- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Generating Three-dimensional Human Granulomas in vitro to Study Mycobacterium tuberculosis-host Interaction
Published: Vol 10, Iss 22, Nov 20, 2020 DOI: 10.21769/BioProtoc.3820 Views: 4786
Reviewed by: Alexandros AlexandratosHsih-Yin Tan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

HIV-CRISPR: A CRISPR/Cas9 Screening Method to Identify Genes Affecting HIV Replication
Ferdinand Roesch and Molly OhAinle
May 5, 2020 9900 Views

From 3D to 2D: Harmonization of Protocols for Two-dimensional Cultures on Cell Culture Inserts of Intestinal Organoids from Various Species
David Warschkau [...] Frank Seeber
Jan 20, 2022 5835 Views

An Ex Vivo Lung Histoculture Model for Studying Pulmonary Infection and Immune Response With SARS-CoV-2 as an Example of RNA Virus
Elena V. Maryukhnich [...] Elena J. Vasilieva
Dec 20, 2025 740 Views
Abstract
Granulomas are organized multicellular structures that constitute the hallmark of an infection by the human pathogen Mycobacterium tuberculosis (Mtb). A better understanding of the complex host-Mtb interactions within the granuloma’s environment may lead to new therapeutic or preventive tools to improve the control of the tuberculosis pandemic. To date, several in vitro models that are able to mimic human nascent granulomas have been reported. Here we describe a protocol in which Mtb-infected human peripheral blood mononuclear cells (PBMCs) are embedded within a collagen matrix leading to the formation of three-dimensional micro-granulomas. Subsequently, PBMCs and Mtb can be retrieved allowing multiparametric readouts from both the host and the pathogen. In addition to the incorporation of a physiological extracellular matrix, this model has the singular advantage of recapitulating dormant-like Mtb features, as well as reproducing Mtb resuscitation observed under immunomodulatory treatments, which have not been reported in other published protocols to generate in vitro granulomas.
Keywords: MycobacteriumBackground
Tuberculosis (TB) is an air-borne disease that encompasses pulmonary and extra-pulmonary infections by the human pathogen Mycobacterium tuberculosis (Mtb). Causing an estimated 1.5 million deaths in 2019 (WHO, 2019), TB remains the world’s deadliest infectious disease. The hallmark of the immunopathogenesis of TB is the formation of structurally organized, multicellular clusters called granulomas (Gengenbacher and Kaufmann, 2012). These structures mainly consist of a core of infected and non-infected macrophages surrounded by a rim of lymphocytes. The hostile environment within granulomas pushes Mtb to enter a slow- or non-replicating dormant state likely associated with a latent form of the disease. Consequently, Mtb dormancy leads to increased tolerance to antibiotics targeting metabolic pathways active during mycobacterial replication.
New strategies are urgently needed in order to lessen the TB death toll. Such strategies may arise from a better understanding of the infection within the granuloma’s environment. However, the complex pathophysiology of Mtb infection makes the development of relevant preclinical models particularly challenging. Despite the widespread use of animal models in the TB field, the establishment of latency and granulomatous lesions comparable to the ones seen in humans can only be observed in non-human primates. Yet, drawbacks inherent to this animal model, such as its high cost or ethical concerns, have favored the development of in vitro models (Guirado and Schlesinger, 2013). In that context, several independent groups have reported in vitro models based on the infection of human peripheral blood mononuclear cells (PBMCs) with virulent Mtb (Kapoor et al., 2013; Guirado et al., 2015; Agrawal et al., 2016; Arbués et al., 2020; Tezera et al., 2017). Despite lacking some relevant cell types (e.g., neutrophils) and the full tissue conditions (such as lung structure or vascularization), such models are able to mimic the organization of human nascent granulomas, orchestrated by relevant cytokines.
The protocol described here notably incorporates two major advantageous features. On the one hand, Mtb-infected PBMCs are embedded in a three-dimensional (3D) matrix of collagen and fibronectin, main components of the lung extracellular matrix (Stek et al., 2018). On the other hand, Mtb displays features associated with dormancy such as accumulation of intracellular triacylglycerides into lipid inclusions, loss of acid-fastness, transcriptional changes leading to a shift in carbon and energy metabolisms, and increase in antibiotic tolerance (Kapoor et al., 2013). Remarkably, this model was also able to reproduce the differential rate of latent TB reactivation observed in the clinic upon treatment with different TNF-α-neutralizing biologics (Arbués et al., 2020). Some particular limitations of this model to take into account are the difficulty of adding new immune cells for dynamic studies or its relatively low throughput.
In conclusion, the in vitro granuloma model described hereunder is particularly relevant for studies focusing on mycobacterial dormancy as well as further understanding the mechanisms involved in Mtb resuscitation.
Materials and Reagents
Filtered micropipette tips (various volumes) (e.g., Clearline, catalog numbers: 713115 , 713117 and 713118 )
Serological pipettes (various volumes) (e.g., Falcon, catalog numbers: 357543 and 357551 )
1.5/2.0 ml microtubes (e.g., Sarstedt, catalog numbers: 72.692.005 and 72.693.005 )
15/50 ml conical centrifuge tubes (e.g., Falcon, catalog numbers: 352096 and 352070 )
Syringe (e.g., BD, catalog number: 300865 )
0.20 μm syringe filter (e.g., Sarstedt, catalog number: 83.1826.001 )
Stericup filter unit, Millipore Express PLUS high flow rate membrane (Millipore, catalog number: S2GPU01RE )
Tissue culture plates, 24-well, flat bottom (e.g., Falcon, catalog number: 353047 )
Single-cell suspension of Mycobacterium tuberculosis (Mtb) prepared according to published methods (bacterial concentration determined by colony forming unit quantification)
Note: Mtb H37Rv (Sébastien Gagneux’s strain collection) was used in our published study (Arbués et al., 2020). Different strains or mycobacterial species could also be used.
Human peripheral blood mononuclear cells (PBMCs) isolated from whole blood or buffy coats following a standard protocol ( Vogel et al., 2018 ) (samples stored in liquid nitrogen can be used, when viability ≥ 95% assessed by trypan blue exclusion)
RPMI-1640 medium, with L-glutamine and sodium bicarbonate (e.g., Sigma-Aldrich, catalog number: R8758 )
Fetal bovine serum (FBS) (e.g., Gibco, catalog number: 10270 ), heat inactivated
Benzonase, E. coli recombinant (BioVision, catalog number: 7680 )
Human serum off-the-clot, type AB (PAN Biotech, catalog number: P40-2701 ), heat inactivated and filtered (Stericup)
Trypan blue solution (e.g., Sigma-Aldrich, catalog number: T8154 )
PureCol bovine type I collagen solution, 3 mg/ml (Advanced BioMatrix, catalog number: 5005 )
Fibronectin from human plasma, 0.1% solution (Sigma-Aldrich, catalog number: F0895 )
Dulbecco’s Phosphate Buffered Saline (PBS) 10x (Sigma-Aldrich, catalog number: D1408 )
Sodium hydroxide solution (NaOH) 1 N for cell culture (Sigma-Aldrich, catalog number: S2770 )
Collagenase type IV, sterile filtered (Sigma-Aldrich, catalog number: C1889 )
Calcium chloride dihydrate (CaCl2·2H2O) (PanReac AppliChem, catalog number: A4689 )
Triton X-100 (e.g., Sigma-Aldrich, catalog number: T8787 )
Millipore double distilled water, sterile
Thawing medium (see Recipes)
Resting medium (see Recipes)
Complete medium (see Recipes)
Extracellular Matrix (ECM) solution (see Recipes)
Collagenase solutions (see Recipes)
0.1% Triton X-100 (see Recipes)
Equipment
Class II biosafety cabinet
5% CO2 incubator, set at 37 °C
Cell counting chamber
Inverted bright-field/phase-contrast microscope (e.g., Leica DM IL LED)
Note: We routinely use a 20x objective to count the PBMCs and a 10x objective to monitor granuloma formation (Procedure A, Steps 1f and 2i, respectively)
Centrifuge for 15/50 ml conical tubes, maximum RCF ≥ 3,000 x g (e.g., Beckman Coulter Allegra X-15R)
Microcentrifuge for 1.5/2.0 ml microtubes, maximum RCF ≥ 6,000 x g (e.g., Eppendorf 5424R)
Procedure
Note: This protocol was developed for infection with virulent Mtb; therefore, all work involving handling of Mtb should be performed in a biosafety level 3 laboratory under a class II biosafety cabinet. However, the protocol has also been successfully applied by our group to class 1 (M. bovis BCG) or class 2 (Mtb H37Ra) organisms in a standard biosafety level 2 laboratory.
Generation of Mtb-induced 3D granulomas
Resting of PBMCs prior to infection
Thaw one or two vials of PBMCs yielding the adequate amount of cells anticipated for the design of the experiment (2.5 x 106 PBMCs/well) and transfer them into conical centrifugation tubes.
Wash the cells by adding 10 ml (the first 3-5 ml dropwise) of pre-warmed (37 °C) thawing medium and mix gently. Centrifuge for 5 min at 400 x g and discard the supernatant by pipetting.
Repeat this washing step.
Resuspend the cell pellet in 5 ml of pre-warmed (37 °C) resting medium.
Place the tube, with the lid loosened, into a 5% CO2 incubator at 37 °C and allow the cells to rest for at least 6 h up to overnight.
Count the rested PBMCs and assess cell viability by trypan blue dye exclusion method.
Note: Cell viability should be ≥ 95%. Lower cell viabilities can lead to Mtb-independent formation of cell aggregates.
Transfer the required amount of PBMCs into a conical centrifuge tube. Pellet the cells by centrifugation at 400 x g for 5 min and discard the supernatant by pipetting.
Resuspend the PBMCs at 107 cells/ml in complete medium. Place the cells, with the lid of the tube loosened, into a 5% CO2 incubator at 37 °C until use.
Mtb infection of PBMCs and embedding (Figure 1A)
Prepare the required volume of extracellular matrix (ECM) solution according to Recipe 4, except for the 1 N NaOH. Keep at 4 °C/on ice until use.
Infect the PBMCs with the volume of Mtb single-cell suspension required to reach the aimed multiplicity of infection (MOI). Mix thoroughly by pipetting up and down 10-15 times in order to obtain a homogeneous suspension of PBMCs and Mtb.
Note: Our experiments are routinely performed using a standard MOI of 1:200 (Mtb:PBMC). However, adjustments in the MOI may be required when using a new batch of single-cell suspension (or different mycobacterial strains or species). The optimal MOI can be experimentally assessed by monitoring the 3D granuloma formation in a dose-response setup (using various donors and uninfected PBMCs as a control).
Distribute 250 μl per well of the Mtb-infected PBMCs in 24-well plates.
Add the required amount of NaOH to the ECM solution (according to Recipe 4) and mix slowly and thoroughly by pipetting up and down 10-15 times.
Note: A thorough homogenization of the ECM solution after addition of NaOH is critical to reach the correct pH and, consequently, a proper setting of the matrix. If the NaOH is homogeneously incorporated into the ECM solution, its mixture with PBMCs (Step A2e) should result in a pale, yellowish pink color. However, if the ECM components are not properly mixed, a color gradient from dark pink to light yellow may be observed across the wells due to pH variation. A too basic pH (dark pink) is detrimental for cell and bacterial viability and prevents the formation of granuloma-like structures.
Add 250 μl of the ECM solution prepared in the previous step (1:1 ratio, v/v) to each well and mix by pipetting up and down 3-4 times.
Place the plate into a 5% CO2 incubator at 37 °C and allow the ECM to set for 45 min.
Top up the set ECM with 500 μl per well of complete medium.
Note: If the full plate is not going to be used, fill up the surrounding empty wells with sterile PBS or water to minimize evaporation.
Place the plate back into the 5% CO2 incubator. Incubate at 37 °C for the desired time periods.
Granuloma formation can be monitored using an inverted bright-field/phase-contrast microscope.
Note: A marked donor-to-donor variability may be observed in terms of granuloma formation, in both number and size. This phenomenon has also been described in other in vitro granuloma models (Agrawal et al., 2016) .
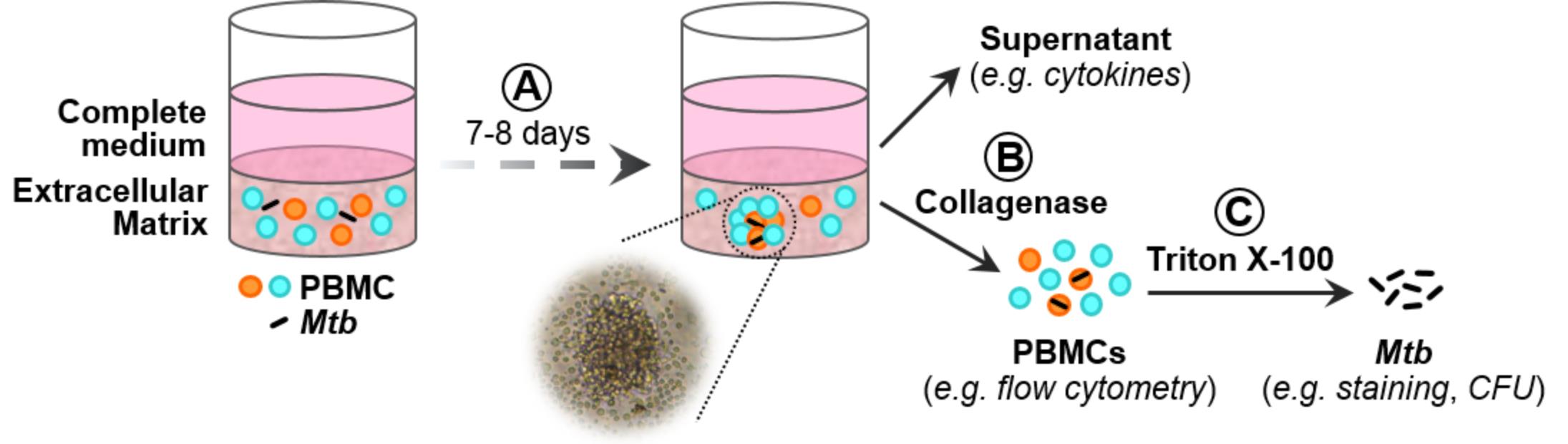
Figure 1. Schematic representation of the 3D in vitro granuloma model. A. Mtb-infected PBMCs are embedded in an extracellular matrix (ECM). Formation of granuloma-like structures can be observed after 7-8 days. B. PBMCs can be released from the ECM by collagenase digestion. C. Subsequent treatment with Triton X-100 allows the retrieval of Mtb.
Release of PBMCs from the ECM (Figure 1B)
At the selected time points, prepare the required volume of collagenase (1 mg/ml) according to Recipe 5.
Remove the supernatant from the wells to be processed. Discard, or filter sterilize and store at -80 °C if analysis of soluble factors is envisaged.
Add 250 μl per well of collagenase and incubate the plate in a 5% CO2 incubator at 37 °C for 40-45 min. Every 15 min mix the liquid fraction by pipetting up and down to facilitate the digestion of the ECM.
Once the ECM liquefied completely, transfer the content of the wells into microtubes.
Pellet the cells by centrifugation at 400 x g for 5 min and discard the supernatant by pipetting.
Released cells can then be used to subsequently recover Mtb (according to Procedure C below), or to analyze host cell parameters, for example by flow cytometry (following standard staining procedures).
Retrieval of Mtb (Figure 1C)
Resuspend the cell pellet obtained in the previous section in 200 μl of 0.1% Triton X-100 by vortexing for a few seconds or pipetting up and down a few times.
Incubate for 20 min at room temperature to lyse the host cells.
To retrieve the Mtb pellet, centrifuge for 5 min at 6,000 x g and discard the supernatant by pipetting.
Various Mtb readouts can be obtained from the recovered bacilli as described in the Data analysis section.
Data analysis
Detailed information concerning data processing and analysis can be found in our original research article ( Arbues et al., 2020 ). Briefly, secreted cytokines can be measured by ELISA or multiplexed bead arrays from 0.2 µm-filtered supernatants. Host cell populations can be characterized by flow cytometry after staining and fixation of the released PBMCs. RNA analysis from both host and Mtb can be performed following standard procedures subsequently to TRI-reagent® treatment. Mtb load can be quantified by determination of colony forming units (CFU) on Middlebrook 7H11 agar supplemented with 10% OADC (0.05% oleic acid, 5% bovine albumin fraction V, 2% dextrose and 0.004% catalase). Importantly, dual auramine/Nile red staining can be used as a proxy of the metabolically active or dormant-like status of the recovered bacilli, respectively. As a general rule, we use at least three independent donors to generate every experimental readout.
Recipes
Thawing medium
RPMI supplemented with 10% FBS and 12.5 U/ml benzonase
Resting medium
RPMI supplemented with 10% FBS
Complete medium
RPMI supplemented with 20% human serum
Extracellular Matrix (ECM) solution
Recipe per ml:
950 μl of collagen solution (3 mg/ml)
4 μl of human fibronectin (1 mg/ml)
50 μl of PBS 10x
10 μl of 1 N NaOH
Calculate the volume of ECM solution required for the particular experiment to be performed: number of wells to be embedded + ≥ 2 extra wells x 250 μl
On the day of the experiment, mix the collagen, fibronectin and 10x PBS and keep the mixture at 4 °C/on ice until use
Immediately before use, add the NaOH and mix slowly and thoroughly by pipetting up and down. The final pH of the solution should be ~7.0
Collagenase solutions
Collagenase buffer: PBS supplemented with 0.42 mM CaCl2
Prepare a 0.5 M CaCl2 solution (dissolve 1.47 g of CaCl2·2H2O in 20 ml of Milli-Q water) and sterilize (autoclave or filter)
To prepare 50 ml of buffer, add 42 μl of 0.5 M CaCl2 to 49.958 ml of PBS
Collagenase stock solution (10 mg/ml)
Dissolve 50 mg of sterile-filtered collagenase in 5 ml of collagenase buffer
Prepare aliquots of convenient volume to prevent repeated freezing-thawing cycles and store at -20 °C (stable for several months)
Collagenase working solution (1 mg/ml)
Calculate the volume of diluted collagenase needed: number of wells to be processed + 1 extra well x 250 μl
On the day of the experiment, dilute the collagenase stock tenfold in collagenase buffer to prepare the working solution (stable at 37 °C for at least 5 h)
0.1% Triton X-100
Prepare a 10% stock solution by mixing 5 ml of Triton X-100 with 45 ml of Milli-Q water
Filter sterilize the stock solution (stable at room temperature for several months)
To prepare 50 ml of working solution, add 500 μl of 10% stock to 49.5 ml of sterile Milli-Q water (stable at room temperature for several months)
Acknowledgments
We would like to thank Christian Schindler for statistical support and script writing of the R code used for the generalized linear mixed-effects model. DP received the funding to implement this model from Novartis AG under a research agreement with the Swiss Tropical and Public Health Institute. This protocol was used to generate the data published in Arbués et al. (2020), and was adapted from Kapoor et al. (2013).
Competing interests
DP received the funding to implement this model from Novartis AG. The funders initiated the study design, but had no role in data collection and analysis. MK is a full-time employee of Novartis.
Ethics
Human PBMCs were isolated from buffy coats obtained from the Interregionale Blutspende SKR AG (Bern, Switzerland). All donors provided informed consent that included information on the use of blood products for research purposes.
References
- Agrawal, N., Bhattacharyya, C., Mukherjee, A., Ullah, U., Pandit, B., Rao, K. V. S. and Majumder, P. P. (2016). Dissecting host factors that regulate the early stages of tuberculosis infection. Tuberculosis(Edinb) 100: 102-113.
- Arbués, A., Brees, D., Chibout, S. D., Fox, T., Kammüller, M. and Portevin, D. (2020). TNF-alpha antagonists differentially induce TGF-beta1-dependent resuscitation of dormant-like Mycobacterium tuberculosis. PLoS Pathog 16(2): e1008312.
- Gengenbacher, M. and Kaufmann, S. H. (2012). Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev 36(3): 514-532.
- Guirado, E., Mbawuike, U., Keiser, T. L., Arcos, J., Azad, A. K., Wang, S. H. and Schlesinger, L. S. (2015). Characterization of host and microbial determinants in individuals with latent tuberculosis infection using a human granuloma model. mBio 6(1): e02537-02514.
- Guirado, E. and Schlesinger, L. S. (2013). Modeling the Mycobacterium tuberculosis Granuloma- the Critical Battlefield in Host Immunity and Disease. Front Immunol 4: 98.
- Kapoor, N., Pawar, S., Sirakova, T. D., Deb, C., Warren, W. L. and Kolattukudy, P. E. (2013). Human granuloma in vitro model, for TB dormancy and resuscitation. PLoS One 8(1): e53657.
- Stek, C., Allwood, B., Walker, N. F., Wilkinson, R. J., Lynen, L. and Meintjes, G. (2018). The Immune Mechanisms of Lung Parenchymal Damage in Tuberculosis and the Role of Host-Directed Therapy. Front Microbiol 9: 2603.
- Tezera, L. B., Bielecka, M. K., Chancellor, A., Reichmann, M. T., Shammari, B. A., Brace, P., Batty, A., Tocheva, A., Jogai, S., Marshall, B. G., Tebruegge, M., Jayasinghe, S. N., Mansour, S. and Elkington, P. T. (2017). Dissection of the host-pathogen interaction in human tuberculosis using a bioengineered 3-dimensional model. Elife 6: e21283.
- Vogel, G., Cuénod, A., Mouchet, R., Strauss, A., Daubenberger, C., Pflüger, V. and Portevin, D. (2018). Functional characterization and phenotypic monitoring of human hematopoietic stem cell expansion and differentiation of monocytes and macrophages by whole-cell mass spectrometry. Stem Cell Res 26: 47-54.
- WHO. (2019). WHO global tuberculosis report 2019. (World Health Organization).
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Arbués, A., Kammüller, M. and Portevin, D. (2020). Generating Three-dimensional Human Granulomas in vitro to Study Mycobacterium tuberculosis-host Interaction. Bio-protocol 10(22): e3820. DOI: 10.21769/BioProtoc.3820.
Category
Immunology > Host defense > Human
Microbiology > Microbe-host interactions > In vitro model
Cell Biology > Cell-based analysis > Bacterial infection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link