC is used mostly for the concomitant growth in double layer agar plates). A critical point that affects predation efficiency (and therefore predatory bacteria growth) is the Ca2+ and Mg2+ concentration and pH of the buffer/broth of culture. Typically, B. bacteriovorus is cultivated at a pH of 7.2 to 7.6 and at a concentration of 2-3 mM Ca2+ (CaCl2) and 1-3 mM Mg2+ (either MgSO4 or MgCl2). Other broth components, such as peptone and yeast extract, seem to increase B. bacteriovorus viability and durability.
Day 1:
Streak out E. coli prey cells on a LB 1% agar plate to get single colonies* and incubate overnight at 37 °C.
Day 2:
Pour 2 ml YT broth in a 15 ml tube. Inoculate this broth with 4 separately picked E. coli colonies using an inoculation loop* and incubate at 37 °C under shaking conditions (~170 rpm) overnight.
Day 3:
Excise a portion of top layer agar from the B. bacteriovorus plaque area (the top agar has a soft gel-like consistency and can be easily taken up using a 1,000 μl pipet set to ≥ 250 μl) and transfer the cells by pipetting up and down into the tube containing the overnight prey culture*. Incubate 1 to 2 days at 29 °C under shaking conditions (~170 rpm) until the culture is clear (lysed).
Pour 5 ml YPSC broth in a 50 ml tube. Inoculate this broth with 4 separately picked E. coli colonies using an inoculation* loop and incubate at 37 °C with shaking at ~170 rpm overnight.
Day 4:
When the B. bacteriovorus culture appears clear, check under a phase contrast microscope for the presence of B. bacteriovorus and lysis of prey cells.
Note: For this purpose, 5 μl of the culture can be used for examination at the microscope. B. bacteriovorus cells can be distinguished from other bacteria by their smaller size and high motility, while bdelloplasts (B. bacteriovorus growing inside preys) are round shaped.
Add 100 μl of the B. bacteriovorus culture to the E. coli overnight culture* and incubate for 24 h at 29 °C under shaking conditions (~170 rpm).
Pour 5 ml YPSC broth in a 50 ml tube. Inoculate this broth with 4 separately picked E. coli colonies using an inoculation loop* and incubate at 37 °C with shaking at ~170 rpm overnight.
Day 5:
Check the B. bacteriovorus culture under a phase contrast microscope for the presence of B. bacteriovorus and lysis of prey cells.
Add 100 μl of the B. bacteriovorus culture (from Day 4) to the E. coli overnight culture* and incubate for 24 h at 29 °C under shaking conditions (~170 rpm).
Inoculate the required volume of YPSC with 4 separately picked E. coli colonies using an inoculation loop* and incubate at 37 °C with shaking at ~170 rpm overnight. When preparing this culture, consider the volume necessary to obtain around 2 ml per antimicrobial to be tested (i.e., 2 ml of culture for each master plate).
Day 6:
Check the B. bacteriovorus culture under a phase contrast microscope for the presence of B. bacteriovorus and lysis of prey cells.
Add 1 ml of B. bacteriovorus culture (from Day 5) to the flask containing the E. coli overnight culture* and incubate for 24 h at 29 °C under shaking conditions (~170 rpm).
Inoculate the required volume of YPSC broth with 4 separately picked E. coli colonies using an inoculation loop* and incubate at 37 °C with shaking at ~170 rpm overnight. Calculate the required volume as follows:
volume E. coli culture = number of master plates * 10 ml + 10 ml
e.g., for 3 plates a volume of E. coli culture equals to 3 * 10 ml + 10 ml = 40 ml is required
Antimicrobial susceptibility testing (Figure 2, left panel)
Day 7:
Prepare antimicrobial two-fold serial dilutions along the rows of clear 96-well plates in 100 μl Ca/Mg/HEPES buffer including controls without antimicrobial (one antimicrobial per plate)*.
Note: Ideally, the MIC point should fall in the middle of the antimicrobial concentration range. As a starting point, the researcher can use the known MIC values for other bacteria species. If the MIC at a specific initial PPR falls outside the range of values or too close to one end, the test should be repeated with the adjusted concentration range.
Check the B. bacteriovorus culture under a phase contrast microscope for the presence of B. bacteriovorus and lysis of prey cells.
Dilute the B. bacteriovorus culture (from Day 6) by a factor of 25 in Ca/Mg/HEPES buffer (this dilution is necessary to ensure an initial PPR in the assay within the range of 0.2-0.005)*.
Divide this B. bacteriovorus dilution in two separate 50 ml tubes*. One part of this dilution will be inactivated and used as a diluent for preparing the serial dilutions. The other part with active B. bacteriovorus will be used for preparing the serial dilutions and for enumeration. Calculate the required volumes as follows:
Volume active bacteria dilution = number of master plates * 4 ml + 10 ml
Volume inactivated bacteria dilution = number of master plates * 13 ml + 10 ml
Inactivate one part of the B. bacteriovorus dilution by incubation in a water bath at 100 °C for 5 min.
Prepare B. bacteriovorus two-fold serial dilutions (from Step C3) in a sterile deep 96-well plate along the column line, including controls (no viable B. bacteriovorus present)*.
Transfer 50 μl from each well of this plate (Step C6) to the corresponding wells of master plates (Step C1, plates with the antimicrobial dilution series)*.
Add 50 μl of the E. coli overnight culture to each plate well*.
Measure the blank at a wavelength of 600 nm in a plate reader.
Seal master plates with breathable film* and incubate for 24 h at 29 °C under shaking conditions (300 rpm, 3 mm diameter orbit (or throw)).
In a spectrophotometer, measure the absorbance at 600 nm of the E. coli overnight culture (measurements have to be performed at different dilutions in fresh broth to be in the linear range of values).
Day 8:
Measure the absorbance of the master plate(s) (from Day 7) at a wavelength of 600 nm in a plate reader.
Enumeration on double layer agar plates
Note: Plating of predatory bacteria for enumeration has to be performed on the same day of the antimicrobial test as cell viability decrease over time and may result in a lower estimate.
Plating on double layer agar plates (or plaque assay) is the most used method to enumerate, separate or identify bacteriophages and predatory bacteria in a sample. Both predatory bacteria and phages lysate prey/host bacteria and form plaques on agar plates. Double layer plates are cast in two steps as a thick layer of agar at the bottom (i.e., bottom agar) and a thin layer on the top (i.e., top agar) containing prey and predatory bacteria. The bottom agar helps to maintain the top agar moist and soft, buffers against pH changes and provides nutrients to prey bacteria. The top agar has a low agar content (to increase the efficiency of plaque formation) and is poured as a thin layer so that plaques form on the surface of the agar (thus increasing visibility) and to prevent plaques from forming on top of each other.
Day 7:
Unscrew the lid of a top agar bottle and melt the agar in a microwave.
Incubate the bottle in a water bath at 55 °C until it reaches this temperature (Note 1).
Supplement the top agar with CaCl2 and MgSO4 to a final concentration of 2 mM and 1 mM, respectively*.
Centrifuge 5 ml of an E. coli overnight culture at 5,000 x g for 6 min and resuspend it in the exhausted broth of culture (which was used to grow cells) to a final OD600 of ~10*.
Prepare ten-fold serial dilutions in Ca/Mg/HEPES buffer of the 25x diluted B. bacteriovorus culture (from Step C3)*.
Note: Place eight 1.5 ml centrifuge tubes in a rack and label them from -1 to -8. Pipet 900 μl of Ca/Mg/HEPES in each tube, then add 100 μl of the B. bacteriovorus 25x diluted culture to the first tube (10-1 dilution). Replace the tip of the pipet with a new one, mix the content of the first tube and transfer 100 μl of this dilution to the second tube (10-2 dilution). Repeat the latter step to ten-fold dilute the 10-2 dilution and so forth for all remaining tubes.
Typically, 10-4 to 10-6 dilutions of a B. bacteriovorus culture result in the formation of single plaques upon plating (Lambert et al., 2008). Duplicate plates are not needed.
To a test tube, add 100 μl of a serial dilution, and 500 μl of the concentrated E. coli overnight culture as well as 5 ml of the top agar*.
After capping the tube with a sterile lid, gently mix by inversion of the tube for 2 times (avoiding the formation of bubbles)*.
Quickly pour the agar onto the Petri dish containing the bottom agar and distribute it evenly by gently swirling the Petri dish*. Leave the dish under the laminar flow hood for the agar to solidify with the lid semi-open.
Seal the plate-lid and plate with parafilm to prevent it from drying out*.
Incubate for 3 to 4 days at 29 °C until the formation of clear plaques in the top agar (due to E. coli lysis).
Estimate the number of B. bacteriovorus cells in the initial inoculation culture by counting plaque forming units (PFU; the equivalent of colony forming units for non-predatory bacteria. The growth of a single B. bacteriovorus cell generates a single plaque).
Optional: pictures of plaques can be acquired by the aid of a plaque visualization chamber (Marine et al., 2020: Figure S9).
Antimicrobial susceptibility testing of prey bacteria (Figure 3, left panel)
Day 1:
Inoculate YPSC broth with 4 separately picked E. coli colonies using an inoculation loop* and incubate at 37 °C with shaking at ~170 rpm overnight.
Day 2:
Prepare two-fold serial antimicrobial dilutions in 100 μl Ca/Mg/HEPES along the rows of deep 96-well plates including controls without antimicrobial (2 antimicrobials per plate can be tested as triplicates)*.
Centrifuge the E. coli overnight culture at 5,000 x g for 6 min, discard the supernatant and re-suspend in Ca/Mg/HEPES buffer (wash step)*.
Centrifuge it at 5,000 x g for 6 min and re-suspend in Ca/Mg/HEPES buffer to a final OD600 of 0.024*.
Add 50 μl of this E. coli suspension and 50 μl of fresh YPSC broth to the wells of master plates (plates with the antimicrobial dilution series)*.
Measure the blank at a wavelength of 600 nm in a plate reader.
Seal plates with breathable film* and incubate for 24 h at 29 °C under shaking conditions (300 rpm, 3 mm throw).
Day 3:
Measure the absorbance of the master plate(s) (from Day 2) at a wavelength of 600 nm in a plate reader.
Data analysis
Data analysis of B. bacteriovorus antimicrobial test results (Figure 2, right panel)
Note: B. bacteriovorus antimicrobial test should be repeated at least three times for each antimicrobial. Only process the data from three experiments (plates) in which both B. bacteriovorus and E. coli show growth in the wells without antibiotic (control). An antibiotic of known efficacy (e.g., trimethoprim or aztreonam; Marine et al., 2020) can be tested to verify the correctness of the procedure.
Estimate from B. bacteriovorus enumerations and E. coli absorbance readings the initial PPRs, by dividing the number of B. bacteriovorus to the number of E. coli cells.
Subtract the blanks from the corresponding absorbance values (including controls).
Subtract the resulting values from control values of E. coli without B. bacteriovorus at the same antimicrobial concentrations.
Convert these absorbance Δ values to growth percentage values.
Plot these data in a graph against the logarithm of antimicrobial concentrations.
Download the GraphPad file for the Gompertz modified equation from the webpage.
Open the file and fit B. bacteriovorus growth inhibition curves to the Gompertz modified equation (Lambert and Pearson, 2000). Apparent MIC values are calculated by the program as the intersection of the line tangential to the lower asymptote with the equation of the line tangential to the inflexion point: MIC = 10(M + 1/A); where A is the distance between the upper and lower asymptote and M is the logarithmic concentration at the inflexion point (Lambert and Pearson, 2000).
For each antimicrobial, plot apparent MIC values against the logarithm of initial PPRs.
Determine the reference MIC value at a PPR of 0.02 by linear regression (within the linear range of values).
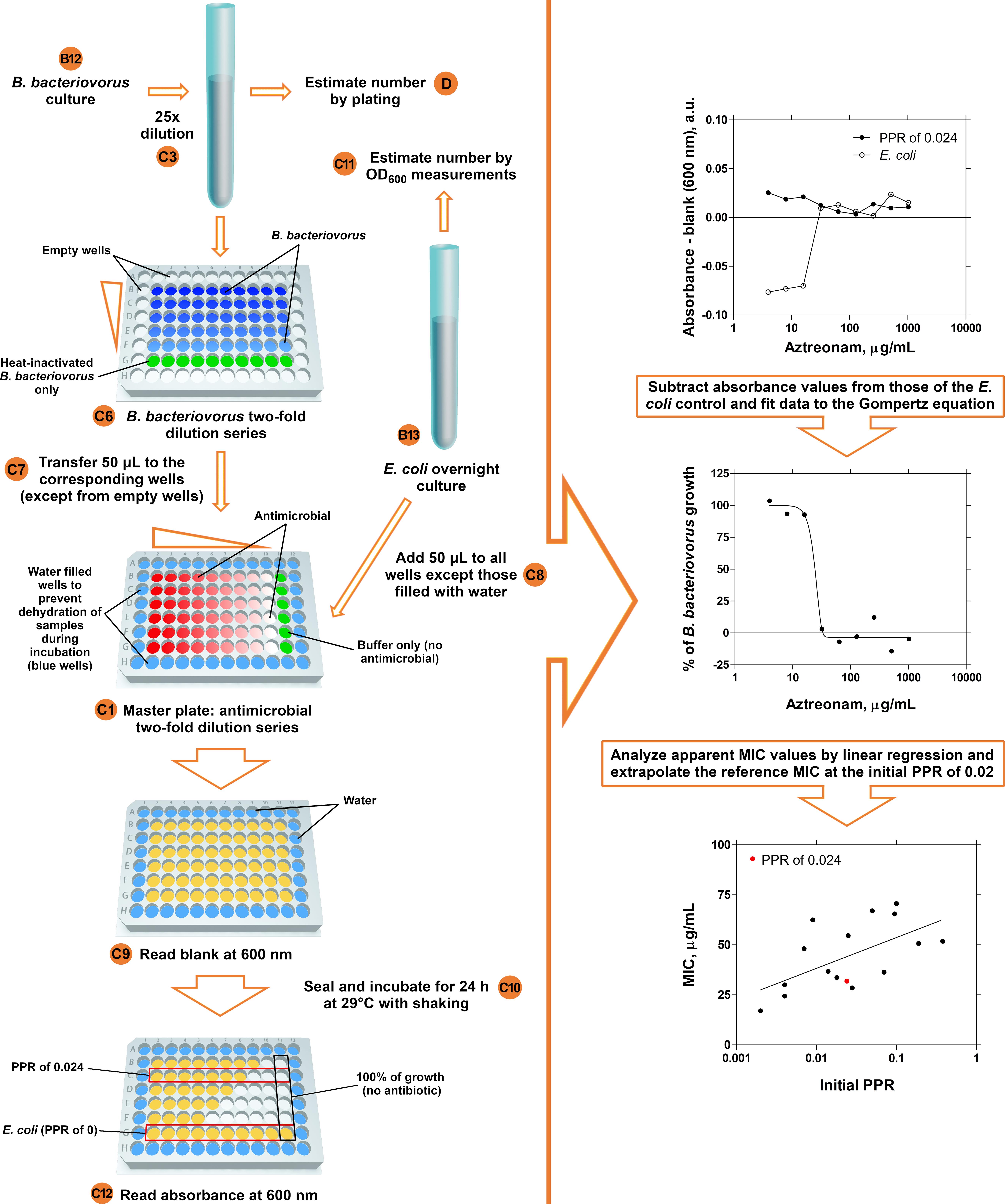
Figure 2. MIC determination of antimicrobials for B. bacteriovorus. Antimicrobial sensitivity test scheme (left panel). The calculation of the MIC of Aztreonam at the initial PPR of 0.024 is shown as an example (right panel). Alphanumeric characters enclosed by circles indicate the corresponding steps in the text.
Data analysis of E. coli antimicrobial test results (Figure 3, right panel)
Note: E. coli antimicrobial test should be repeated at least three times for each antimicrobial. Only process the data from three experiments (plates) in which E. coli shows growth in the wells without antibiotic (control). An antibiotic of known efficacy (e.g., aztreonam; Marine et al., 2020) can be tested to verify the correctness of the procedure.
Subtract the blanks from the corresponding absorbance values (including controls).
Convert these absorbance values to growth percentage values.
Plot these data in a graph against the logarithm of antimicrobial concentrations.
Fit E. coli growth inhibition curves to the Gompertz modified equation (Lambert and Pearson, 2000).
Compare results obtained for B. bacteriovorus and for E. coli by dividing B. bacteriovorus MIC values for each antimicrobial to the corresponding E. coli MIC value. A ratio higher than 1 indicates a higher resistance of B. bacteriovorus against an antimicrobial in comparison to E. coli and vice versa.
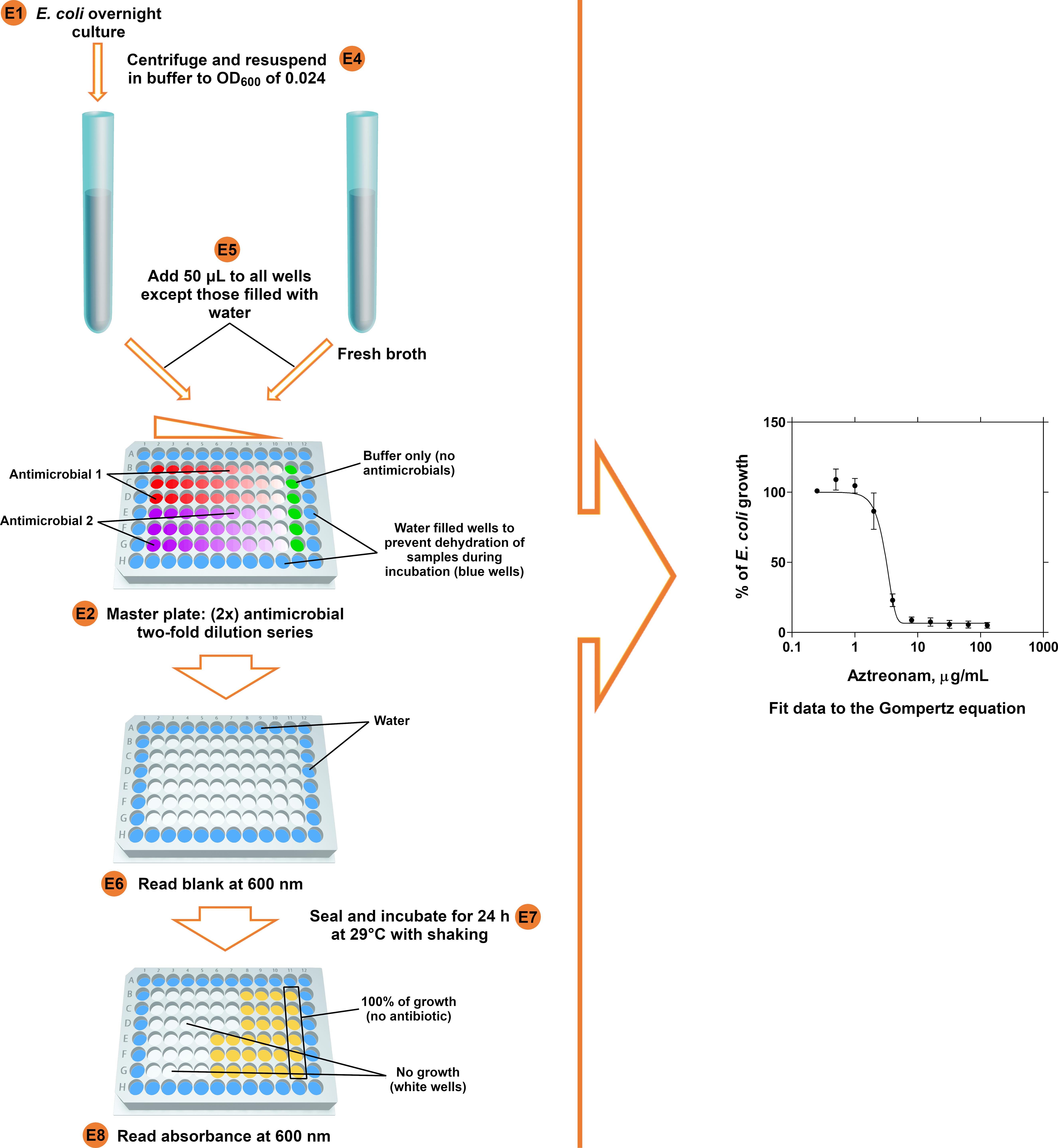
Figure 3. MIC determination of antimicrobials for E. coli. Antimicrobial sensitivity test scheme (left panel). The calculation of the MIC of Aztreonam is shown as an example (right panel). Alphanumeric characters enclosed by circles indicate the corresponding steps in the text.
Notes
To speed up the cooling of melted agar, sealed bottles can be held manually at a 45° angle under running tap water while rotating along the long axis and mildly shaking them for a short time. Keeping the bottles in the water bath ensures, after a reasonable period of time, that they reach the set temperature of 55 °C (which can be determined by a thermometer placed in the water bath). The use of a higher temperature is not recommended as it kills bacteria. On the other hand, too low a temperature will cause the agar to solidify and result in an uneven surface of the cast agar.
Recipes
Note: Sterilize all buffers and broths by autoclaving.
CaCl2 stock solution
200 mM CaCl2
MgSO4 stock solution
100 mM MgSO4
YT broth
5 g/L yeast extract
10 g/L tryptone
5 g/L NaCl
2 mM CaCl2
1 mM MgSO4
Dissolve ingredients except CaCl2 and MgSO4 and adjust pH of the medium to 7.6 with NaOH. After autoclaving add CaCl2 and MgSO4 from 200 mM and 100 mM sterile stock solutions
YPSC broth
1 g/L yeast extract
1 g/L peptone
0.5 g/L anhydrous sodium acetate
2 mM CaCl2
1 mM MgSO4
Dissolve ingredients except CaCl2 and MgSO4 and adjust pH of the medium to 7.6 with NaOH. After autoclaving add CaCl2 and MgSO4 from 200 mM and 100 mM sterile stock solutions
Ca/Mg/HEPES buffer
25 mM HEPES pH 7.6
2 mM CaCl2
1 mM MgSO4
Bottom agar
25 mM HEPES pH 7.6
1% agar
Top agar
25 mM HEPES pH 7.6
0.6% agar
LB agar
5 g/L yeast extract
10 g/L tryptone
5 g/L NaCl
1% agar
Acknowledgments
This work was supported by funding from DFG-EXC115 (Cluster of Excellence Macromolecular Complexes at the Goethe University Frankfurt). E.M. was supported by a stipend from the FAZIT Foundation, Frankfurt am Main, Germany. This protocol was adapted from previous work (Marine et al., 2020). We thank Prof. Sockett RE, Lambert C, Milner D and their research group for the transfer of knowledge on B. bacteriovorus culture methods.
Competing interests
The authors declare no competing interests.
References
- Im, H., Choi, S. Y., Son, S. and Mitchell, R. J. (2017). Combined application of bacterial predation and violacein to kill polymicrobial pathogenic communities. Sci Rep 7(1): 14415.
- Lambert, R. J. and Pearson, J. (2000). Susceptibility testing: accurate and reproducible minimum inhibitory concentration (MIC) and non-inhibitory concentration (NIC) values. J Appl Microbiol 88(5): 784-790.
- Lambert, C. and Sockett, R. E. (2008). Laboratory maintenance of Bdellovibrio. Curr Protoc Microbiol Chapter 7: Unit 7B 2.
- Madhusoodanan, J. (2019). Inner Workings: Probing predatory bacteria as an antibacterial remedy. Proc Natl Acad Sci U S A 116(46): 22887-22890.
- Marine, E., Milner, D. S., Lambert, C., Sockett, R. E. and Pos, K. M. (2020). A novel method to determine antibiotic sensitivity in Bdellovibrio bacteriovorus reveals a DHFR-dependent natural trimethoprim resistance. Sci Rep 10(1): 5315.
- Pasternak, Z., Njagi, M., Shani, Y., Chanyi, R., Rotem, O., Lurie-Weinberger, M. N., Koval, S., Pietrokovski, S., Gophna, U. and Jurkevitch, E. (2014). In and out: an analysis of epibiotic vs periplasmic bacterial predators. ISME J 8(3): 625-635.
- Rendulic, S., Jagtap, P., Rosinus, A., Eppinger, M., Baar, C., Lanz, C., Keller, H., Lambert, C., Evans, K.J., Goesmann, A., Meyer, F., Sockett, R.E. and Schuster, S.C. (2004). A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303(5658): 689-92.
- Slonczewski, J., Foster, J. and Zinser, E. (2020). Microbiology: An evolving science. 5th Ed. W.W Norton& Company p. 131.
- Tyson, J. and Elizabeth Sockett, R. (2017). Nature knows best: employing whole microbial strategies to tackle antibiotic resistant pathogens. Environ Microbiol Rep 9(1): 47-49.
- Wiegand, I., Hilpert, K. and Hancock, R. E. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration(MIC) of antimicrobial substances. Nat Protoc 3(2): 163-175.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Marine, E. and Pos, K. M. (2020). Antimicrobial Sensitivity Assay for Bdellovibrio bacteriovorus. Bio-protocol 10(24): e3865. DOI: 10.21769/BioProtoc.3865.
Category
Microbiology > Antimicrobial assay > Antibacterial assay
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









