- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Salivary Charged Metabolites Using Capillary Electrophoresis Time-of-flight-mass Spectrometry
Published: Vol 10, Iss 20, Oct 20, 2020 DOI: 10.21769/BioProtoc.3797 Views: 4619
Reviewed by: Rakesh BamChing Yao YangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

ATAC Sequencing Protocol For Cryopreserved Mammalian Cells
Juan Manuel Caravaca [...] Bao Tran
Jan 20, 2022 5219 Views

Detection and Quantification of Calcium Ions in the Endoplasmic Reticulum and Cytoplasm of Cultured Cells Using Fluorescent Reporter Proteins and ImageJ Software
Shunsuke Saito and Kazutoshi Mori
Aug 20, 2023 2850 Views

A Protocol for Laser-Assisted Microdissection and tRF & tiRNA Sequencing in Lung Adenocarcinoma
Zi Wang [...] Feng Jiang
Apr 5, 2025 2997 Views
Abstract
Salivary metabolomics have provided the potentials to detect both oral and systemic diseases. Capillary electrophoresis time-of-flight-mass spectrometry (CE-TOFMS) enables the identification and quantification of various charged metabolites. This method has been employed to biomarker discoveries using human saliva samples, especially for various types of cancers. The untargeted analysis contributes to finding new biomarkers. i.e., the analysis of all detectable signals including both known and unknown metabolites extends the coverage of metabolite to be observed. However, the observed data includes thousands of peaks. Besides, non-linear migration time fluctuation and skewed peaks are caused by the sample condition. The presented pretreatment protocols of saliva samples enhance the reproducibility of migration time drift, which facilitates the matching peaks across the samples and also results in reproducible absolute concentrations of the detected metabolites. The described protocols are utilized not only for saliva but for any liquid samples with slight modifications.
Keywords: MetabolomicsBackground
Saliva is one of the biofluids suitable for monitoring the systemic conditions. The non-invasive availability of saliva samples enables frequent, timely, and cost-effective diagnostics, which would contribute to realizing personalized medicine. Therefore biomarker discoveries for various diseases have been reported (Wang et al., 2017). Saliva contains microorganisms and also a wide variety of components, such as genome, coding and non-coding RNA, proteins, and metabolites (Bonne and Wong, 2012; Yoshizawa et al., 2013). Traditionally, the profiling of these components has been utilized for the diagnostic of oral diseases (Dawes and Wong, 2019; Martina et al., 2020). Recently, accumulated evidence has revealed the potential to monitor systematic health conditions (Sugimoto et al., 2013; Kaczor-Urbanowicz et al., 2017).
Among various omics technologies, metabolomics has been utilized to discover the biomarkers for metabolic diseases, including cancers (Trezzi et al., 2015). The metabolic aberrance in cancer cells, such as the Warburg effect (Warburg, 1956), potentially reflects metabolite profiles in biofluids. Metabolomics-based liquid biopsy is therefore intensively developed (Armitage and Ciborowski, 2017). In comparison to the blood and urine-based diagnostics, the use of saliva is an emerging approach while salivary metabolic profiles showed the potential to detect various cancers (Sugimoto et al., 2010a).
The salivary diagnostics of oral cancer are the most reported among various cancers (Washio and Takahashi, 2016; Chattopadhyay and Panda, 2019). We previously confirmed the consistently elevated metabolites in saliva and oral cancer tissues (Ishikawa et al., 2016). The effect of saliva collection after various fasting duration on these markers was also evaluated (Ishikawa et al., 2017). The evaluation of the specificity of oral cancer-specific markers against various diseases in the oral cavity, e.g., periodontal diseases, is also conducted (Mikkonen et al., 2016). Discrimination of oral leukoplakia, oral lichen planus, and oral cancer is practically useful in clinical settings (Sridharan et al., 2019; Ishikawa et al., 2020). The storage condition of the saliva samples affected the discrimination abilities of metabolite markers (Wang et al., 2014), which requires the standard of the protocol to deal with the saliva samples for realizing reproducible diagnostics.
Recently, salivary metabolite biomarkers to diagnose the cancers far from the oral cavity have been reported. The quantified polyamines in saliva were utilized for the detection of breast cancers (Takayama et al., 2016; Murata et al., 2019) using liquid chromatography-mass spectrometry (LC-MS). These analyses were conducted using triple-quadrupole-MS (MS) for targeted analyses. We utilized capillary electrophoresis time-of-flight-MS (CE-TOFMS) to conduct untargeted analyses to quantify hundreds of salivary metabolites for pancreatic cancer detections (Asai et al., 2018). MS requires the ionization of metabolites to be detected and therefore various separation system, detection, and their interface are available (Monton and Soga, 2007). Among these systems, CE-MS is one of a powerful tool that can analyze the charged metabolites using only two modes, cation and anion, i.e., positively and negatively charged metabolites (Soga et al., 2003; Soga, 2007). Here, the metabolite profiling protocol for saliva samples using CE-TOFMS is described.
Materials and Reagents
- Pipette tips 100 μl, 10 μl (Eppendorf, catalog numbers: 0030.073.428, 0030 073.363)
- 1.5 ml reaction tube (Greiner-Bio-One, catalog number: 616201 )
- Nanosep Centrifugal Devices With Omega Membrane 3K, Gray (PALL, catalog number: OD003C34 )
- Polypropylene vial (Agilent, catalog number: 5190-3155 )
- Snap Cap (Agilent, catalog number: 5042-6491 )
- Nitrile gloves (TOP, catalog number: 15731 )
- 2 ml Crimp/Snap Top Vials & Caps (Agilent, catalog number: 5182-9697 )
- Fused Silica Capillary (Polymicro Technologies, catalog number: TSP050375 )
- COSMO(+)Capillary (NACALAI TESQUE, catalog number: 07584-44 )
- L-Methionine sulfone, 99+% (Thermo Fisher Scientific Inc, catalog number: A17027 )
- 3-Aminopyrrolidine dihydrochloride (Sigma-Aldrich, catalog number: 404624 )
- 2-(N-Morpholino)ethanesulfonic Acid (MES) (FUJIFILM Wako Pure Chemical, catalog number: 341-01622 )
- D-Camphor-10-sulfonic Acid Sodium Salt (CSA) (FUJIFILM Wako Pure Chemical, catalog number: 037-01032 )
- 1,3,5-Benzenetricarboxylic Acid (Trimesate) (FUJIFILM Wako Pure Chemical, catalog number: 206-03641 )
- Spermidine (Merk/Sigma-Aldrich, catalog number: S2626 )
- Spermine (FUJIFILM Wako Pure Chemical, catalog number: 194-09813 )
- N1-Acetylspermine (FUJIFILM Wako Pure Chemical, catalog number: 014-20421 )
- N1-Acetylspermidine (Sigma-Aldrich Japan, 01467)
- N8-Acetylspermidine (Fluka, catalog number: A3658 )
- DL-a-Amino-n-butyric acid (2AB) (Sigma-Aldrich, catalog number: A1754 )
- Methanol (FUJIFILM Wako Pure Chemical, catalog number: 134-14523 )
- Ammonium Acetate (FUJIFILM Wako Pure Chemical, catalog number: 019-02835 )
- Hexakis(2,2-difluoroethoxy)phosphazene (Synquest Laboratories, catalog number: 8H79-3-02 )
- Formic Acid Formate (FUJIFILM Wako Pure Chemical, catalog number: 063-05895 )
- Ammonium Formate (Kanto Kagaku, catalog number: 01294-00 )
- Acetic Acid (FUJIFILM Wako Pure Chemical, catalog number: 012-00245 )
- Milli-Q water (MERCK, Tokyo, Japan, Milli-Q IQ 7000 ICP-MS)
- Internal standards mixture (see Recipes)
- Sheath liquid (see Recipes)
- Run buffer (see Recipes)
- Standard mixture (see Recipes)
Equipment
- Measuring flask 5 ml (IWAKI, catalog number: 5640FK5S )
- Laboratory Bottles 100 ml (SCHOTT/DURAN, catalog number: 017200-100A )
- Pipettes 100 μl, and 10 μl (Eppendorf, catalog numbers: 3121 000.074, 3121 000.015)
- Centrifuge (TOMY, model: MX-307 )
- Roter (TOMY, model: AR015-24 )
- MicroMixer (TAITEC, model: E-36 )
- CE system (Agilent Technologies, model: 7100 )
- LC/MSD TOF system (Agilent, catalog number: G1969A )
- Isocratic HPLC pump (Agilent, model: 1260 series )
- CEMS adapter kit (Agilent, catalog number: G1603A )
- CE-ESI-MS sprayer kit (Agilent, catalog number: G1607A )
- Circulation type hand cooler (THOMAS, catalog number: 2241059 )
Software
- Agilent Chemstation software was used for CE, ver. B.02.01.SR1 (Agilent Technologies, Santa Clara, CA, USA)
- Agilent MassHunter software, ver. B.02.01.SR1 (Agilent) for TOF-MS data analyses
- R, ver. 3.4.3 (R Foundation for Statistical Computing, Vienna, Austria)
- JMP, ver. 13.2.0 (SAS Institute Inc., Cary, NC, USA)
- WEKA, ver. 3.6.13 (The University of Waikato, Hamilton, New Sealand)
- GraphPad Prism, ver 7.03 (GraphPad Software Inc., San Diego, CA, USA)
- MasterHands, ver 2.17.3.18 (Keio University, Tsuruoka, Japan)
Procedure
Prepare all solutions and mixtures at room temperature. All sample processing is conducted using Nitrile gloves. Anaerobic condition is not necessary.
- Saliva collection
- Subjects with a primary disease without a history of prior malignancy.
- Subjects who did not receive any prior treatment in the form of chemotherapy, radiotherapy, surgery, or alternative therapy.
- The saliva providers were not allowed to take any food except water intake after 9:00 p.m. on the previous day.
- All samples were collected from 8:00 to 11:00 a.m.
- The subjects were required to brush their teeth without toothpaste on the day of saliva collection and had to refrain from drinking water, smoking, tooth-brushing, and intense exercise 1 h before saliva collection.
- They were required to gargle with water just before saliva collection.
- Approximately 400 μl of unstimulated saliva was collected in a 50 cc polypropylene tube (see Note 1).
- A polypropylene straw 1.1 cm in diameter was used to assist the saliva collection.
- After collection, saliva samples were immediately stored at -80 °C.
- Processing of saliva samples.
The overview of saliva processing is depicted in Figure 1.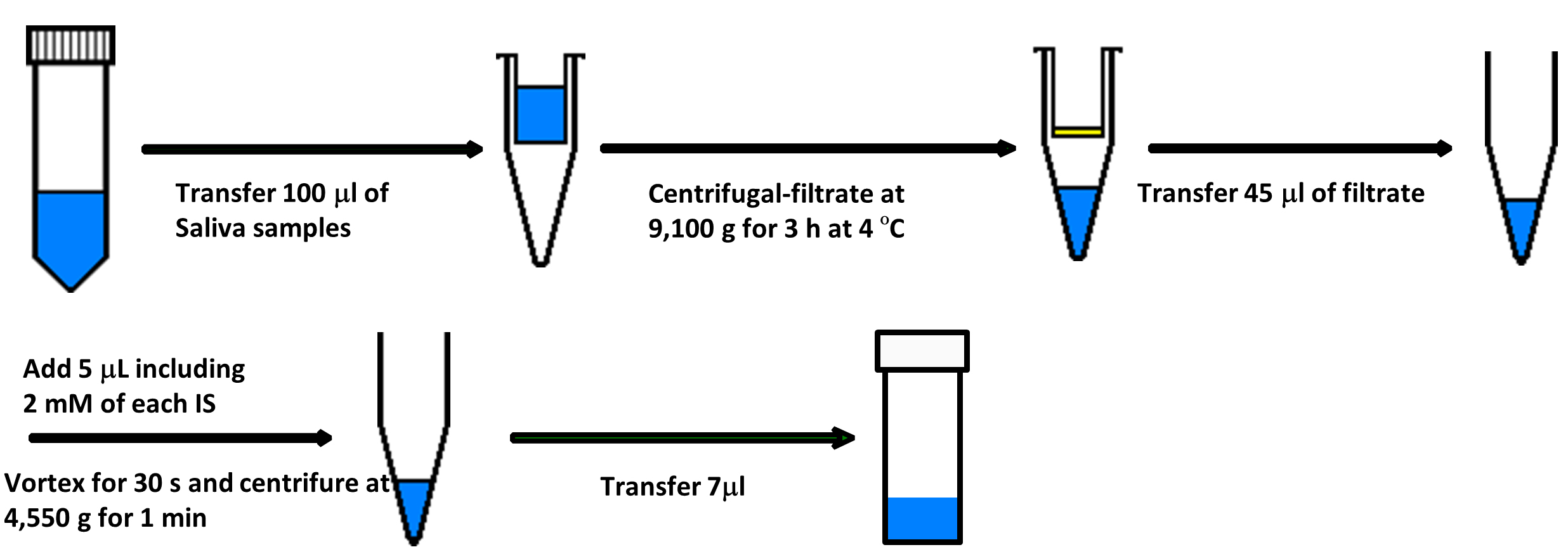
Figure 1. A protocol of saliva processing- A frozen saliva sample is thawed at 4 °C.
- 100 μl of the saliva sample is moved to a 1.5-ml tube with a 5-kDa cutoff filter using a pipet.
- The tube is centrifuged at 9,100 × g for 3 h at 4 °C to eliminate large molecules.
- If the filtrate is not less than 45 μl (Figure 2), the rest of the raw saliva samples should be processed independently (Steps B2 and B3) and the filtrate should be merged.
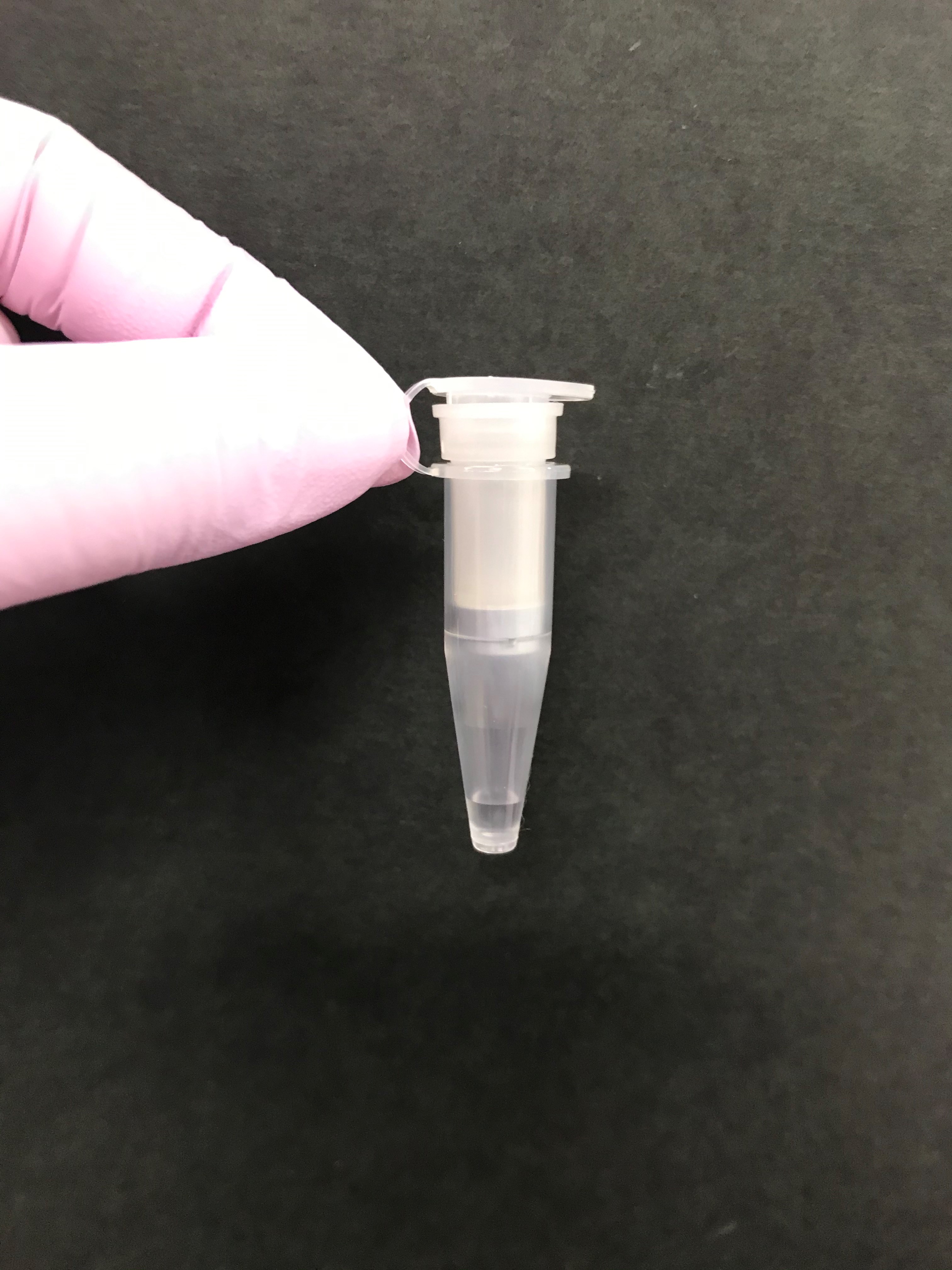
Figure 2. A tube with a filter. The filtrate of 45 μl is shown. - 45 μl of the filtrate is moved to a 1.5 ml of microtube using a pipet.
- 5 μl of water containing an internal standards mixture (Recipe 1) was added to the tube using a pipet, mixed 30 s using vortex, and centrifuged at 4,550 × g for 1 min at 4 °C to mix well.
- 7 μl of the mixture is transferred to a vial and the vial is covered with a snap cap.
- Setup for the cation measurement
- CE
- Capillary: Fused-silica, i.d. 50 μm × 100 cm
- Buffer: 1 M Formic acid
- Voltage: Positive, 30 kV
- Temperature: 20 °C
- Injection: Pressure injection 50 mbar, 5 s (approximately 5 nl)
- TOFMS
- Ion Source: Electrospray ionization (ESI)
- Polarity: Positive
- Capillary voltage: 4,000 V
- Fragmentor: 75 V
- Skimmer: 50 V
- OCT RFV: 500 V
- Drying gas: N2, 10 L/min
- Drying gas temp.: 300 °C
- Nebulizer gas press.: 7 psig
- Sheath liquid: Cation sheath liquid (Recipe 2)
- Flow rate: 10 μl/min
- Lock mass: 2MeOH13Cisotope m/z 66.063061 and Hexakis(2,2-difluoroethoxy)phosphazene m/z 622.028963
- CE
- Setup for the anion measurement
- CE
- Capillary: COSMO(+), i.d. 50 μm × 105 cm
- Buffer: 50 mM Ammonium acetate, pH 8.5
- Voltage: Negative, 30 kV
- Temperature: 20 °C
- Injection: Pressure injection 50 mbar, 30 s (approximately 30 nl)
- Preconditioning: 2 min at 50 mM Ammonium acetate, pH 3.4 and 5 min at run buffer
- TOFMS
- Ion Source: ESI
- Polarity: Negative
- Capillary voltage: 3,500 V
- Fragmentor: 100 V
- Skimmer: 50 V
- OCT RFV: 200 V
- Drying gas: N2, 10 L/min
- Drying gas temp.: 300 °C
- Nebulizer gas press.: 7 psig
- Sheath liquid: Anion sheath liquid (see Recipes)
- Flow rate: 10 μl/min
- Lock mass: 2CH3COOH13C isotope m/z 120.038339, Hexakis(2,2-difluoroethoxy)phosphazene+CH3COOH m/z 680.035541
- ESI needle: Platinum
- Measurement
- Sample preconditioning
- Cation for standard mixtures: 4 min at a cation run buffer (see Recipes).
- Cation for samples: 5 min at Ammonium Formate, 5 min at Milli-Q, and 5 min at an anion run buffer (Recipe 3).
- Anion for standard mixtures and samples: 2 min at 50 mM Ammonium acetate, pH 3.4, and 5 min at a cation run buffer (Recipe 3).
- Three samples and a standard mixture are measured for accessing the quality.
- Evaluate the stability of the total ion electrophoresis (TIE) (Figures 3 and 4). If the TIE is unstable, adjust the ESI sprayer until the intensities of the TIE becomes steady.
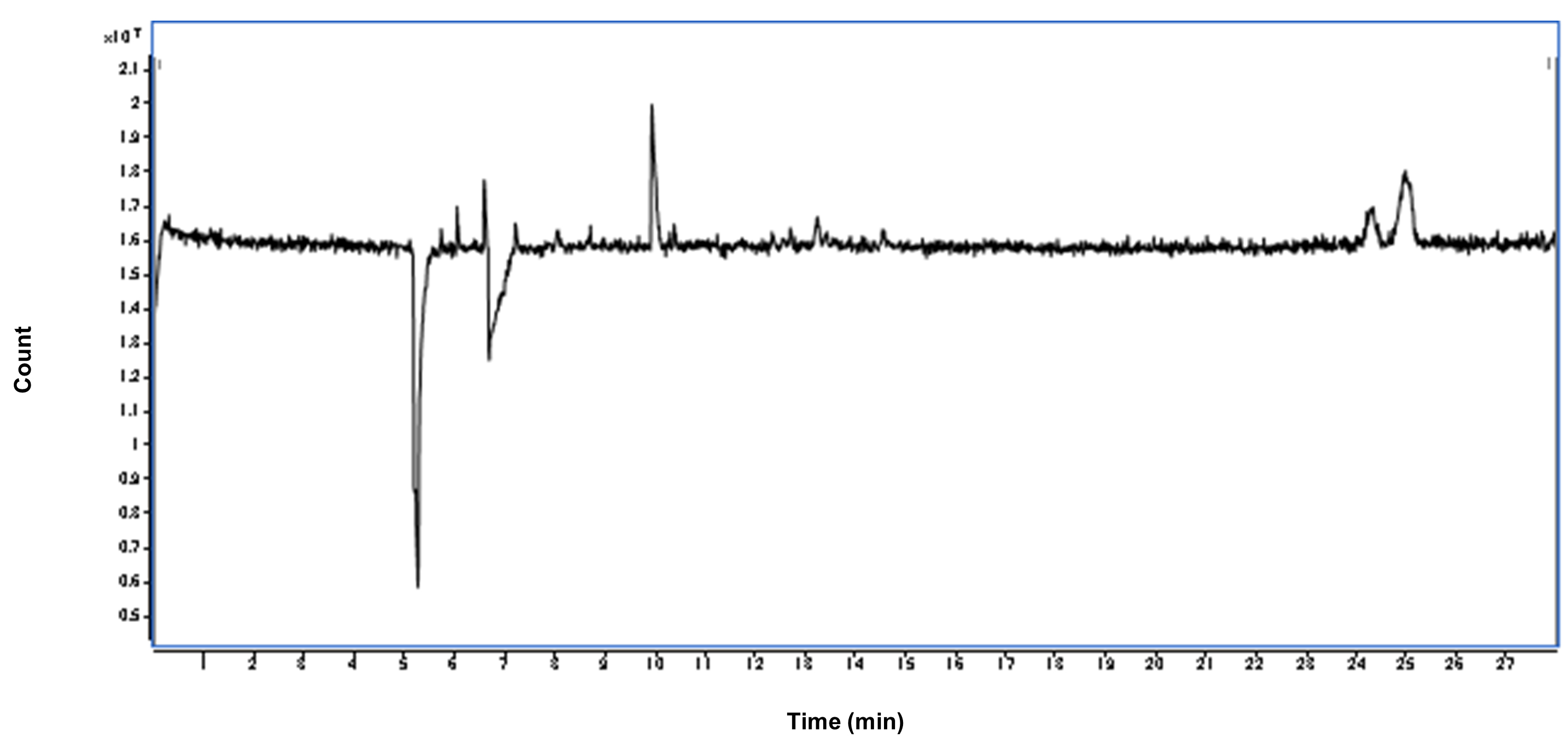
Figure 3. An example of the stable TIE of the cation measurement. The X-axis indicates the migration time. The Y-axis indicates the intensities of TIE, the sum of intensities over the m/z range in the observed data. Under the successful conditioning, this line becomes flat except for a few parts. Two significant drops of the TIE are observed at the left because of the existence of the inorganic cations, such as Na+ and K+. Two large peaks are observed of the existence of neutral molecules. The other small alternations are negligible and the flat of the other part indicates the stability of the conditioning.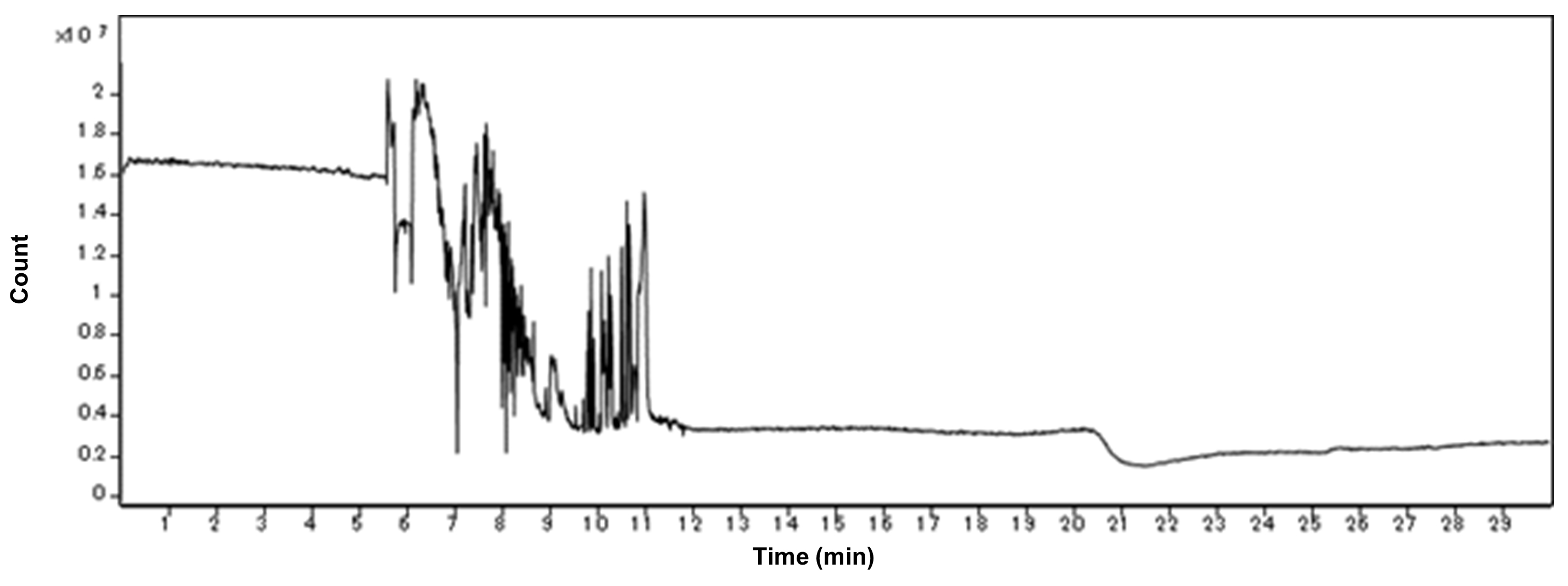
Figure 4. An example of the unstable TIE of the cation measurement. Compared to the TIE line in Figure 3, the line here fluctuates, which indicate the unsuccessful conditioning. Most of the cases, the adjustment of ESI sprayer is not suitable and the electric current fluctuates, which causes such TIE fluctuations. - Evaluate the stability of electric current during the measurement (Figures 5 and 6). Dilute the final solution and measure the sample again if the TIE is unstable.
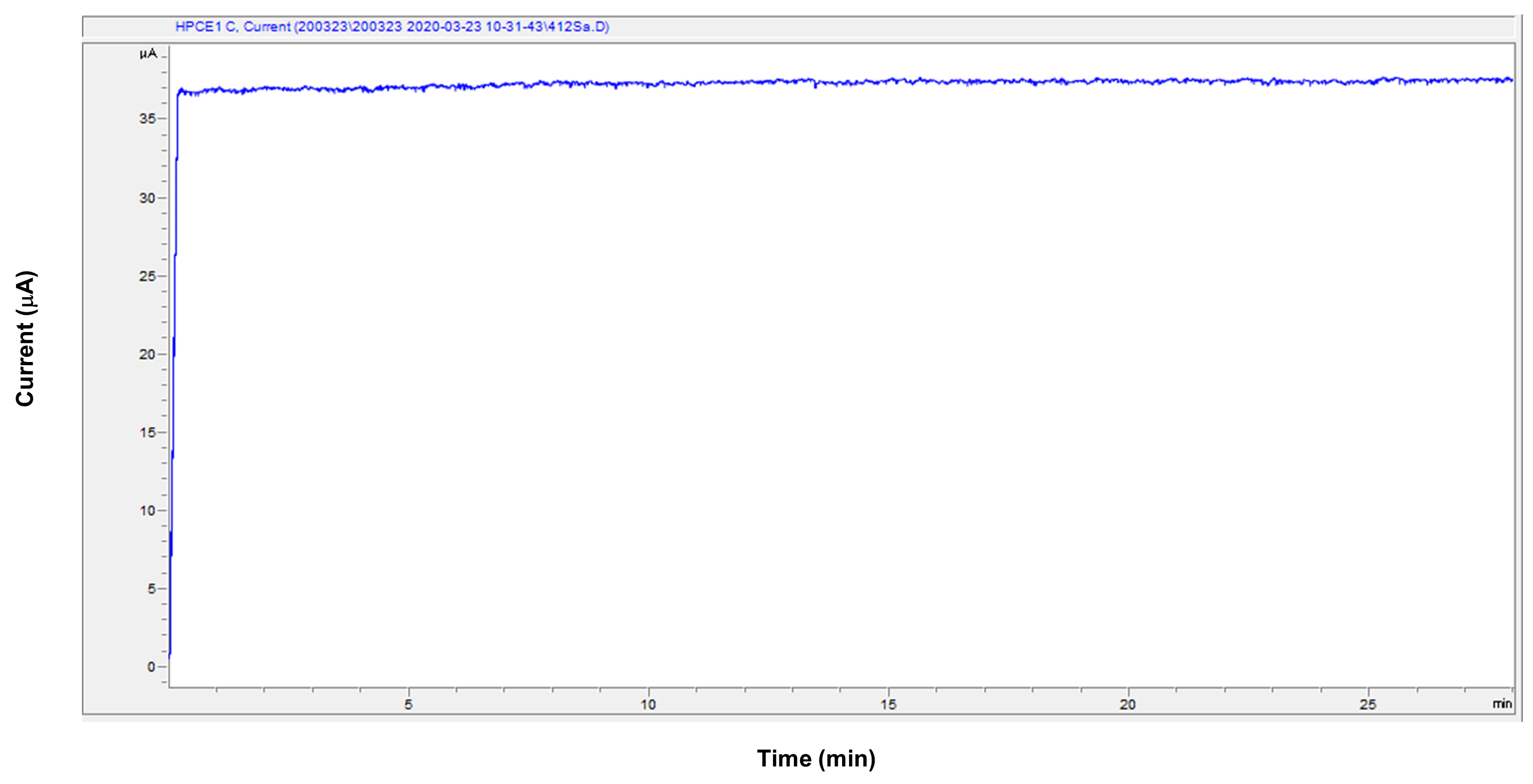
Figure 5. An example of the stable electric current of the cation measurement. The X-axis indicates the migration time. The Y-axis indicates the electric current during the measurement. Under the successful measurement, this line becomes flat except for the start of the measurement.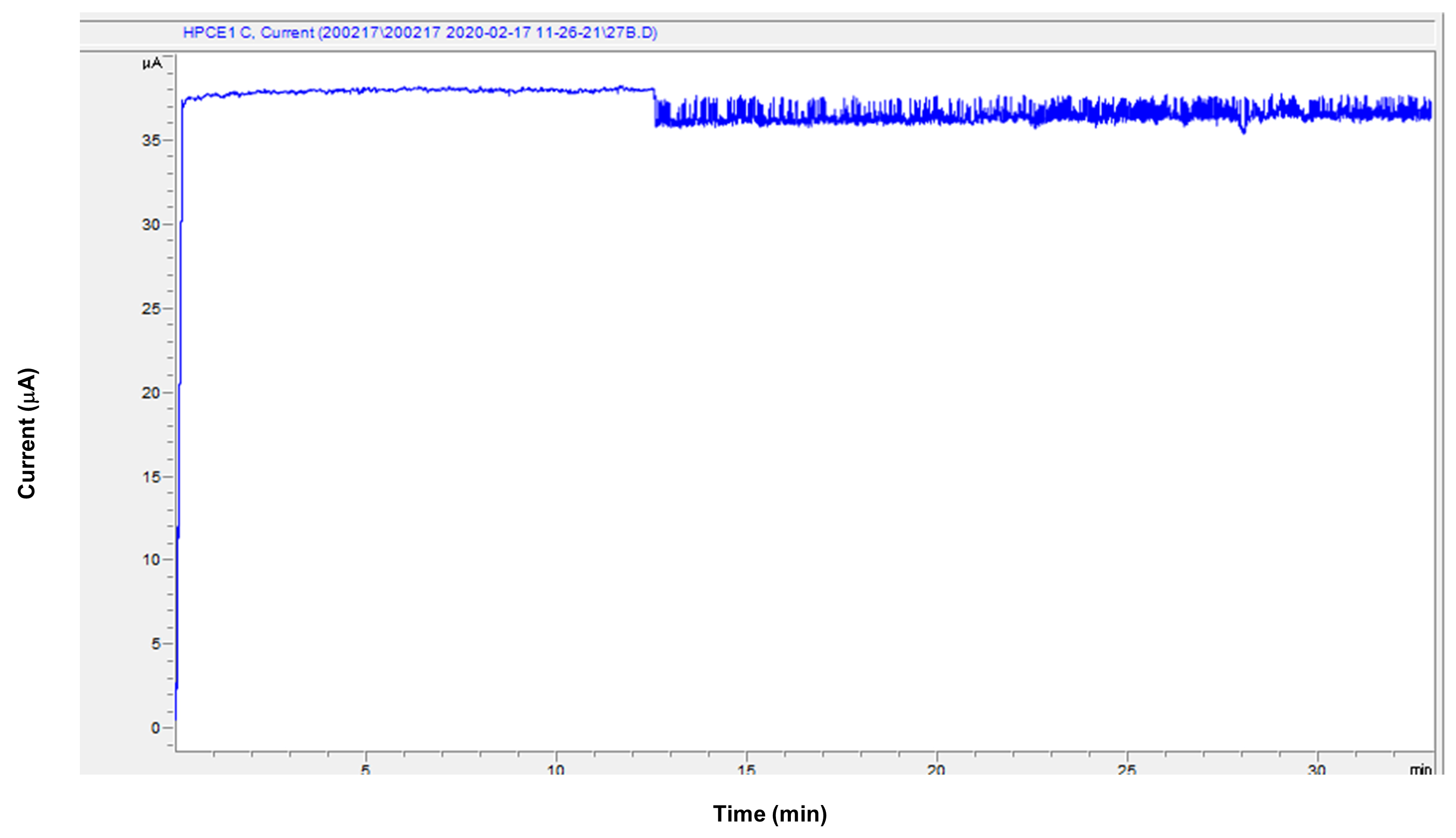
Figure 6. An example of the unstable electric current of the cation measurement. The X-axis indicates the migration time. The Y-axis indicates the electric current during the measurement. This example shows many spike peaks. When the condition of capillary becomes abnormal, the line doesn't stay flat. - Measure saliva samples.
- Sample preconditioning
- CE
Data analysis
Data analysis includes data processing and statistical analysis (Figure 7). The data processing includes data conversion and peak integration for each raw data. The process data were aligned and a data matrix is yielded after filling missing peaks (Sugimoto et al., 2012). This matrix is used for the subsequent data analysis, i.e., statistical analysis.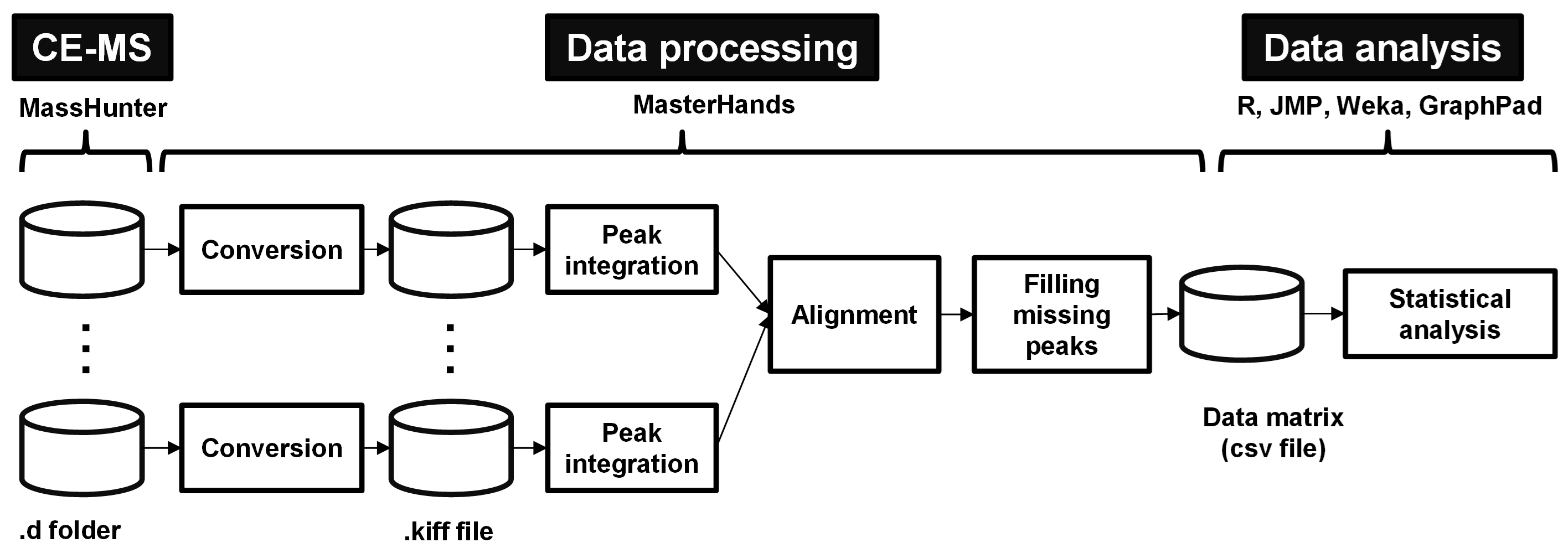
Figure 7. Overview of data processing and data analysis
- For each sample, a data folder (.d extension) is yielded by MassHunter, a vendor-supplied software for CE-MS controls. Data folders of the standard mixture, quality control, and saliva samples are prepared.
- Each data folder is converted to another format file (.kiff extension) using a data conversion function of MasterHands (Sugimoto et al., 2012)
- Subsequent data processing will be conducted using MasterHands
- Detect and integrate peaks with default options of all data files; signal/noise (S/N) radio > 2.0 for peak detection and 50% of the depth between two peaks for peak separation (see Note 2).
- Run alignment function to correct and matching of the peaks across all samples is conducted with default options. If the alignment function is not stable (Figure 8), rerun the alignment with the different gap penalty option (Sugimoto et al., 2010b).
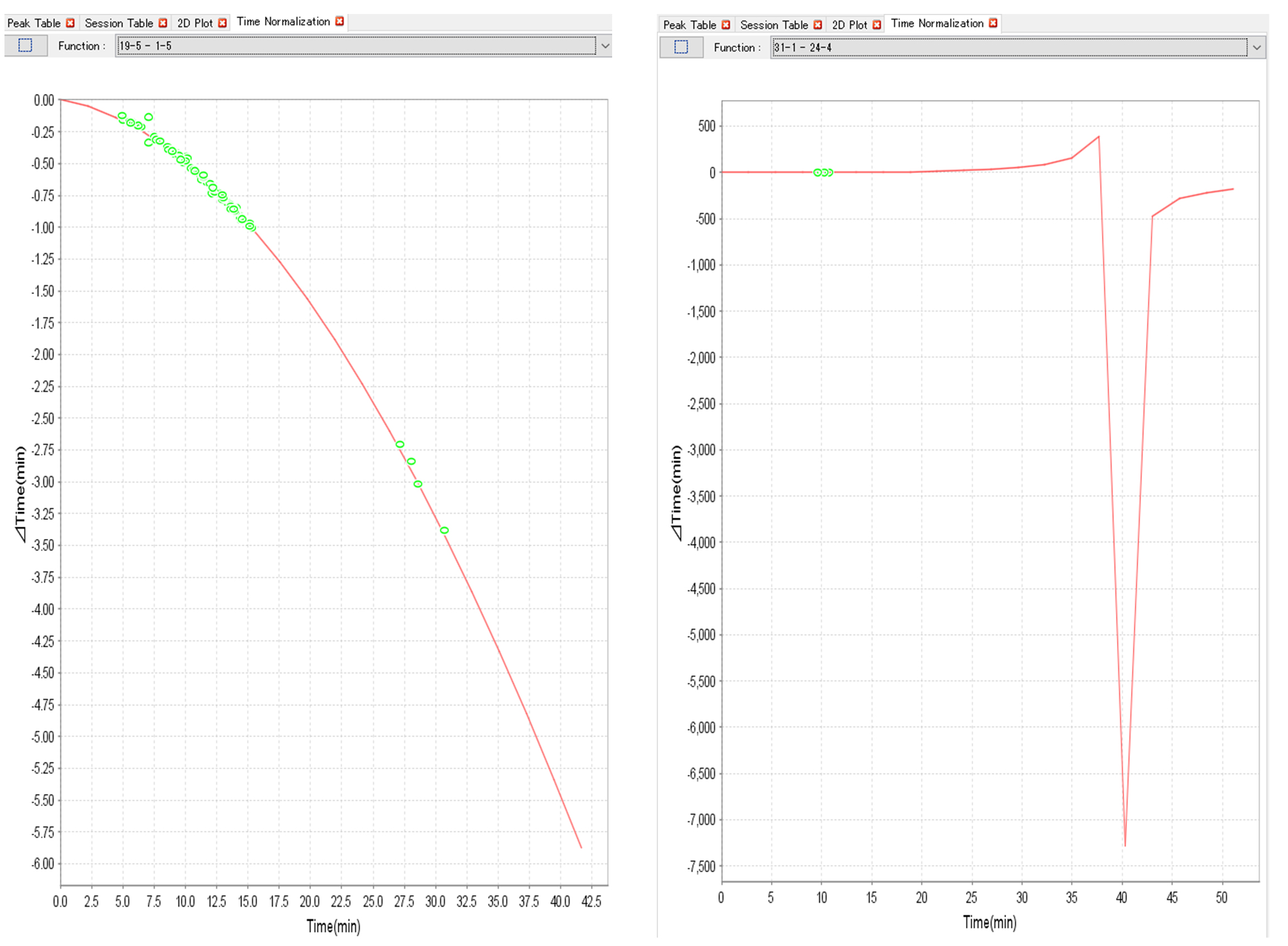
Figure 8. Examples of alignment functions. Stable (left) and unstable (right) cases. - Evaluate the overlaid peaks for internal standards. If the overlaid peaks are not observed, rerun the alignment with the different options (Figure 9).
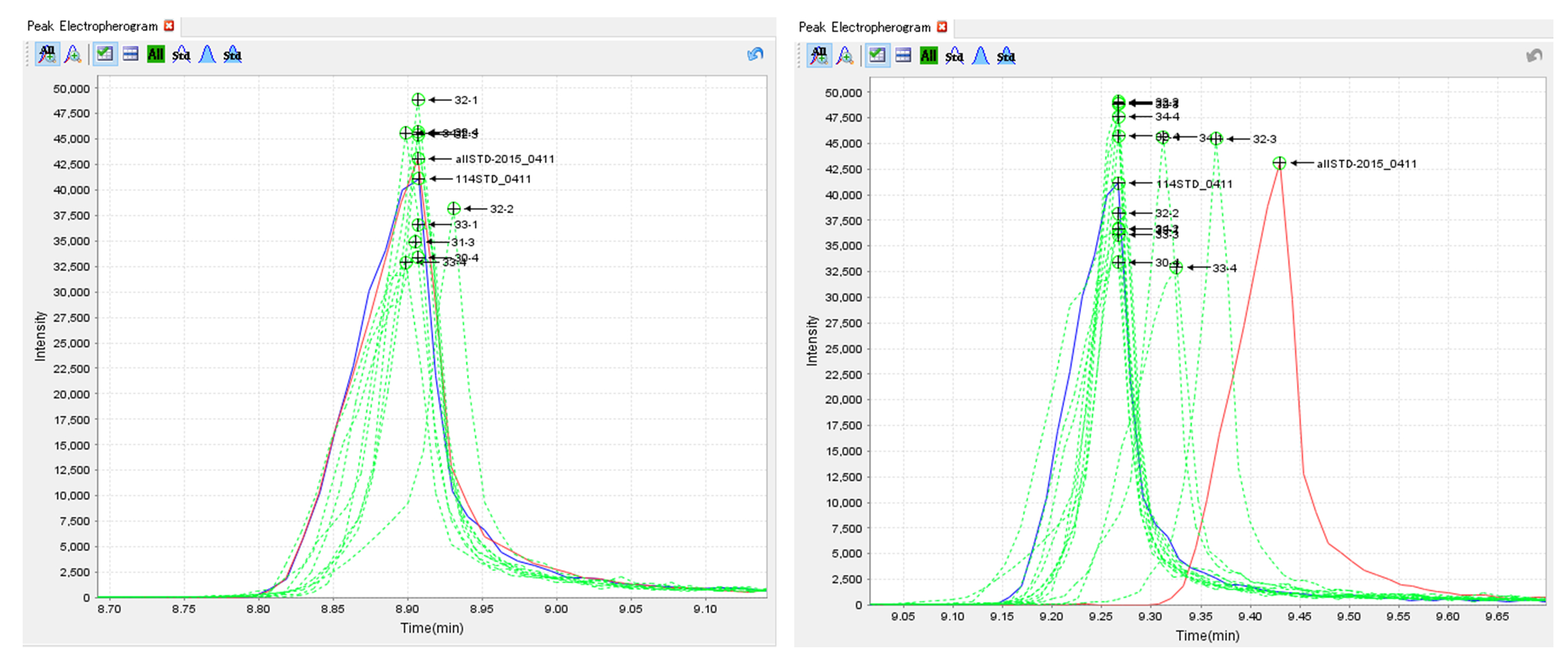
Figure 9. Examples of overlaid peaks. Successful (left) and unsuccessful (right) results. - Run reintegration function to filling missing peaks based on the aligned peak matrix.
- Exchange the peak area less than the lower limit of quantification to 0.
- The peak area of all peaks will be generated and these peak areas are divided by that of internal standard (methionine sulfone for cation and CSA for anion).
- Based on the corrected migration times and m/z values among standard mixture and samples, the metabolite names will be assigned.
- Export the peak matrix as a CSV file.
- Calculate the absolute concentration of the salivary metabolite concentrations.
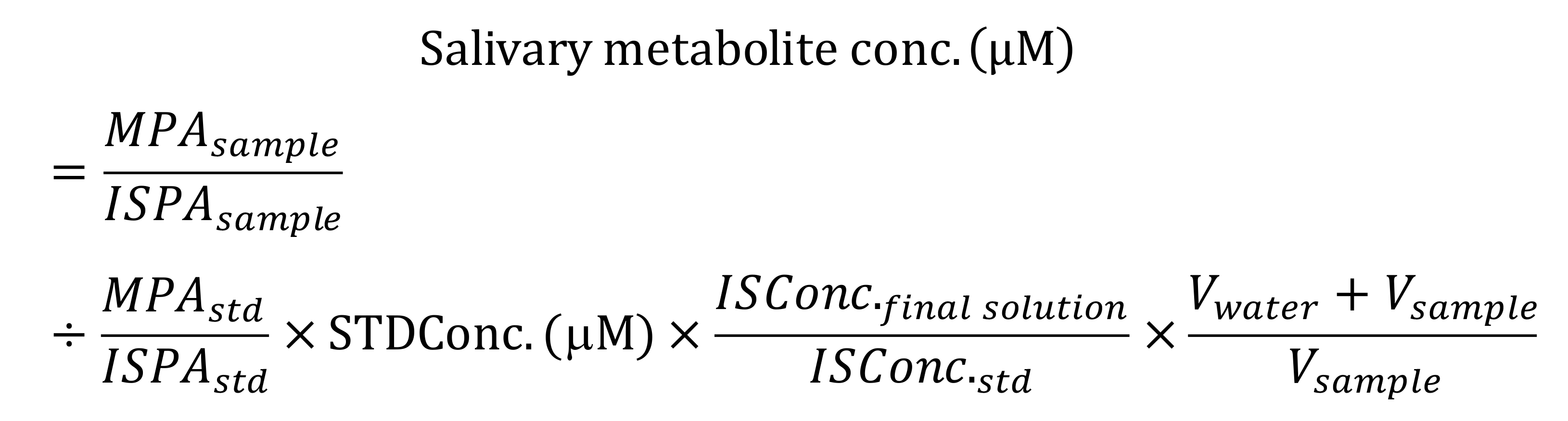
where, M, IS, STD, PA, V indicates metabolite, internal standard, standard, peak area, and volume, respectively.
Statistical analysis
- Conduct principal component analysis (PCA) using JMP to visualize the overall similarity and discrepancy of the metabolite concentration patterns among salivary samples (see Note 3).
- Conduct the Mann–Whitney test using R to access the difference of metabolite concentrations between 2 groups. The R function of wilcox.test() with correct = F options is used (see Note 4).
- Correct P-values using R using false discovery rate (Benjamini and Hochberg methods), considering multiple independent tests. The adjust() method of R vector with method = “fdr” options is used.
- Select only metabolite showing corrected P < 0.05 and fold change (F.C.) > 4.0 of the averaged concentrations between pancreatic patients (PC) and non-PC groups (healthy controls with chronic pancreatitis).
- Select the minimum metabolite set for MLR using JMP. Select the option of mixed selection (the combination of backward and forward selections with P < 0.05) to eliminate multicollinearity. Select [analysis]-[fit model menu] on JMP, and set stepwise options for personality. Select the “P-value Threshold” for stopping rule and select “mixed” for the direction. Input 0.05 for the threshold and press the “Go” button.
- Develop a multiple logistic regression (MLR) model to discriminate patients from non-PC and calculate an area under the receiver operating characteristic curves (AUC). Press the “Run model” button.
- Evaluate the generalization ability of the MLR model using Weka. Run k-fold cross-validations (CVs) to evaluate the generalization ability of the developed MLR. Repeat a CV test with 200 random values and k = 5, 10, and 20 (totally, 600 tests). Run weka.classifiers.functions.Logistic class of weka package with –x k (k is replaced to numerical values).
- Correct AUC values to calculate its mean and 95% confidential intervals. Select [Analyze Data]-[ROC Curve] menu using GraphPad.
Notes
- The use of polypropylene should be evaluated beforehand. Using the presented protocol, we confirmed that no effect of eluted compounds on the quantification quality of the saliva metabolites. However, this confirmation is limited to our standard library. Various condition of storage and solvent was compared (Tomita et al., 2018). Based on the observed fluctuation of these data, we computationally produced noise on the quantification level of the salivary metabolite and evaluated the effect of salivary biomarkers (Asai et al., 2018) on their discrimination abilities. Based on these data, standardization of operating procedures should be defined.
- Calibration ranges for each metabolite should be defined using a standard mixture before the analysis of saliva samples. In our protocol, we confirmed that the metabolites in our standard library showed high linearity between the peak area and the concentration up to 1 mM. Based on these data, the upper quantification range was defined 500 M, and the corresponding peak area was calculated beforehand. The peak areas showing larger than this threshold are considered as saturated peaks. The peaks showing S/N < 1.5 are considered as lower quantification limit.
- For PCA analyses, only frequently observed metabolite data should be used. We used metabolites detected at least 50% per group (Asai et al., 2018), while this threshold depends on the number of samples and the number of metabolites.
- The comparison of multiple groups depends on the study design. The salivary biomarker discovery study to detect pancreatic cancer included control (C), pancreatic cancer (PC), and chronic pancreatitis (CP) (Asai et al., 2018). Usually, a Kruskal-Wallis test should be used for the comparison of three groups. Here, we aimed to find metabolite showing high concentration only in a PC group. Therefore, we treated C and CP as one group and used a Mann-Whitney test to compare PC vs (C + CP) groups.
Recipes
- Internal standards mixture
2 mM of Methionine sulfone
2 mM of MES
2 mM of CSA
2 mM of 3-Aminopyrrolidine
2 mM of Trimesate - Sheath liquid
Cation: Methanol/water (50% v/v) containing 0.1 μM Hexakis (2,2-difluorothoxy) phosphazene
Anion: Ammonium acetate (5 mmol/L) in 50% methanol/water (50% v/v) containing 0.1 μM Hexakis (2,2-difluorothoxy) phosphazene - Run buffer
Cation: 1 M of Formic acid
Anion: 50 mM Ammonium acetate (pH 3.4 and pH 8.5) - Standard mixture
Cation: 200 μM of internal standards and 20 μM of each metabolite
Anion: 200 μM of internal standards and 50 μM of each metabolite
Acknowledgments
This work was supported by a grant from the Program on Open Innovation Platform with Enterprises, Research Institute and Academia (OPERA, JPMJOP1842), and grants from Tsuruoka City and Yamagata Prefecture.
Competing interests
There are no competing interests to be declared.
Ethics
This study was approved by the ethics committee of Tokyo Medical University (approval no. 1560, 30 September 2010). Written informed consent was obtained from all patients and from volunteers who agreed to serve as saliva donors. Our study was carried out following the Helsinki Declaration.
References
- Armitage, E. G. and Ciborowski, M. (2017). Applications of metabolomics in cancer studies. Adv Exp Med Biol 965: 209-234.
- Asai, Y., Itoi, T., Sugimoto, M., Sofuni, A., Tsuchiya, T., Tanaka, R., Tonozuka, R., Honjo, M., Mukai, S., Fujita, M., Yamamoto, K., Matsunami, Y., Kurosawa, T., Nagakawa, Y., Kaneko, M., Ota, S., Kawachi, S., Shimazu, M., Soga, T., Tomita, M. and Sunamura, M. (2018). Elevated polyamines in saliva of pancreatic cancer. Cancers (Basel) 10(2).
- Bonne, N. J. and Wong, D. T. (2012). Salivary biomarker development using genomic, proteomic and metabolomic approaches. Genome Med 4(10): 82.
- Chattopadhyay, I. and Panda, M. (2019). Recent trends of saliva omics biomarkers for the diagnosis and treatment of oral cancer. J Oral Biosci 61(2): 84-94.
- Dawes, C. and Wong, D. T. W. (2019). Role of saliva and salivary diagnostics in the advancement of oral health. J Dent Res 98(2): 133-141.
- Ishikawa, S., Sugimoto, M., Edamatsu, K., Sugano, A., Kitabatake, K. and Iino, M. (2020). Discrimination of oral squamous cell carcinoma from oral lichen planus by salivary metabolomics. Oral Dis 26(1): 35-42.
- Ishikawa, S., Sugimoto, M., Kitabatake, K., Sugano, A., Nakamura, M., Kaneko, M., Ota, S., Hiwatari, K., Enomoto, A., Soga, T., Tomita, M. and Iino, M. (2016). Identification of salivary metabolomic biomarkers for oral cancer screening. Sci Rep 6: 31520.
- Ishikawa, S., Sugimoto, M., Kitabatake, K., Tu, M., Sugano, A., Yamamori, I., Iba, A., Yusa, K., Kaneko, M., Ota, S., Hiwatari, K., Enomoto, A., Masaru, T. and Iino, M. (2017). Effect of timing of collection of salivary metabolomic biomarkers on oral cancer detection. Amino Acids 49(4): 761-770.
- Kaczor-Urbanowicz, K. E., Martin Carreras-Presas, C., Aro, K., Tu, M., Garcia-Godoy, F. and Wong, D. T. (2017). Saliva diagnostics - Current views and directions. Exp Biol Med (Maywood) 242(5): 459-472.
- Martina, E., Campanati, A., Diotallevi, F. and Offidani, A. (2020). Saliva and oral diseases. J Clin Med 9(2).
- Mikkonen, J. J., Singh, S. P., Herrala, M., Lappalainen, R., Myllymaa, S. and Kullaa, A. M. (2016). Salivary metabolomics in the diagnosis of oral cancer and periodontal diseases. J Periodontal Res 51(4): 431-437.
- Monton, M., R., and Soga, T. (2007). Metabolome analysis by capillary electrophoresis-mass spectrometry. J Chromatogr A 168(1-2): 237-246.
- Murata, T., Yanagisawa, T., Kurihara, T., Kaneko, M., Ota, S., Enomoto, A., Tomita, M., Sugimoto, M., Sunamura, M., Hayashida, T., Kitagawa, Y. and Jinno, H. (2019). Salivary metabolomics with alternative decision tree-based machine learning methods for breast cancer discrimination. Breast Cancer Res Treat 177(3): 591-601.
- Soga, T., Ohashi, Y., Ueno, Y., Naraoka, H., Tomita, M., and Nishioka, T. (2003). Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J Proteome Res 2(5): 488-494.
- Soga, T. (2007). Capillary electrophoresis-mass spectrometry for metabolomics. Methods Mol Biol 358:129-137.
- Sridharan, G., Ramani, P., Patankar, S. and Vijayaraghavan, R. (2019). Evaluation of salivary metabolomics in oral leukoplakia and oral squamous cell carcinoma. J Oral Pathol Med 48(4): 299-306.
- Sugimoto, M., Wong, D. T., Hirayama, A., Soga, T. and Tomita, M. (2010a). Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 6(1): 78-95.
- Sugimoto, M., Kawakami, M., Robert, M., Soga, T., Tomita, and M. (2012). Bioinformatics tools for mass spectroscopy-based metabolomic data processing and analysis. Curr Bioinform 7(1): 96-108.
- Sugimoto, M., Saruta, J., Matsuki, C., To, M., Onuma, H., Kaneko, M., Soga, T., Tomita, M. and Tsukinoki, K. (2013). Physiological and environmental parameters associated with mass spectrometry-based salivary metabolomic profiles. Metabolomics 9(2): 454-463.
- Sugimoto, M., Hirayama, A., Ishikawa, T., Robert, M., Baran, R., Uehara, K., Kawai, K., Soga, T. and Tomita, M. (2010b). Differential metabolomics software for capillary electrophoresis-mass spectrometry data analysis. Metabolomics 6(1): 27-41.
- Takayama, T., Tsutsui, H., Shimizu, I., Toyama, T., Yoshimoto, N., Endo, Y., Inoue, K., Todoroki, K., Min, J. Z., Mizuno, H. and Toyo'oka, T. (2016). Diagnostic approach to breast cancer patients based on target metabolomics in saliva by liquid chromatography with tandem mass spectrometry. Clin Chim Acta 452: 18-26.
- Tomita, A., Mori, M., Hiwatari, K., Yamaguchi, E., Itoi, T., Sunamura, M., Soga, T., Tomita, M,, Sugimoto, M. (2018). Effect of storage conditions on salivary polyamines quantified via liquid chromatography-mass spectrometry. Sci Rep 8(1):12075.
- Trezzi, J. P., Vlassis, N. and Hiller, K. (2015). The role of metabolomics in the study of cancer biomarkers and in the development of diagnostic tools. Adv Exp Med Biol 867: 41-57.
- Wang, Q., Gao, P., Wang, X. and Duan, Y. (2014). Investigation and identification of potential biomarkers in human saliva for the early diagnosis of oral squamous cell carcinoma. Clin Chim Acta 427: 79-85.
- Wang, X., Kaczor-Urbanowicz, K. E. and Wong, D. T. (2017). Salivary biomarkers in cancer detection. Med Oncol 34(1): 7.
- Warburg, O. (1956). On respiratory impairment in cancer cells. Science 124(3215): 269-270.
- Washio, J. and Takahashi, N. (2016). Metabolomic studies of oral biofilm, oral cancer, and beyond. Int J Mol Sci 17(6).
- Yoshizawa, J. M., Schafer, C. A., Schafer, J. J., Farrell, J. J., Paster, B. J. and Wong, D. T. (2013). Salivary biomarkers: toward future clinical and diagnostic utilities. Clin Microbiol Rev 26(4): 781-791.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sugimoto, M., Ota, S., Kaneko, M., Enomoto, A. and Soga, T. (2020). Quantification of Salivary Charged Metabolites Using Capillary Electrophoresis Time-of-flight-mass Spectrometry. Bio-protocol 10(20): e3797. DOI: 10.21769/BioProtoc.3797.
Category
Cancer Biology > Cancer biochemistry > Cancer metabolism
Biochemistry > Other compound > Ion
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










