- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Mechanical Characterization of Glandular Acini Using a Micro-indentation Instrument
Published: Vol 10, Iss 23, Dec 5, 2020 DOI: 10.21769/BioProtoc.3847 Views: 4385
Reviewed by: Sourav S PatnaikFereshteh AzediAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Efficient Method for Immortalizing Mouse Embryonic Fibroblasts by CRISPR-mediated Deletion of the Tp53 Gene
Srisathya Srinivasan and Hsin-Yi Henry Ho
Jan 20, 2025 2744 Views

Puromycin Proximity Ligation Assay (Puro-PLA) to Assess Local Translation in Axons From Human Neurons
Raffaella De Pace [...] Saikat Ghosh
Mar 5, 2025 3294 Views

Assay for Site-Specific Homologous Recombination Activity in Adherent Cells, Suspension Cells, and Tumor Tissues
Yuki Yoshino [...] Natsuko Chiba
Apr 5, 2025 2433 Views
Abstract
The linker of nucleoskeleton and cytoskeleton (LINC) complex is responsible for tethering the nucleus to the cytoskeleton, providing a pathway for the cell’s nucleus to sense mechanical signals from the environment. Recently, we explored the role of the LINC complex in the development of glandular epithelial acini, such as those found in kidneys, breasts, and other organs. Acini developed with disrupted LINC complexes exhibited a loss of structural integrity, including filling of the lumen structures. As part of our investigation, we performed a mechanical indentation assay of LINC disrupted and undisrupted MDCK II cells using a micro-indentation instrument mounted above a laser-scanning confocal microscope. Through a combination of force measurements acquired from the micro-indentation instrument and contact area measurements taken from fluorescence images, we determined the average contact pressure at which the acini structure ruptured. Here, we provide a detailed description of the design of the micro-indentation instrument, as well as the experimental steps developed to perform these bio-indentation measurements. Furthermore, we discuss the data analysis steps necessary to determine the rupture pressure of the acini structures. While this protocol is focused on the indentation of individual glandular acini, the methods presented here can be adapted to perform a variety of mechanical indentation experiments for both 2D and 3D biological systems.
Keywords: Bio-indentationBackground
Bio-indentation measurements have emerged as means to measure the material properties of biological systems at length scales ranging from sub-cellular biopolymers to multi-cellular tissue structures. Nano-indentation instruments with micron sized probes and contact areas on the order of 10-100 nm2 have been used to measure the material properties of biopolymers and individual cells (Stolz et al., 2004; Sen et al., 2005; Strasser et al., 2007; Li et al., 2008). Similarly, micro-indentation instrument with millimeter sized indentation tips and contact areas on the order of 10-100 µm2 have been used to study the collective behavior and responses of cellular structures to external mechanical forces (Ahn et al., 2010; Levental et al., 2010; Schulze et al., 2017). Exploring cellular responses to mechanical queues from the micro-environment is crucial toward understanding how cells regulate growth, undergo differentiation or morphological changes, and express various genes (Vogel and Sheetz, 2006).
Transmission of mechanical signals from the cytoplasmic cytoskeleton to the nuclear envelope occurs through an assembly of proteins collectively referred to as the linker of nucleoskeleton and cytoskeleton (LINC) complex (Crisp et al., 2006). In addition to facilitating the mechanotransduction of signals to the nucleus, the LINC complex is responsible for tethering the nucleus to the cytoskeleton and regulating cytoplasmic filament organization (Crisp et al., 2006; Lei et al., 2009; Mellad et al., 2011; Tapley and Starr, 2013). Disruption of the LINC complex has been proposed as a potential pathway in the development of a variety of human diseases including cancerous development in glandular epithelia; a reduction in the expression of LINC complex proteins, including SUN1, SUN2, nesprin-2, and lamin A/C, has been observed in breast cancer tissues (Debnath and Brugge, 2005; Horn et al., 2013; Meinke et al., 2014; Matsumoto et al., 2015). However, the precise role that the loss of the LINC complex plays during malignant transformation is not well understood. Recently, we explored the effect of LINC complex disruption on the development and maintenance of higher-order cellular structures (Zhang et al., 2019). Development of glandular acini were hindered when the LINC complex was disrupted resulting in the loss of structural integrity and the filling of the lumens in MDCK II and MCF-10A acini. As part of our investigation into the role of the LINC complex in acinar development, we performed a bio-indentation assay of MDCK II acini to determine the average contact pressure required to induce rupture of individual acini structures. MDCK II acini with filled lumens resulting from the disruption of the LINC complex were found to require higher contact pressures to induce rupture compared to undisrupted MDCK II acini.
Here, we present the protocol for performing the mechanical indentation assay of individual MDCK II acini that was developed as part of our exploration into the role the LINC complex plays in acinar development. Through a combination of force measurements recorded using a micro-indentation instrument and fluorescence images captured through confocal microscopy, we measure the pressure necessary to mechanically rupture individual glandular acini. We discuss the design and operation of a micro-indentation instrument capable of applying vertical displacements on the order nanometers and measuring normal forces on the order of micro-Newtons. Additionally, we discuss the experimental protocol necessary to perform these indentation measurements and the data analysis necessary to determine the average contact pressure to rupture the acini structure. While this manuscript is focused on the indentation of individual acini, these methods can be adapted to perform a variety of mechanical indentation explorations of both 2D and 3D biological systems.
Materials and Reagents
The following materials and reagents are used as part of this bio-indentation protocol:
8-well chamber slide (Nunc Lab-Tek, catalog number:154534 )
35 mm glass bottom Petri dish, 20 mm well size #0 thickness (Cell Vis: D35-20-0-N)
Matrigel matrix basement membrane (Corning, catalog number: 35623 )
MDCK II cell line (gifted from Jennifer Lippincott-Schwartz)
MCF-10A cell line (ATCC, CRL-10317 ; RRID: CVCL_0598)
DMEM medium w/ 4.5 g/L glucose (Mediatech Cellgro, catalog number: 23-10-013-CM )
Donor bovine serum (DBS) (Gibco, catalog number: 16030074 )
Ethylenediaminetetraacetic acid (EDTA) (Corning, catalog number: 46034CI )
Sodium orthovanadate (Na3VO4) (Sigma Aldrich, catalog number: S6508 )
Sodium fluoride (NaF) (Sigma Aldrich, catalog number: 67414 )
Calcein AM (Invitrogen, catalog number: C3100MP )
0.25% trypsin (Corning, catalog number: 25053CI )
Poly-L-lysine (Sigma Aldrich, catalog number: P4832 )
Phosphate buffered saline (PBS) solution 1x (Fisher Scientific, catalog number: BP243820 )
F-127 pluronic (Sigma Aldrich, catalog number: P2443 )
Millipore water
MDCK cell growth medium (see Recipes)
MDCK acini indentation medium (see Recipes)
Matrigel wash solution (see Recipes)
Acini dye medium (see Recipes)
F-127 pluronic solution (see Recipes)
Equipment
The following equipment is used as part of this bio-indentation protocol:
Micro-indentation Instrument:
Sapphire indentation tip (Edmund Optics, catalog number: 48-430 )
Capacitance load cell
Nylon M6 threaded rod (McMaster-Carr, catalog number: 98882A249 )
Nylon M6 hexagonal nut (McMaster-Carr, catalog number: 93800A600 )
Aluminum mount for cantilever beam (custom-built)
Vertical precision Z-positioner (Physik Instrumente, model: P-622.ZCL PIHera )
Aluminum mounting adapter (custom-built)
25 mm compact XY linear translational stage (Thor Lab, model: LX20 )
Aluminum mounting flange (custom-built)
Super glue gel control (Locktite)
Nikon Eclipse Ti2-E Inverted Confocal Microscope:
Nikon Ti2-Eclipse inverted microscope (Nikon)
Nikon C2+ confocal scan head (Nikon)
C2-DU3 detector unit (Nikon)
DAPI filter cube 438/24 (Nikon)
FITC filter cube 525/50 (Nikon)
561 long pass filter (Nikon, model: 561LP )
Reflectance filter cube (Nikon, model: A1-C2 C165305 )
LU-N4 laser unit (Nikon)
4x microscope objective (Nikon, model: MRD00045 )
10x microscope objective (Nikon, model: MRD00101 )
20x microscope objective (Nikon, model: MRD00205 )
Automated translational stage (Nikon, model: 761794 )
perfect focus (Nikon)
Petri dish support plate:
Aluminum Petri dish support plate (custom-built)
Thermocouple (Omega)
Power supply (Velleman, model: PS613U )
Temperature controller (Omega, model: CN7800 )
Nichrome heating wire (Pelican Wire, model: 2130N80DGS )
O2 plasma cleaner (Plasma Etch, model: PE-50 )
Cell incubator
Cell centrifuge
Description of Indentation Instrument
Bio-indentation measurements of individual acini structures are performed using a micro-indentation instrument mounted above an inverted microscope (Figure 1A). The micro-indentation instrument consists of a hemispherical indentation tip, a precision load cell, a vertical Z-positioner, an XY translational stage, and a male dovetail mounting flange (Figure 1B). The indentation instrument is mounted into the condenser slot of the inverted microscope and secured using a hexagonal set screw (Figure 1C). Below, we describe the individual components of the micro-indentation instrument used to perform the indentation assay reported by Zhang et al., 2019 (A detailed CAD rendering of the indentation instrument can be found in the work presented by Schulze et al., 2017). Alternatively, there are several commercially available micro-indentation instruments, including the Bruker Hysitron Biosoft indenter, that provide comparable levels of performance.
The bio-indentation measurements reported by Zhang et al. (2019) were performed using a micro-indentation instrument with a 1.6 mm radius of curvature hemispherical sapphire indentation tip. The load cell of the instrument consists of a double-leaf cantilever beam with a spring stiffness of k = 49.94 N/m, and a capacitive probe with a 100 µm detection range. The normal force of the indentation tip is calculated from the measured vertical deflection of the cantilever beam and the spring stiffness. Calibration of the bio-indentation instrument was performed by hanging weights of known mass from the double-leaf cantilever beam and measuring the resulting displacement using the capacitance probe. The spring stiffness was determined by fitting a line to the resulting force vs displacement curve. To provide an offset from the load cell and to allow for easy removal and cleaning, the indentation tip is attached to a ~15 mm long nylon threaded rod; the indentation tip is superglued to the tip of the mounting rod and a complementary mounting nut is superglued to the load cell. To control the vertical displacement of the indentation tip, the load cell is mounted to a PI piezoelectric translational z-stage capable of a 250 µm travel range with 1 nm resolution. The translational z-stage is subsequently mounted onto a 2-axis linear translational stage, allowing for alignment of the indentation tip in the XY plane. A D3N male dovetail mounting flange is fixed to the top of the XY translational stage to enable the bio-indentation instrument to be mounted into the condenser slot of an inverted microscope. The bio-indentation instrument is controlled through a custom-written Labview program. This program controls the vertical displacement of the piezoelectric stage while recording the signal from the capacitive sensor to calculate the vertical deflection of the double-leaf cantilever beam.
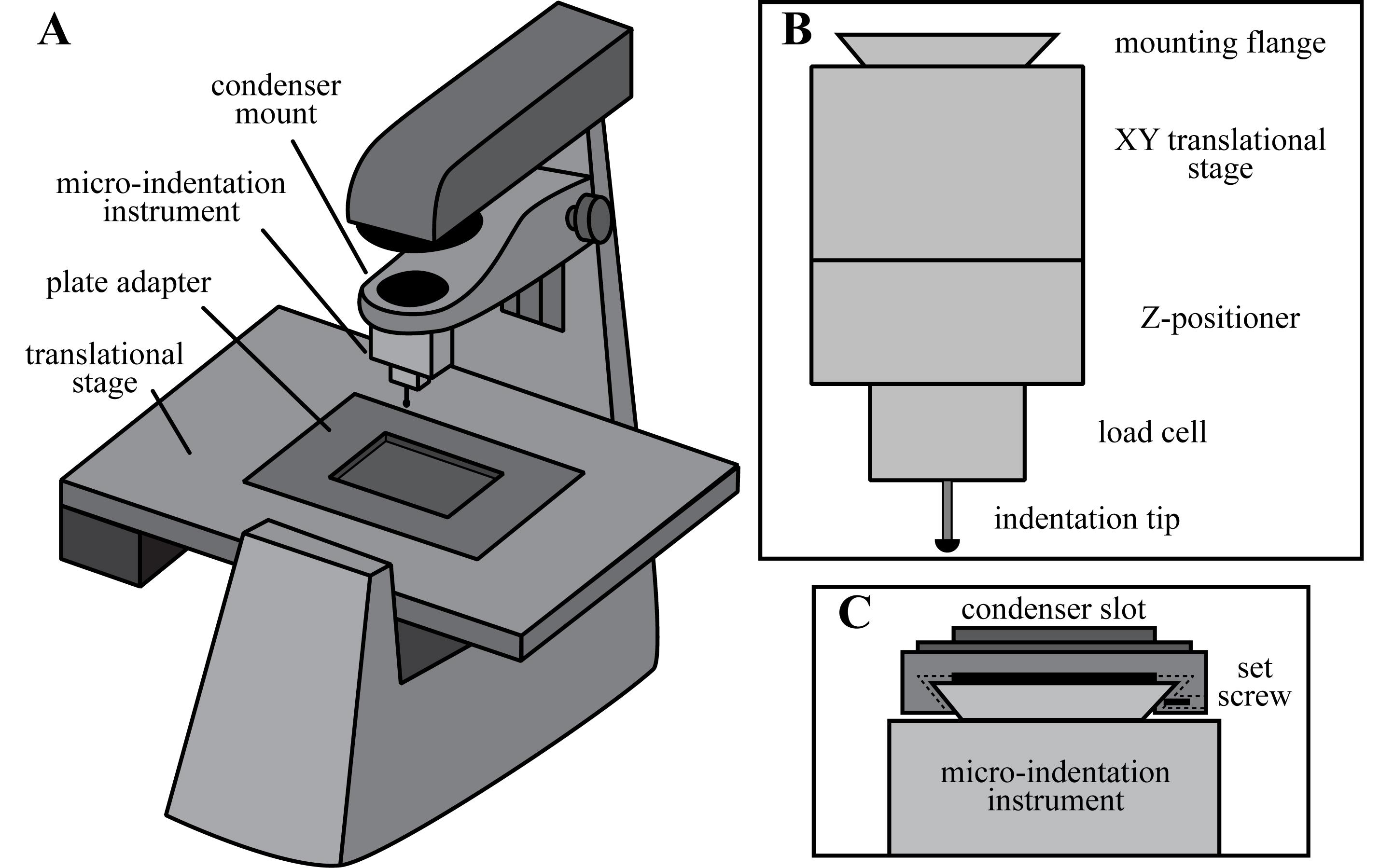
Figure 1. Micro-indentation instrument. A. Bio-indentation measurements are performed using a micro-indentation instrument mounted above an inverted microscope. B. The micro-indentation instrument consists of a hemispherical indentation tip, a load cell to measure the normal force, a vertical Z-positioner with nanometer resolution, a manual XY translational stage, and a dovetail mounting flange. C. The indentation instrument is mounted into the condenser slot of the inverted microscope and secured in place with a hexagonal set screw.
The bio-indentation instrument is mounted on a Nikon Eclipse Ti2-E inverted microscope equipped with a C2+ confocal imaging system and a motorized translational stage. The microscope is configured to capture FITC fluorescence images using the 488 nm excitation laser and a 525/50 dichroic filter cube (Figure 2A). Additionally, confocal reflectance microscopy is used to align the indentation tip directly above the microscope objective prior to the indentation measurement. For reflectance imaging, the 488 nm laser is used as a light source and a reflectance filter cube is installed in the detector unit allowing all light to pass through to the detector channel (Figure 2B).
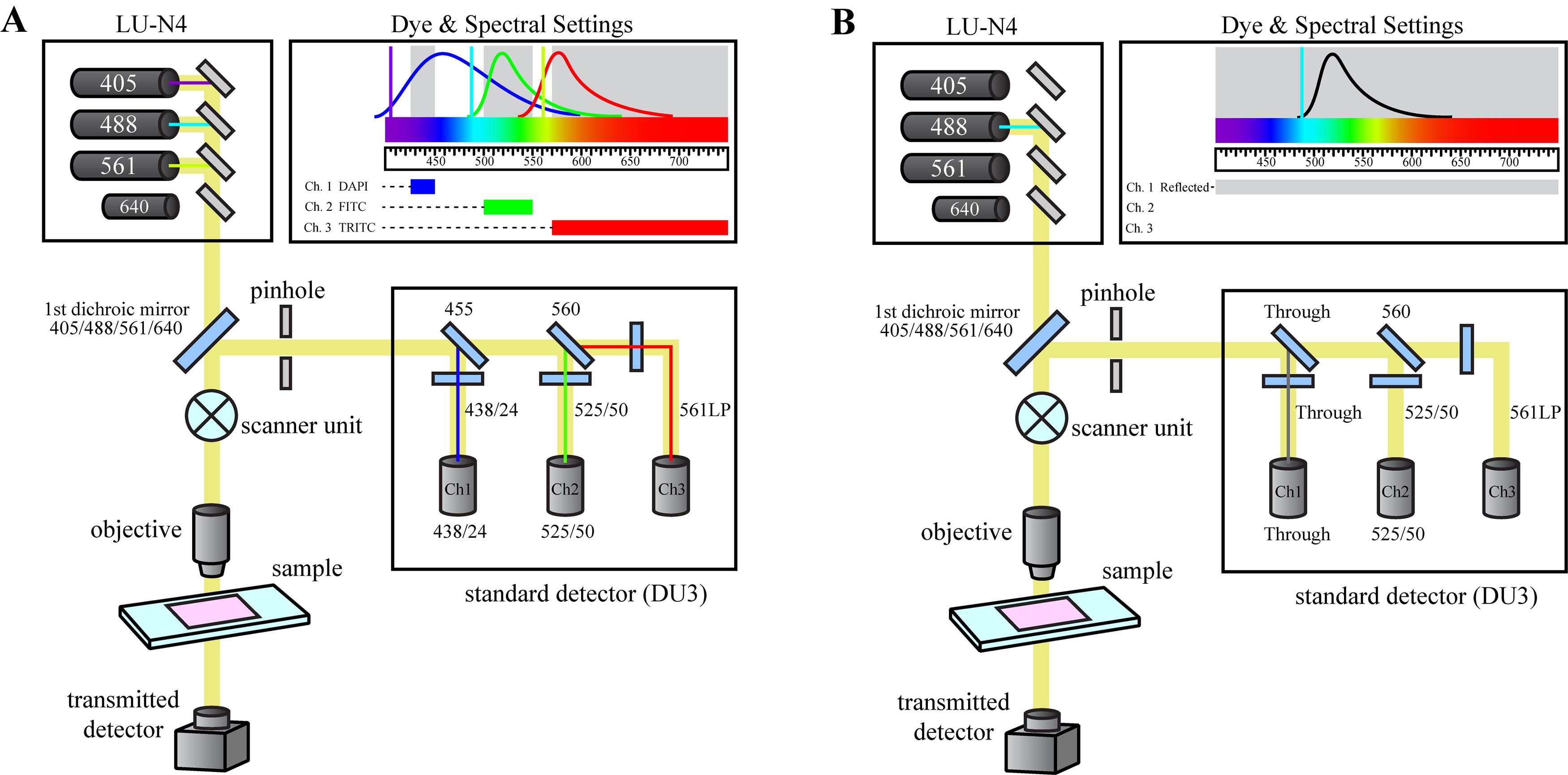
Figure 2. Microscope optical configurations. The Nikon Eclipse Ti2-E Confocal microscope is configured to allow for either (A) fluorescence imaging of the acini structure during the indentation measurement, or (B) reflectance microscopy during the instrumental set-up to determine the center of the indentation tip.
Glass bottom Petri dishes are secured during the indentation measurements using a custom-built Petri dish support plate that fits within the plate adapter of the Nikon Eclipse Ti-2 microscope (Figure 3). The glass bottom Petri dish is positioned over the objective hole in the center of the support plate (1) and is firmly secured using two mounting arms (2). The mounting arms apply pressure to the Petri dish lid to mitigate movement during the indentation measurement. A hole is drilled in the lid of the Petri dish to provide an opening for the bio-indentation instrument. To regulate the temperature during the indentation measurement, the plate is equipped with a thermocouple (3) and nichrome heating wire (4) that are connected to an on-off temperature controller. The power voltage and current settings of the power supply are set to maintain a desired temperature of 37 °C while minimizing the frequency of on-off cycles.
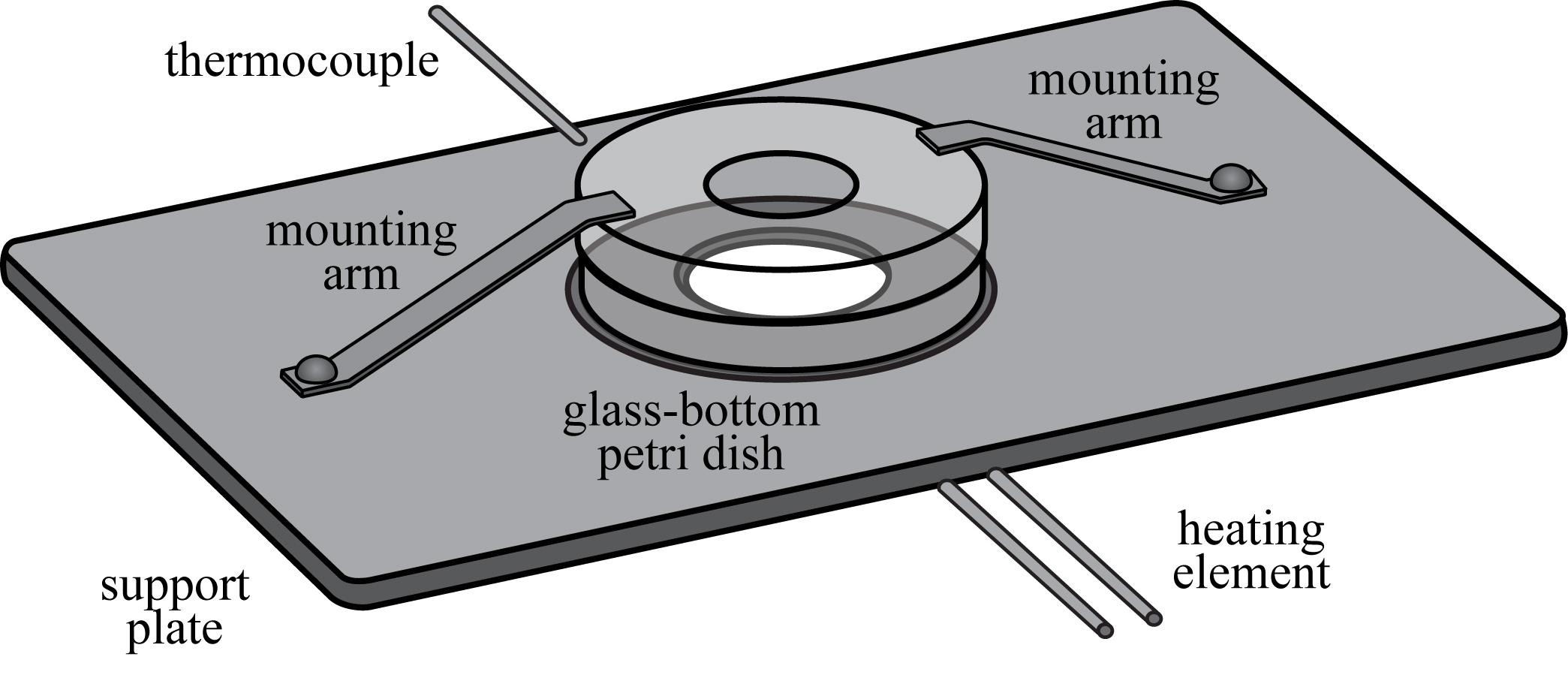
Figure 3. Petri dish support plate. Glass bottom Petri dishes are secured during the indentation measurement using a custom-built support plate that fits into the plate adapter of the microscope’s translational stage. The temperature of the Petri dish is regulated using a temperature controller, a thermocouple, and a nichrome heating element.
Software
The following software is used as part of this bio-indentation protocol and data analysis process:
Nikon NIS-Elements AR imaging software (Version 4.30.02)
National Instruments LabView (Version 2014)
FIJI ImageJ (https://imagej.net/Fiji) (Version 1.52g)
Microsoft Excel (Office 365)
OriginPro graphing software (version 8.5)
Procedure
Cell Culture and Acini Growth
Note: A complete description of cell culture protocols, cell line development, and growth of acini structures are reported by Zhang et al. (2019).
Culture and maintain MDCK II cells following standard cell culture protocols for thawing, feeding, and passaging. Incubate cells at 37 °C in a humidified, 5% CO2 environment. Cell culture medium ingredients and their corresponding concentrations are provided in the Recipes section of this protocol.
Induce acinar formation of MDCK II cells in an 8-well chamber slide one week prior to performing the mechanical indentation measurement.
Coat an 8-well Nunc Lab-Tek chamber slide with Matrigel by spreading 40-45 µl of Matrigel in each well of the chamber slide. Incubate for at least 30 min at 37 °C or until the gels have completely solidified.
Trypsinize MDCK II cells following standard cell culture procedures. Centrifuge the cells at 125 x g for 15 min and remove the supernatant. Resuspend the MDCK II cells into MDCK acini indentation medium. MDCK acini indentation medium ingredients and their corresponding concentrations are provided in the Recipes section of this protocol.
Seed the MDCK II cells into Matrigel coated 8-well chamber slide at a seeding density of ~5,000 cells/well with MDCK indentation medium. Incubate the chamber slides at 37 °C in a humidified 5% CO2 environment for approximately 7 days or until the MDCK cells have formed glandular acini structures.
Transfer the acini structures into a glass bottom Petri dish for the indentation measurement.
Coat the glass bottom Petri dish with 0.01 wt% poly-L-lysine. Place 200 µl of 0.01 wt% poly-L-lysine into glass bottom Petri dish and let sit for 1 hr. Remove the poly-L-lysine solution and rinse with 1x PBS solution. The poly-L-lysine should prevent any slippage of the acini structure during the indentation process.
Remove the acini from the Matrigel growth matrix. Wash the acini in Matrigel coated 8-well chamber slide with a 1x cold PBS solution at 4 °C. Incubate the Matrigel coated 8-well chamber slide containing the grown acini in a 1x PBS solution supplemented with 5 mM EDTA, 1 mM NaVO4, and 1.5 mM NaF to remove the Matrigel growth matrix from the acini structure. Isolate the acini structures from the Matrigel support matrix by centrifuging at 4 °C for 15 min at 125 x g. Remove the supernatant and rinse the resulting pellet with a 1x PBS solution prepared at 4 °C. Centrifuge the acini structure again for 15 min at 125 x g and remove the supernatant. Resuspend the pellet into a 1x PBS solution at 4 °C and transfer the acini into poly-L-lysine coated glass bottom Petri dishes at a seeding density of ~1,000 acini/Petri dish. It is important to keep the acini density relatively low in the glass bottom Petri dishes to avoid contacting multiple acini structures during the indentation experiment.
Dye the acini structures with calcein AM to check the viability of individual acini and enable fluorescence confocal microscopy of the acini during the indentation process. Replace the PBS solution with the appropriate cell growth medium supplemented with 1:2,000 parts calcein AM cell dye. Incubate the acini for 20 min at 37 °C in a humidified, 5% CO2 environment. Remove the medium and calcein AM solution and replace with fresh cell culture medium. Continue to incubate the acini at 37 °C in a humidified, 5% CO2 environment until it is time to perform the indentation experiment.
Instrument Set-Up
Mount the bio-indentation instrument and Petri dish support plate onto the confocal microscope. The bio-indentation instrument mounts onto the confocal using the condenser slot; slide the mounting flange of the bio-indentation instrument into the condenser slot and tighten the mounting screw until the bio-indentation instrument is securely mounted. Place the Petri dish support plate into the plate adapter on the confocal microscope’s translational stage. Connect the Petri dish support plate to the temperature controller and power supply unit, and adjust the power settings to maintain a temperature of 37 °C.
Coat the indentation tip with F-127 pluronic to mitigate the effects of adhesion between the glass indentation tip and the acini structure. Remove the indentation tip from the double-leaf cantilever beam by unfastening the mounting screw from the mounting nut. Plasma clean the indentation tip using the O2 plasma cleaner for 1 min. Immediately after plasma cleaning, submerge the indentation tip into a 0.1 wt% F-127 pluronic solution for 10 min. Rinse the indentation tip with a 1x PBS solution before remounting the indentation tip onto the double-leaf cantilever beam.
Adjust the height of the capacitance probe to ensure the double-leaf cantilever beam is within the detection range of the capacitance probe. The capacitance probe should be close enough to detect the cantilever beam but far enough away that the deflection of the cantilever beam during the indentation stays within the detection limits of the capacitance probe.
Align the indentation tip over the center of the microscope objective. Beginning with the lowest magnitude objective, lower the indentation tip and adjust the objective focal point until the indentation tip is within focus. Using the XY linear translational stage, adjust the location of the indentation tip to align the center of the indentation tip with the center of the microscope’s field of view. Install the reflectance filter cube into the confocal microscope’s detector unit and load the reflectance imaging settings in the NIS-Elements software (Figure 2B). Adjust the focal plane of the confocal microscope to find the apex of the indentation tip. Continue to align the center of the indentation tip with the center of the microscope’s field of view.
Repeat the alignment steps using progressively higher objective magnifications until the indentation tip is centered directly above the 20x microscope objective. It is recommended to use the 1.5x optical zoom feature during the indentation. Once the indentation is aligned over the microscope objective, remove the reflectance cube from the confocal microscope’s detector unit, re-install the 438/24 filter cube, and select the DAPI/FITC/TRITC imaging settings in the NIS-Elements software (Figure 2A). It is recommended at this time to check the indentation tip for any debris that may cause irregular surface contact between the indentation tip and the acini structure.
Secure the glass bottom Petri dish on the confocal microscope’s translational stage. Place the glass bottom Petri dish containing the acini structures onto the Petri dish support plate. Secure the Petri dish to the support plate using the two mounting arms.
Determine the location of the glass surface of the Petri dish and zero the indentation instrument’s force and displacement values. Navigate to a region in the center of the Petri dish in which there are no acini present in the microscope’s field of view. Initiate the indentation software and zero the force and displacement measurement. Drive the indentation tip 100 µm toward the surface of the Petri dish using the piezoelectric z-stage; at this time, the indentation tip should not be in contact with the surface and the force measurements should be fluctuating about 0 N. Manually lower the indentation instrument using the focal adjustment of the condenser stage until the indentation tip comes into contact with the glass surface as evident by a sudden and drastic increase in the measured force along with a decrease in the noise in the force measurement. Once contact with the surface has been made, slowly raise the indentation instrument using the focal adjustment of the condenser stage until the indentation tip is no longer in contact with the surface and the measured force returns to 0 N. Raise the indentation tip 100 µm using the piezoelectric z-stage. The indentation tip should now be located ~100 µm above the glass surface of the Petri dish – high enough to avoid contact with acini structures when moving the confocal microscope’s translational stage, but low enough to achieve contact during the indentation measurement.
Mechanical Indentation of Acini Structure
Mechanical indentations of glandular acini structures are performed to determine the average applied pressure at which the acini structures mechanically fail. The indentation tip is slowly brought down into contact with an individual acini structure at a fixed indentation rate while the applied force is recorded with simultaneous fluorescence imaging of the cells. As the indentation force exceeds the background level of noise in the measured force, the acini structures are observed to flatten out. Further increasing the applied force results in continued deformation of the acini until there is a structural collapse resulting in the formation of cellular blebs and/or the bursting of cells (Figure 4A). These rupture events are captured in the fluorescence images of the deforming acini enabling us to pinpoint the time at which the first rupture event occurs (Figure 4B). After the initial rupture, the indentation tip continues to push into the acini structure until the measured applied load reaches the specified maximum indentation force. Once the maximum indentation force is achieved, the indentation tip pulls off the structure at the designated indentation rate, returning to the starting z-position. To perform the mechanical indentation of an acini structure:
Locate an isolated acini structure and align it in the center of microscope’s field of view, directly beneath the indentation tip. It is important that the acini structure is isolated within the microscope’s field of view to avoid contact with other acini structures. It is recommended to use brightfield imaging while locating the acini structures to avoid prolonged exposure of the acini structures to the confocal laser and photobleaching of the fluorescence dye.
Misalignment of the acini structure can lead to irregular surface contact between the indentation tip and the acini resulting in improper force measurements as the indentation tip comes into contact with the glass substrate prior to the acini structure rupturing. A sudden, near vertical increase in the force vs displacement curve is a strong indicator that the acini structure is not properly aligned with the indentation tip.
Navigate the focal plane of the confocal microscope to the desired imaging plane for the indentation measurement. It is recommended to choose a focal plane that is close to the bottom of the acini structure to fully capture the deformation during the indentation process (Figure 4A); a focal plane chosen too high may result in the acini structure deforming out of the imaging plane prior to the rupture event occurring.
Note: It is optional to perform a confocal z-scan at this time to capture the shape of the acini structure prior to the indentation measurement.
Prepare the confocal microscope for the indentation measurement. Initiate perfect focus in the confocal software and adjust the region of interest and the capture delay of the confocal software to achieve the fastest framerate possible. A higher framerate during the indentation measurement will result in an improved temporal resolution when determining the rupture pressure of the acini structure.
Prepare the indentation instrument for the indentation measurement. Within the indentation instrument software, set the parameters for the indentation rate and maximum indentation force. We recommend using an indentation rate of 0.1 µm/s and a maximum indentation force of 100 µN. Faster indentation rates will reduce the temporal resolution of the fluorescence images captured throughout the measurement. A slower indentation rate will improve the temporal resolution when determining the rupture pressure but will increase the experimental time and laser exposure of the cells. The maximum indentation force should be set sufficiently high enough that the acini structure ruptures before the maximum force is reached. From our experience, a 100 µN maximum indentation load was sufficiently high enough to induce rupture of the acini; however, a higher maximum indentation force will not alter the results of the measurement.
Simultaneously initiate the indentation measurement of the acini structure and the imaging time-lapse on the confocal microscope. Record any discrepancies in start times of the indentation measurement and imaging time-lapse.
Data analysis
Once the indentation has been completed, the rupture pressure of the acini structure is determined from the normal contact force measured by the indentation instrument and the contact area measured from the fluorescence images.
Determine the time at which the acini structure first ruptures using the time-lapse images captured using the confocal microscope (Figure 4B). Correct the time for any discrepancies between the start times of the indentation measurement and the confocal time-lapse.
Measure the contact area of the acini structure at moment in which the rupture event is first observed (Figure 4B). This can be accomplished using either the NIS Elements software or FIJI ImageJ. The contact area can be measured by drawing the best-fit circle around the acini structure at the moment of rupture.
Determine the normal contact force experience by the acini structure at the time of rupture. This step can be accomplished using either Microsoft Excel or OriginLabs graphing software. Load the force vs time data into the graphing software and record the indentation force at the previously determined time of rupture.
Calculate the rupture pressure of the acini structure from the measured contact area and the measured normal contact force. The average contact pressure for Hertzian contact of a sphere is given as <P> = F/A, where F is the measured normal force from the indentation instrument and A is the measured contact area from the confocal images.
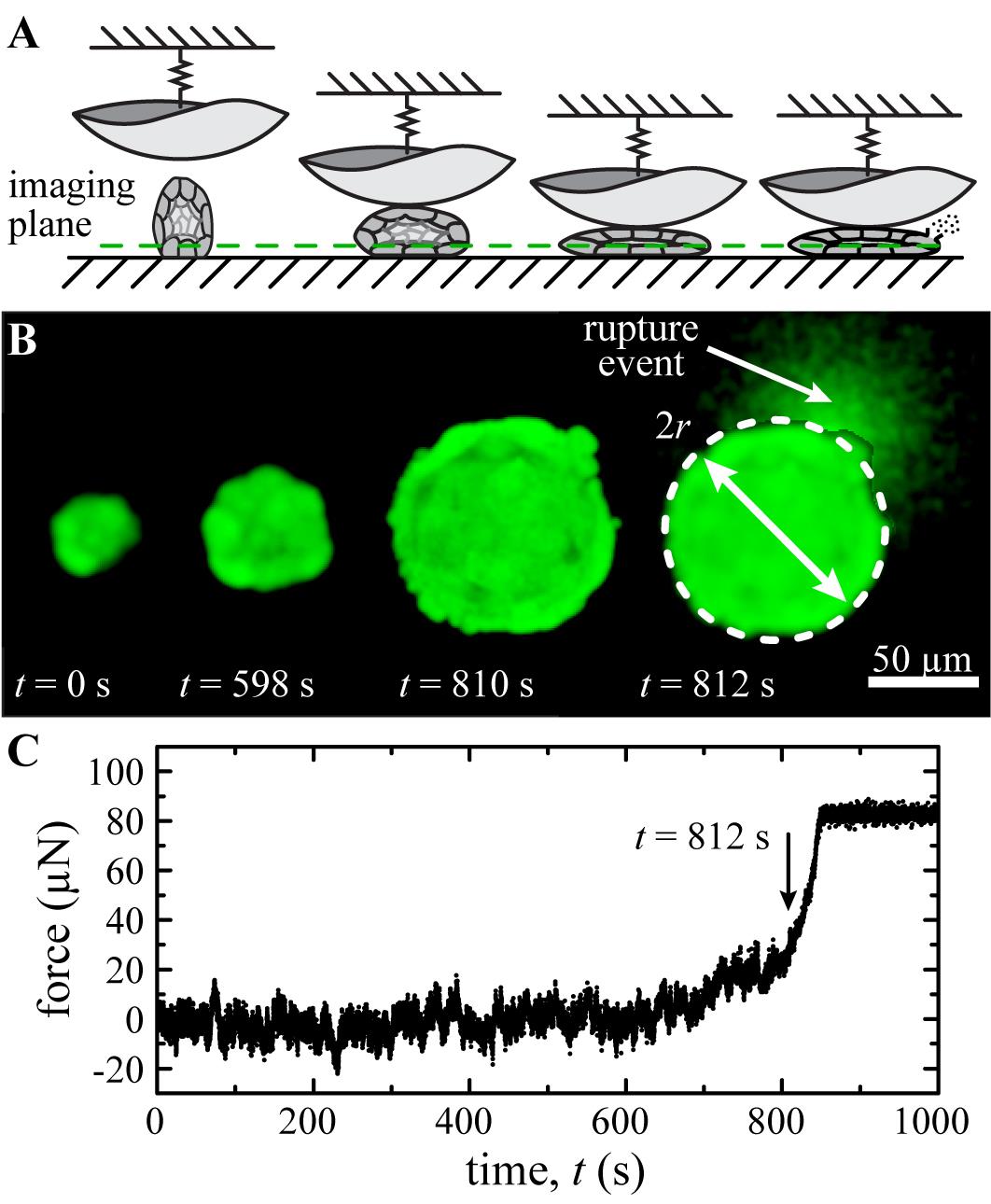
Figure 4. Data analysis from micro-indentation measurements. A. Schematic illustration of the bio-indentation experiment. The imaging plane is chosen close to the bottom of the acini structure to capture the full deformation during the indentation measurement. B. Fluorescence images captured through confocal microscopy show the deformation of the acini structure throughout the indentation. As the force is increased, the formation of cellular blebs and/or the bursting of cells can be observed between subsequent frames indicating the rupturing of the acini structure. In the presented example, we observe the rupture of the acini at t = 812 s as evident by rupture of a cell and the release of the calcein dye into the surrounding media. The contact area is determined by manually fitting a circle to the acini structure at the instance in rupture is observed. C. The indentation force at the time of rupture is determined from the force measurements of the bio-indentation instrument. (µN: micro-Newton)
Discussion
Bio-indentation provides a means to explore both the material properties of biological materials, as well as the collective behavior of cellular systems to externally applied forces. By coupling mechanical indentation with fluorescence images, we directly observe the deformation and rupture of individual glandular acini structures resulting from an externally applied mechanical force. This protocol was developed as part of our exploration into the role the LINC complex plays in the development and maintenance of glandular acini structures (Zhang et al., 2019). Acini formed from MDCK II cells with disrupted LINC complexes failed at higher applied pressures than those formed with intact LINC complexes. These results appear consistent with the observations of filled lumens in acini structures with disrupted LINC complexes. Furthermore, treating LINC-disrupted acini with Y-27632 resulted in the recovery of the rupture pressure to undisrupted levels.
Although the protocol presented here is focused on performing bio-indentation measurements on individual acini structures, similar indentation protocols can be developed for exploring a variety of biological systems. For example, the elastic modulus and permeability of MDCK monolayers, and the minimum pressure necessary to rupture the nuclear envelope of 3T3 fibroblast cells have been investigated using similar micro-indentation protocols (Schulze et al., 2017; Halfmann et al., 2019). Furthermore, the effect of shear stresses and friction on biological systems can be studied with the addition of a second load cell to measure lateral forces (Urueña et al., 2018). Continued exploration into the response of biological systems to external forces and mechanical queues will be crucial for understanding collective cell behavior and developmental biology.
Recipes
Below are the recipes for the various solution used for culturing the MDCK cells and performing the bio-indentation protocol:
MDCK cell growth medium
DMEM medium
4.5 g/L glucose
10% v/v DBS
MDCK acini indentation medium
DMEM medium
4.5 g/L glucose
10% v/v donor bovine serum
2 % v/v Matrigel matrix basement membrane
Matrigel wash solution
1x PBS solution
5 mM EDTA
1 mM Na3VO4
1.5 mM NaF
Acini dye medium
cell growth medium
Calcein AM (1:2,000)
F-127 pluronic solution
Water
0.1 wt% F-127 pluronic
Acknowledgments
The indentation protocols presented here were developed as part of the research work of Zhang et al. (2019). This work was supported by the NIH (grant R01 EB014869) to T.P. Lele, and the NSF (grant DMR-1352043) to T.E. Angelini. The authors would like to acknowledge Dr. Kyle D. Schulze and Dr. W. Gregory Sawyer for their contribution in developing the micro-indentation instrument used in this protocol. The authors would also like to thank Dr. Jennifer Lippencott-Schwart for graciously donating the MDCK II cells.
Competing interests
The authors have no financial conflicts of interest to declare.
References
- Ahn, B. M., Kim, J., Ian, L., Rha, K. H. and Kim, H. J. (2010). Mechanical property characterization of prostate cancer using a minimally motorized indenter in an ex vivo indentation experiment. Urology 76(4): 1007-1011.
- Crisp, M., Liu, Q., Roux, K., Rattner, J., Shanahan, C., Burke, B., Stahl, P. D. and Hodzic, D. (2006). Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol 172(1): 41-53.
- Debnath, J. and Brugge, J. S. (2005). Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer 5(9): 675-688.
- Halfmann, C. T., Sears, R. M., Katiyar, A., Busselman, B. W., Aman, L. K., Zhang, Q., O’Bryan, C. S., Angelini, T. E., Lele, T. P. and Roux, K. J. (2019). Repair of nuclear ruptures requires barrier-to-autointegration factor. J Cell Biol 201901116.
- Horn, H. F., Brownstein, Z., Lenz, D. R., Shivatzki, S., Dror, A. A., Dagan-Rosenfeld, O., Friedman, L. M., Roux, K. J., Kozlov, S. and Jeang, K. T. (2013). The LINC complex is essential for hearing. J Clin Invest 123(2): .
- Lei, K., Zhang, X., Ding, X., Guo, X., Chen, M., Zhu, B., Xu, T., Zhuang, Y., Xu, R. and Han, M. (2009). SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Nati Acad Sci 106(25): 10207-10212.
- Levental, I., Levental, K., Klein, E., Assoian, R., Miller, R., Wells, R. and Janmey, P. (2010). A simple indentation device for measuring micrometer-scale tissue stiffness. J Phys Condens Matter 22(19): 194120.
- Li, Q., Lee, G. Y., Ong, C. N. and Lim, C. T. (2008). AFM indentation study of breast cancer cells. Biochem Biophys Res Commun 374(4): 609-613.
- Matsumoto, A., Hieda, M., Yokoyama, Y., Nishioka, Y., Yoshidome, K., Tsujimoto, M. and Matsuura, N. (2015). Global loss of a nuclear lamina component, lamin A/C, and LINC complex components SUN 1, SUN 2, and nesprin-2 in breast cancer. Cancer Med 4(10): 1547-1557.
- Meinke, P., Mattioli, E., Haque, F., Antoku, S., Columbaro, M., Straatman, K. R., Worman, H. J., Gundersen, G. G., Lattanzi, G. and Wehnert, M. (2014). Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genetics 10(9): e1004605.
- Mellad, J. A., Warren, D. T. and Shanahan, C. M. (2011). Nesprins LINC the nucleus and cytoskeleton. Curr Opin Cell Biol 23(1): 47-54.
- Schulze, K., Zehnder, S., Urueña, J., Bhattacharjee, T., Sawyer, W. and Angelini, T. (2017). Elastic modulus and hydraulic permeability of MDCK monolayers. J Biomech 53: 210-213.
- Sen, S., Subramanian, S. and Discher, D. E. (2005). Indentation and adhesive probing of a cell membrane with AFM: theoretical model and experiments. Biophys J 89(5): 3203-3213.
- Stolz, M., Raiteri, R., Daniels, A., VanLandingham, M. R., Baschong, W. and Aebi, U. (2004). Dynamic elastic modulus of porcine articular cartilage determined at two different levels of tissue organization by indentation-type atomic force microscopy. Biophys J 86(5): 3269-3283.
- Strasser, S., Zink, A., Janko, M., Heckl, W. M. and Thalhammer, S. (2007). Structural investigations on native collagen type I fibrils using AFM. Biochem Biophys Res Commun 354(1): 27-32.
- Tapley, E. C. and Starr, D. A. (2013). Connecting the nucleus to the cytoskeleton by SUN–KASH bridges across the nuclear envelope. Curr Opin Cell Biol 25(1): 57-62.
- Urueña, J. M., Hart, S. M., Hood, D. L., McGhee, E. O., Niemi, S. R., Schulze, K. D., Levings, P. P., Sawyer, W. G. and Pitenis, A. A. (2018). Considerations for biotribometers: cells, gels, and tissues. Tribology Letters 66(4): 141.
- Vogel, V. and Sheetz, M. (2006). Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol 7(4): 265-275.
- Zhang, Q., Narayanan, V., Mui, K. L., O’Bryan, C. S., Anderson, R. H., Birendra, K., Cabe, J. I., Denis, K. B., Antoku, S. and Roux, K. J. (2019). Mechanical Stabilization of the Glandular Acinus by Linker of Nucleoskeleton and Cytoskeleton Complex. Curr Biol 29(17): 2826-2839. e2824.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
O'Bryan, C. S., Zhang, Q., Lele, T. and Angelini, T. E. (2020). Mechanical Characterization of Glandular Acini Using a Micro-indentation Instrument. Bio-protocol 10(23): e3847. DOI: 10.21769/BioProtoc.3847.
Category
Biophysics > Bioengineering
Biophysics > Microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








