- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ribosome Purification from an α-proteobacterium and rRNA Analysis by Northern Blot
Published: Vol 10, Iss 23, Dec 5, 2020 DOI: 10.21769/BioProtoc.3835 Views: 3323
Reviewed by: Alka MehraKenji SugiyamaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Glucose Starvation, Magnesium Ion Starvation, and Bile Stress Assays
Aryashree Arunima and Mrutyunjay Suar
Sep 20, 2021 2961 Views

Analysis of Heterocyst and Akinete Specific Glycolipids in Cyanobacteria Using Thin-layer Chromatography
Ritu Garg [...] Iris Maldener
Mar 20, 2022 2451 Views

Detecting Photoactivatable Cre-mediated Gene Deletion Efficiency in Escherichia coli
Yuta Koganezawa [...] Miki Umetani
Jun 5, 2023 1932 Views
Abstract
Ribosomes are an integral part of cellular life. They are complex molecular machines consisting of multiple ribosomal proteins and RNAs. To study different aspects of ribosome composition, many methods have been developed over the decades. Here, we describe how to purify ribosomes from the α-proteobacterium Rhodopseudomonas palustris. Following this protocol, RNA can be extracted from either purified ribosomes or directly from cell cultures, and ribosomal RNAs quantified using Northern blot. This protocol gives an example of studying ribosomes in a bacterium other than the commonly used E. coli. The challenge of performing Northern blots with rRNA is also addressed in detail.
Keywords: Ribosome profileBackground
The fate of bacterial cells is closely linked to their ribosomes. Our recent study showed that active ribosomes play an important role in the survival mechanisms of nutrient-deprived R. palustris cells (Yin et al., 2019). The ribosomes are purified via a series of ultracentrifugations with a protocol optimized from the classical methods (Lawrence et al., 2016). Ribosomal RNA populations are detected with Northern blot employing a less frequently used capillary transfer system. The state of ribosomes can be greatly affected by the details of purification steps. These methods, described here in detail, should be of broad interest to researchers who study the translation apparatus in a wide variety of bacteria.
Materials and Reagents
Note: All reagents can be made de novo or purchased from different vendors as long as they are RNase free.
Centrifuge tubes, RNase-free (ThermoFisher AM12400 or equivalent). Use ultracentrifuge compatible tube for ultracentrifugation (please refer to the manual of reader’s ultracentrifuge for more information).
Screw-cap tube, 2 ml (Sigma-Aldrich, BR780758 or equivalent)
Costar Sterile Disposable Reagent Reservoirs (Fisher Scientific, catalog number: 07-200-128 )
HEPES-KOH, 1 M, pH 7.5 (Boston Bioproduct, catalog number: BBH-75-K )
MgCl2, 1 M (Boston Bioproduct, catalog number: BM-670 )
NaCl, Molecular Biology Grade (Promega Corp, catalog number: H5271 )
Recombinant RNasin Ribonuclease Inhibitor (Promega Corp, catalog number: N2511 )
miRNeasy Mini Kit (Qiagen, catalog number: 217004 )
Agarose, molecular biology grade
UltraPure DNase/RNase-Free Distilled Water (Invitrogen, catalog number: 10977-023 )
Amersham Hybond-XL membrane (GE Life Sciences)
10x Tris-borate-EDTA (TBE) buffer (Thermo Fisher Scientific, catalog number: AM9863 )
20x SSC buffer (Sigma-Aldrich, catalog number: S6639 )
DIG RNA Labeling Mix (Sigma-Aldrich, catalog number: 11277073910 )
DIG Easy Hyb (Sigma-Aldrich, catalog number: 11603558001 )
DIG Wash and Block Buffer Set (Sigma-Aldrich, catalog number: 11585762001 )
Anti-Digoxigenin-AP, Fab fragments (Sigma-Aldrich, catalog number: 11093274910 )
CDP-Star (Sigma-Aldrich, catalog number: 11685627001 )
RNA Gel Loading Dye (2x) (ThermoFisher, catalog number: R0641 )
Buffer A1 (10x) (see Recipes)
Lysis buffer (see Recipes)
Equipment
Note: The working bench as well as electrophoresis, transfer and Northern blot systems should be cleaned with RNaseZap or equivalent to decontaminate RNase before use. Items below that do not have a catalog number listed can be obtained from any reliable supplier.
Mini-Beadbeater-24 (Bio Spec Products Inc., catalog number: 112011 )
Zirconia/Silica Beads, 0.1 mm dia (Bio Spec Products Inc., catalog number: 11079101z )
Beckman TL-1000 ultracentrifuge (discontinued model, can be substituted with equivalent ultracentrifuge with temperature-control). Please make sure to use compatible centrifuge tubes for ultracentrifugation.
Temperature-controlled tabletop centrifuge
Electrophoresis system suitable for agarose gel
A spectrometer capable of measuring UV absorbance, as well as matching quartz cuvettes
A short wavelength UV light
Pipette as well as matching pipetting tips. Any kind of pipette (manual, automatic, serological, etc.) works as long as they meet the requirement and are fit for use
Liquid nitrogen tank (and liquid nitrogen supply) or -80 °C freezer for storing bacterial strains
Incubator for bacterial growth
Procedure
Preparation of -80 °C cell pellets
Grow R. palustris cultures as required by the experimental design. Typical growth conditions that we use are given in Kim and Harwood (1991). However, the composition of the cultivation medium is not important for the protocol we describe here. It also does not matter if cells are grown aerobically or anaerobically. For one typical sample, we usually use 50-70 ml of culture at Abs660 ~1 (medium is used as blank).
Add 10 ml of RNase-free water to one 50-ml centrifuge tube. Place the tube horizontally in a freezer (any sub-zero °C freezer would work) to form a thin sheet of ice (we usually do this a few days in advance to make sure ice is formed). One tube with ice sheet would be used collect ~35 ml of cell culture. Prepare as many tubes as required in advance.
When the culture reaches the desired growth state, pour the culture directly into the tube containing the ice sheet and place the tube on ice. The purpose of this step is to minimize the time required for the culture to cool down and thus capture the native form of ribosomes. It is also referred as “fast-cool” in some literature ( Lawrence et al., 2016 ).
Invert the tubes with culture gently from time to time to facilitate the cooling down of the culture. We usually transfer the now cold culture into a pre-chilled centrifuge tube. This process usually takes 5-10 min in our experience.
Spin at ~5,000 x g for 10 min at 4 °C to collect the cell pellets.
Discard the supernatant and freeze the pellets with liquid nitrogen. The frozen cell pellets can now be stored at -80 °C. In our experience, the ribosome profile of the sample can be preserved for 6 weeks at least.
Note: For each sample, we usually collect cells from ~70 ml culture and store the pellets in two 50-ml centrifuge tubes. This can be easily scaled up or down depending on the need of sampling and the availability of equipment.
Purification of ribosomes
Prepare two screw-capped 2 ml centrifuge tubes for each sample (assuming the pellets are collected in two 50 ml centrifuge tubes, see the Note in Procedure A). Add ~500 µl Zirconia/Silica beads to each tube and autoclave all of the tubes.
Resuspend the cell pellet in 1.2 ml cold lysis buffer in each 50 ml centrifuge tube. Use pipet to mix well and transfer the resuspended sample to the 2 ml screw-cap tube containing Zirconia/Silica beads.
In a 4 °C room, lyse the resuspended cells with a Biospec Mini-Beadbeater-24 at 3,500 rpm for 1 min and then chill them on ice for 1min. Repeat 3 more times, 4 times in total.
Note: The setup of beadbeater cycle, speed and time may need to be adjusted according to the species of interest.
Centrifuge for 10 min at 20,000 x g with a tabletop microcentrifuge at 4 °C to remove the beads and cell debris.
Collect 1 ml of supernatant from each tube. Combine supernatants from the same sample if needed (2 ml of supernatant for each sample in our setting, see the Note in Procedure A). Centrifuge for 30 min at 30,000 x g at 4 °C with an ultracentrifuge to further clarify the lysates. Collect the supernatant.
Centrifuge the supernatant from last step at 100,000 x g with an ultracentrifuge for four hours at 4 °C to pellet the ribosomes.
After the spin, carefully remove the supernatant by pipetting without disturbing the pelleted ribosomes. The pelleted ribosomes, when abundant, visually resemble a contact lens.
Without disturbing the pellets, carefully add 200-500 µl Buffer A1 to the centrifuge tube containing the pelleted ribosomes. Leave the samples on ice overnight to allow the ribosomes to dissolve in the buffer.
On the next day, gently mix the dissolved ribosomes. Take a sample and determine the absorbance at 260 nm (A260). The volume of the sample taken depends on the concentration and the setup of spectrometer. The sample can also be diluted if necessary. In our experience, a typical yield from one sample is 400 µl of 20 relative units (R.U.) of A260 in total. Note that the yield can vary significantly depending on the experiment.
Adjust all the samples to the same concentration. The minimum concentration of ribosome for the following analysis is 50 R.U. of A260/ml. Freeze the purified ribosomes with liquid nitrogen and store at -80 °C for further analysis. The purified ribosomes are suitable for further assays such as separation on a sucrose gradient or for isolation and analysis of rRNA associated with ribosomes.
Analyze rRNA by electrophoresis and Northern blot.
This procedure can be applied to total RNA isolated from cultures or from purified ribosomes. RNA is purified following either a QIAzol-based method (see Yin and Harwood, 2020) or the commercially available protocol for the miRNeasy Mini Kit (Qiagen). For the QIAzol-based method, we typically start with 400 µl of an 0.1 absorbance (RU of A260) preparation of ribosomes and the method yields a 15 µl volume of approximately 500 ng RNA/µl.
An optional step is to use an RNA ScreenTape System (Agilent) to get a snapshot of the rRNA profile. To specifically detect rRNA by Northern blot, rRNA has to be first separated by electrophoresis on an agarose gel. Since agarose gels are usually too thick to be used in the commonly used electro-transfer system, this calls for a capillary transfer system (upward or downward) to transfer RNA from the gel to a membrane for subsequent Northern blot detection. This procedure will be described here in detail. For Northern blot detection, we use a non-radioactive system and follow its commercially available manual (DIG). This greatly saves time and effort as described in detail in the accompanying protocol “Charging state analysis of transfer RNA from an α-proteobacterium” (Yin and Harwood, 2020). The principle of designing and generating a DIG-labeled probe is also described there.
In a 50-ml centrifuge tube, add 0.2 g agarose to 20 ml 1x TBE buffer. Melt agarose in a microwave and pour the gel.
Mix 1 µg total RNA with 2x loading dye. Heat at 70 °C for 10 min. Immediately chill on ice for 3 min. Spin briefly.
Analyze the sample on an agarose gel with a running buffer of 1x TBE buffer. For a gel of ~10 cm in length, a one-hour run at 100 V usually achieves satisfactory separation. After electrophoresis with a setting like this,16S rRNA is usually in the middle of the gel.
After the electrophoresis, immerse the gel in 20x SSC twice, each time for 10 min.
Assemble the transfer apparatus as follows:
Top
Light weight such as a pipet tip box
1 Whatman filter paper connecting to the reservoir, immersed in 20x SSC
1 Whatman filter paper, slightly bigger than the gel, immersed in 20x SSC
Agarose gel, the side with RNA facing down
Membrane cut to gel size, pre-immersed in 20x SSC
2 Whatman filter papers, slightly bigger than the gel, immersed in 20x SSC
Dry paper towels (~20 sheets at least)
Bottom
Note: This describes the transfer apparatus for downward transfer. Slightly modified setup can be used for upward transfer.
Set up the capillary transfer system as shown in Figure 1. Leave on bench overnight.
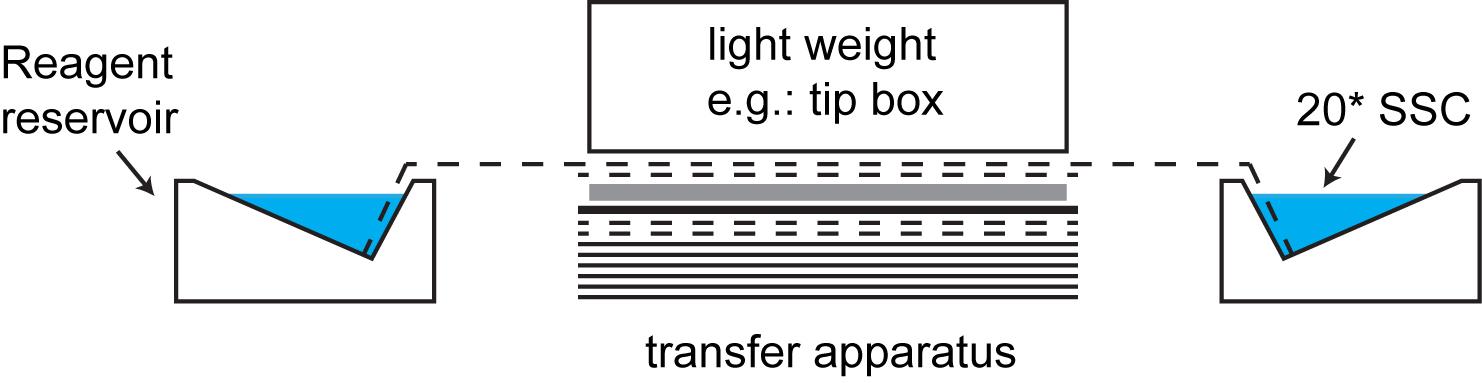
Figure 1. Capillary transfer system (RNA transferred downward from gel to membrane)The membrane can be cross-linked the next morning. One option to cross-link is to place the membrane 1-2 cm from a shortwave UV light, the side with sample facing UV light, and expose it to UV for 10 min. Next, follow commercially available protocol for DIG system for Northern blot (Sessions in Hybridization and Immunological detection in https://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Roche/Bulletin/1/12039672910bul.pdf).
Data analysis
An example of 16S rRNA Northern blot is shown in Figure 2. Total RNA isolated from wild-type (WT) and a mutated (mutant) R. palustris are analyzed by RNAtape and Northern blot. The RNAtape result shows an extra band immediately above the 16S rRNA from the mutant. This band was subsequently identified as an intermediate product of 16S rRNA synthesis, as it is identified in Northern blot for 16S rRNA. Detailed analysis of rRNA Northern blots can be found in Figures 6 and 7 fromYin et al. (2019).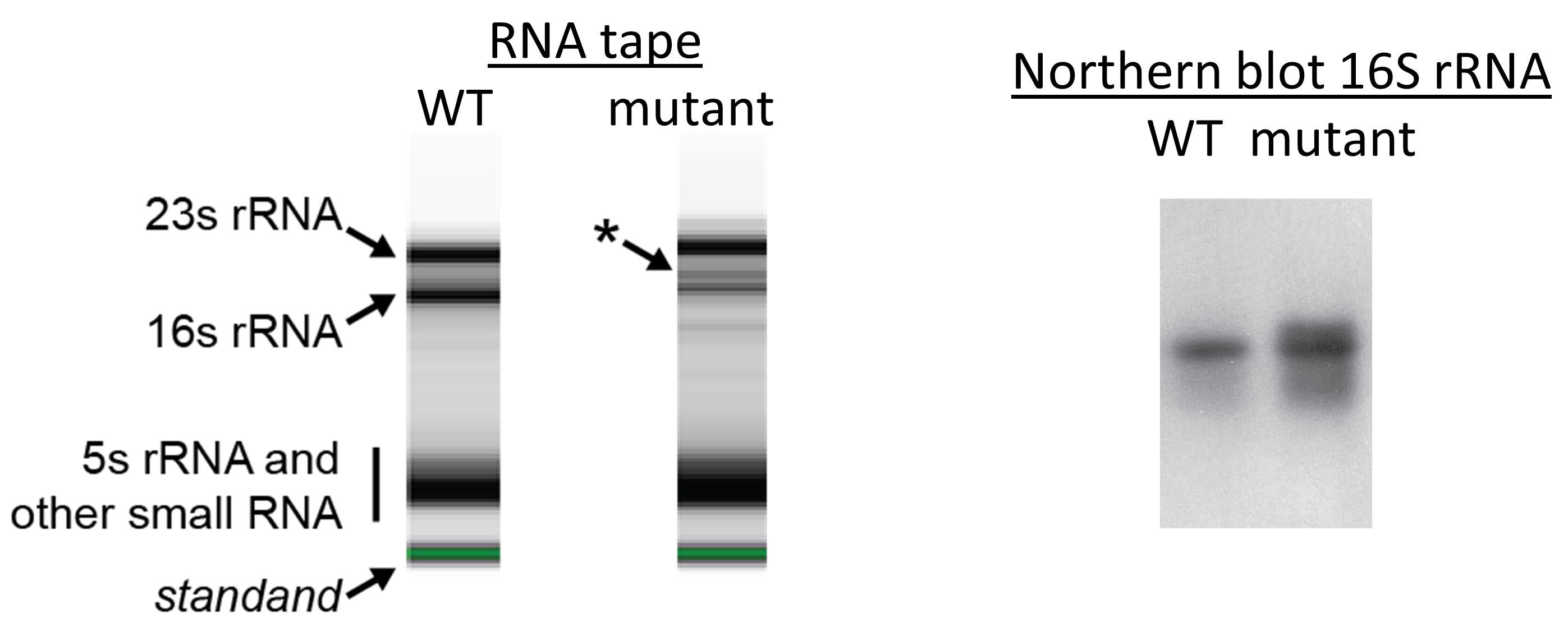
Figure 2. An example of 16S rRNA Northern blot. RNA isolated from wild type (WT) on RNA tape shows typical 23S rRNA, 16S rRNA and other RNA species. RNA isolated from mutant with defective ribosome biosynthesis on RNA tape shows an extra band immediately above 16S rRNA (asterisk) on RNA tape. Northern blot using a probe targeting 16S rRNA detects the 16S rRNA in WT and the mutant, as well as the extra band immediately above 16S rRNA. The result is consistent with the hypothesis that the extra band is an intermediate product of 16S rRNA biosynthesis. Note that in the figure above, our purpose was to determine the nature of irregular band labeled with asterisk, hence more total RNA from the mutant was analyzed in Northern blot comparing to the WT. Same amount of total RNA was analyzed on RNA tape in the figure above.
Recipes
Note: The reagents listed here below are RNase free. The working bench should be cleaned with RNaseZap or equivalent to decontaminate RNase before use.
Buffer A1 (10x)
200 mM HEPES-KOH, pH 7.5
300 mM NaCl
80 mM MgCl2
Notes:
Buffer A1 is one of the most critical items in ribosome purification. When analyzed in vitro, ribosome profiles are very sensitive to the buffer components. For example, if MgCl2 is reduced to 60 mM in 10x Buffer A1, the population of 70S ribosome will be greatly reduced in sucrose gradient analysis. Higher MgCl2 concentrations are also sometimes used to better preserve the population of 100S ribosome. Therefore, one should be cautious about interpreting results from assays such as sucrose gradients, especially with organisms that are less commonly studied. A useful control is to purify ribosomes from E. coli with the buffers to be used, and then examine whether they behave as expected.
On the same note, additional consideration should be given to the enzyme activities needed to be preserved if purified ribosomes are to be used in assays such as in vitro translation. The recipe here is sufficient to preserve the profile of ribosomes during the purification process, namely the population of 30S, 50S, 70S and 100S ribosomes. We encourage the reader to do a preliminary study as well as a broader literature review to determine the optimal buffer components for their organism, system and question of interest (Lawrence et al., 2016; Basu and Yap, 2017).
Lysis buffer
Add RNasin into Buffer A1 (1x) to a final concentration of 1 µl/ml
Acknowledgments
This work was supported by grant W911NF-15-1-0150 to C.S.H. from the U.S. Army Research Office. We thank Prof. David Morris, University of Washington, for his help with ribosome profiling. This protocol is derived from our previous work published in 2019 (Yin et al., 2019).
Competing interests
The authors claim no financial or non-financial competing interests.
References
- Basu, A. and Yap, M. N. (2017). Disassembly of the Staphylococcus aureus hibernating 100S ribosome by an evolutionarily conserved GTPase. Proc Natl Acad Sci U S A 114(39): E8165-E8173.
- Kim, M. K. and Harwood, C. S. (1991). Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiology Letters 83: 199-203.
- Lawrence, M. G., Shamsuzzaman, M., Kondopaka, M., Pascual, C., Zengel, J. M. and Lindahl, L. (2016). The extended loops of ribosomal proteins uL4 and uL22 of Escherichia coli contribute to ribosome assembly and protein translation. Nucleic Acids Res 44(12): 5798-5810.
- Yin, L., Ma, H., Nakayasu, E. S., Payne, S. H., Morris, D. R. and Harwood, C. S. (2019). Bacterial longevity requires protein synthesis and a stringent response. mBio 10(5).
- Yin, L. and Harwood, C. S. (2020). Charging State Analysis of Transfer RNA from an α-proteobacterium. Bio-protocol 10(23): e3834.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yin, L. and Harwood, C. S. (2020). Ribosome Purification from an α-proteobacterium and rRNA Analysis by Northern Blot. Bio-protocol 10(23): e3835. DOI: 10.21769/BioProtoc.3835.
Category
Microbiology > Microbial physiology > Adaptation
Microbiology > Microbial biochemistry > RNA
Biochemistry > RNA > RNA structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










