- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Staining and Quantitative Analysis of Myelinating Oligodendrocytes in the Mouse Grey Matter
Published: Vol 10, Iss 20, Oct 20, 2020 DOI: 10.21769/BioProtoc.3792 Views: 4660
Reviewed by: Ehsan KheradpezhouhMiao HeNarayan Subramanian

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Systematic Analysis of Smooth Muscle and Cartilage Ring Formation during Mouse Tracheal Tubulogenesis
Haoyu Wu [...] Wenguang Yin
Jul 5, 2023 1534 Views

Protocol for the Implantation of Scaffolds in a Humanized Mouse Cutaneous Excisional Wound Healing Model
Dina Gadalla [...] David G. Lott
Sep 20, 2024 1451 Views

Dissection and Whole-Mount Immunofluorescent Staining of Mouse Hind Paw Muscles for Neuromuscular Junction Analysis
Rebecca L. Simkin [...] James N. Sleigh
May 20, 2025 3909 Views
Abstract
Oligodendrocytes generate distinct patterns of myelination throughout the CNS. Variations in myelination along axons may enable neurons to fine-tune conduction velocities and alter signal synchronisation. Here we outline a staining protocol permitting the assessment of the number and length of myelin sheaths formed by oligodendrocyte in the mouse grey matter. This protocol enables the investigation of myelination without the need for reporter mice or technically challenging protocols, aiding the investigation of factors influencing myelin production in the brain.
Keywords: OligodendrocyteBackground
Oligodendrocytes generate the insulating myelin sheaths around axons in the central nervous system (CNS). Myelin sheaths accelerate axonal conduction velocities through the concentration of voltage-dependent sodium channels at the small unmyelinated gaps between sheaths –the nodes of Ranvier (Huxley and Stampfli, 1949; Rushton, 1951; Waxman, 1997). Although oligodendrocytes are located throughout the CNS, not all axons are myelinated suggesting that the formation and size of myelin sheaths may precisely permit the fine-tuning of action potential speeds and neuronal synchronisation (Pajevic et al., 2014). Understanding what influences the amount of myelin generated by oligodendrocytes (i.e., the number and size of sheaths formed by one cell) is therefore important for understanding how myelin alters neuronal function.
A number of techniques have been developed to analyze the complex morphologies of oligodendrocytes in vivo. Initially, the studies of Pio del Rio-Hortega used silver carbonate staining to identify and distinguish oligodendrocytes based on the number and length of the myelin sheaths formed (Reviewed by Perez-Cerda et al., 2015). Common methods now involve the generation and breeding of transgenic mice expressing fluorescent reporters downstream of oligodendrocyte-specific gene promoters (Rivers et al., 2008; Chong et al., 2012; Young et al., 2013; Hill et al., 2018; Hughes et al., 2018). Further, oligodendrocyte morphologies can be labelled through infection with viruses harbouring genes encoding fluorescent proteins (Osanai et al., 2017). While, these strategies enable the visualisation of oligodendrocytes throughout the CNS, the generation of transgenic reporter mice, in combination with genetic knockouts, can be a very time-consuming and costly process, while viral labelling is a technically challenging procedure and invasive.
To circumvent the challenges of existing techniques, we developed a system for the immunofluorescence staining and analysis of the detailed morphology of individual oligodendrocytes in the mouse grey matter. Targeting one of the most abundant mature oligodendrocyte proteins, CNPase, enables labelling of the entire oligodendrocyte: cell soma, fine processes and myelin sheaths. This allows the complex individual morphologies of oligodendrocytes to be traced in the sparsely myelinated grey matter of the mouse cortex enabling the assessment of myelin sheath number and length. This method can overcome the need to generate transgenic reporter mice and specialised labelling of oligodendrocytes, allowing the assessment of myelination following various interventions such as in genetic knockout studies.
Materials and Reagents
- 7 ml Bijou tubes (Greiner Bio-One Inc., catalog number: 189170 )
- Tinfoil
- Double Edge Razor Blades FEATHER® Brand (Electron Microscopy Sciences, catalog number: 72002-01 )
- Small paintbrush (e.g., Electron Microscopy Sciences, catalog number: 66100-00 )
- 24-well plates
- 1.5 ml Eppendorf tubes
- Superfrost plus slides (VWR, catalog number: 631-0108 )
- Glass coverslips (VWR, catalog number: 631-0146 )
- Mice
Note: Oligodendrocytes are continuously generated in the mouse brain and therefore add new myelin to the sparsely myelinated upper cortical layers. Identification and analysis of individual oligodendrocytes therefore becomes more challenging in older animals due to the increase in myelin. Postnatal days 21, 35 and 90 have been used to confidently assess myelination. - Superglue
- Phosphate buffered saline (PBS)
- 4% Paraformaldehyde (PFA) in PBS
- Sodium azide
- Low gelling temperature agarose (Sigma-Aldrich, catalog number: A9414 )
- Embedding peel-a-way mould (Polysciences, catalog number: 18986-1 )
- Tri-sodium citrate (Sigma-Aldrich, catalog number: S4641 )
- Tween 20 (Sigma-Aldrich, catalog number: P1379 )
- Goat serum (Sigma-Aldrich, catalog number: G6767 )
- Triton X-100 (Sigma-Aldrich, catalog number: X100 )
- CNPase primary antibody (Atlas Antibodies, catalog number: AMAb91072 )
- Alexa fluorophore-conjugated secondary mouse antibody
- Nuclear marker, e.g., Hoechst (Thermo Fischer Scientific, catalog number: 62249 )
- Fluoromount-G (Cambridge Bioscience, catalog number: 0100-01 )
- Other tested antibodies:
- 2% Low gelling temperature agarose in H2O (see Recipes)
- Antigen retrieval buffer (see Recipes)
- Blocking buffer (see Recipes)
Equipment
- Dissection tools required for standard mouse perfusion, e.g.,
Straight iris scissors (Fine Science Tools, catalog number: 14058-11 )
Standard tweezers (Fine Science Tools, catalog number: 11027-12 )
Student Vannas Spring Scissors (Fine Science Tools, catalog number: 91501-09) - Microwave
- Vibratome (Leica Biosystems)
- Heat block
- Confocal microscope
Software
- ImageJ, Fiji (https://imagej.net/Fiji)
- Simple neurite tracer (Longair et al., 2011) (Included with Fiji, https://imagej.net/Simple_Neurite_Tracer)
- Microsoft Excel (https://products.office.com/en-gb/excel)
Procedure
- Preparation of Tissue
- Prepare a 4% PFA solution in PBS.
- Fix mouse tissue by cardiac perfusion with 4% PFA solution.
- Carefully dissect out the brain and transfer to a 7 ml bijou.
- Post-fix in 4% PFA overnight at 4 °C.
- Wash the brain in PBS 3 times for 10 min each
Note: Brains can be stored in PBS at 4 °C for a period of weeks. For long term storage at 4 °C 0.01% sodium azide should be added.
- Vibratome Cutting
- Prepare a 2% agarose solution in H2O.
- Heat the 2% agarose solution in a microwave until in solution and allow to cool until bottle can be held comfortably while the solution is still liquid.
- Position the brain in an embedding peel-a-way mould or homemade mould made from tinfoil (see Figure 1A).
- Pour the cooled agarose solution over the brain and reposition.
- Leave for at least 5 min until the agarose solution has set.
- Trim down excess agarose using a blade and glue the brain in agarose onto the vibratome specimen holder positioned so that the angle of the vibratome blade will cut flat coronal sections (see Figures 1B-1C).
- Secure the specimen holder onto the vibratome (see Figure 1C).
- Fill the buffer tray with PBS until the agarose is covered.
- Cut 100 µm coronal sections of the desired brain regions using a speed of 5 and frequency of 7.
- Carefully collect sections with a paintbrush and transfer to the wells of 24-well plate containing PBS (see Figure 1D).
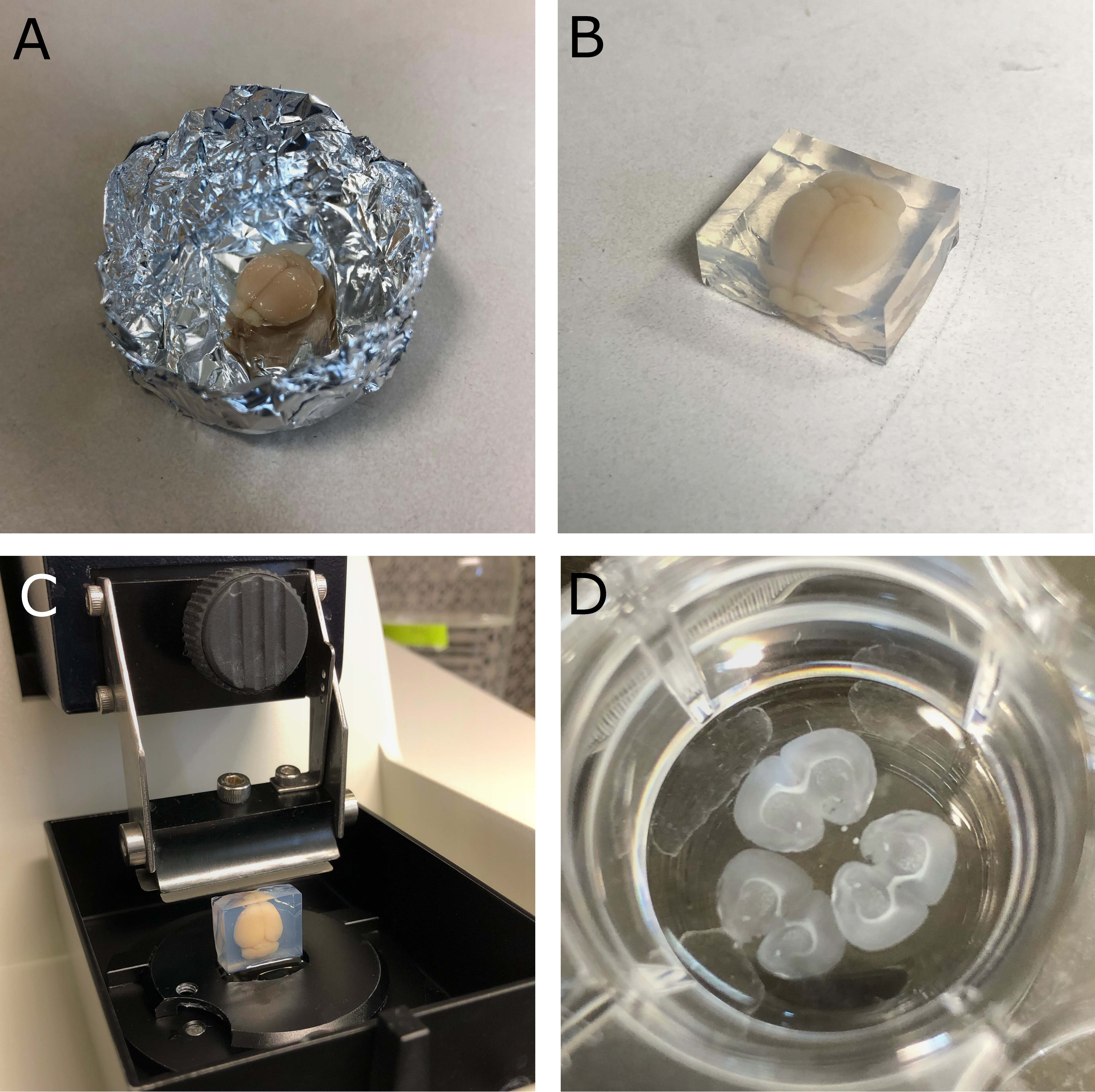
Figure 1. Cutting coronal mouse brain sections. A. Mouse brain placed in a homemade foil mould. B. Mouse brain embedded in 2% agarose with excess cut off. C. Embedded brain glued to specimen holder and secured in place on the vibratome. D. 100µm coronal brain sections in a 24-well plate.
- Immunofluorescence
- Using a paintbrush carefully transfer the brains sections to Eppendorf tubes contacting 1 ml of antigen retrieval buffer (10 mM citrate, 0.05% Tween, pH 6).
- Transfer tubes to a preheated heat block set at 95 °C for 20 min.
- Carefully remove sections from the tubes and transfer back to PBS in a 24-well plate.
- Using a pastette or P100 pipette carefully remove the PBS and add 250 µl blocking buffer (10% goat serum, 0.25% triton, in PBS) to each well. Leave for 2 h at room temperature on a shaker.
- Remove blocking buffer and add primary CNPase antibody (1:2,000) in blocking buffer for 36-48 h at 4 °C on a shaker.
Note: Other primary antibodies can be used in conjunction with CNPase, see materials and reagents section for suggestions. - Remove primary antibody and wash sections with PBS 3 x 1 h at room temperature on a shaker.
- Remove PBS and add Alexa fluorophore-conjugated mouse secondary antibody (1:1,000) in blocking buffer overnight at 4 °C on a shaker.
- Remove secondary antibodies and wash sections in PBS for 3 x 1 h at room temperature.
- Stain with a nuclear marker for 20 min at room temperature.
- Mount sections on to superfrost slides using fluoromount G and secure with cover glass.
- Analysis of Myelin Sheath Number and Length
- Using a confocal microscope at 63x magnification and a z step size of 0.5 µm, image individual CNPase positive oligodendrocytes in the sparsely myelinated layers II-III of the mouse cortex (Figure 2).
Note: Prior to imaging, slides and images should be suitably blinded, e.g., using the filename randomiser plugin on Fiji.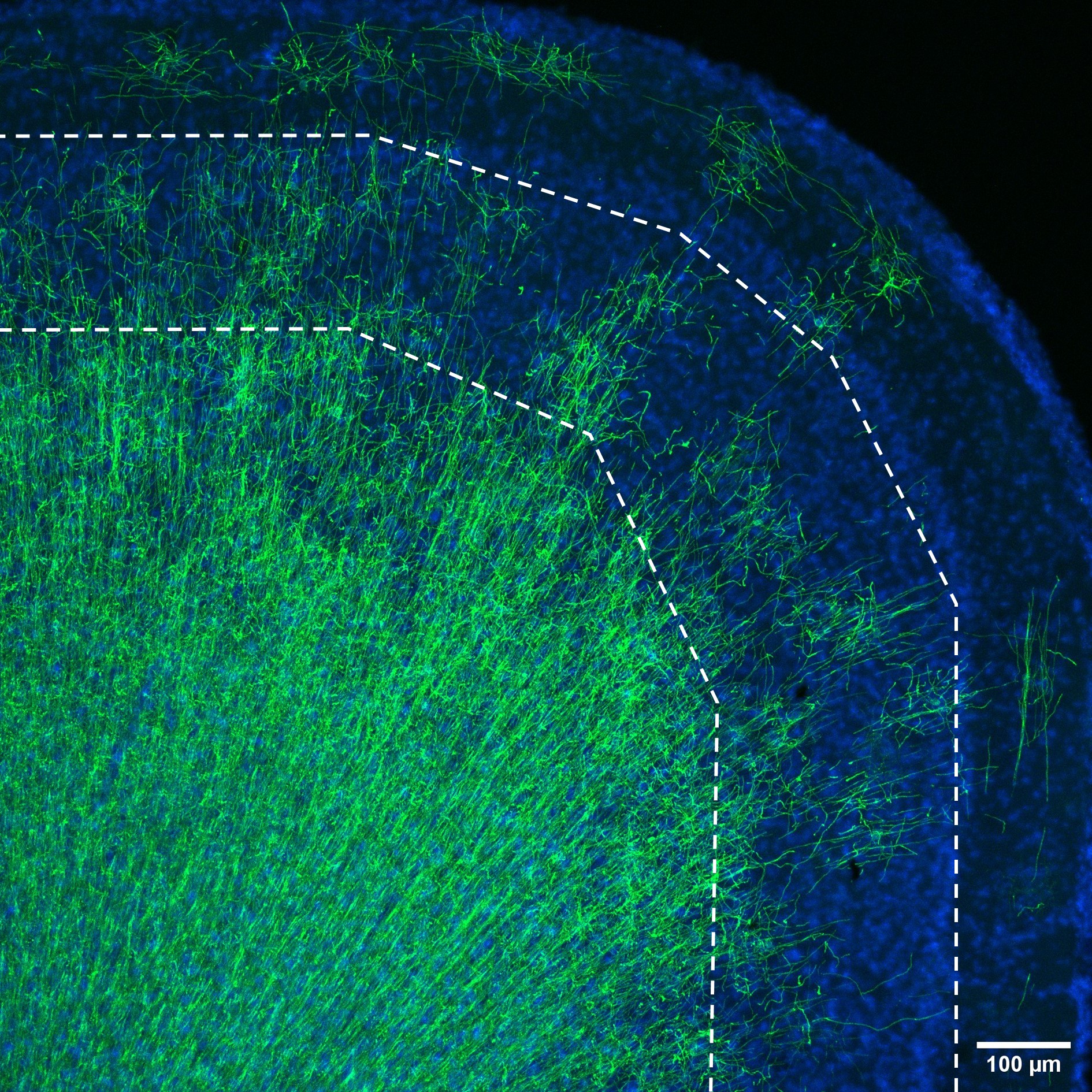
Figure 2. The sparsely myelinated upper layers of the mouse grey matter. Layers II-III and IV (dashed area) of the mouse cortex is sparsely myelinated. Due to this oligodendrocyte morphologies, the number and length of myelin sheaths generated, can be assessed. In the deeper layers of the cortex and white matter, the number of oligodendrocytes does not allow confident analysis of myelination. CNPase–green, Hoechst–blue. Scale bar = 100 µm. - Open ImageJ–Fiji. Select Plugins ➔ Segmentation ➔ Simple neurite tracer.
Note: Check here for more detailed instructions on how to use simple neurite tracer. - Using simple neurite tracer move through the z-stack and follow the thin processes connecting the cell body to the myelin sheaths–signal intensity is much brighter in myelin sheaths compared to the fine processes (Figure 3).
Note: Only analyze oligodendrocytes with all complete myelin sheaths present within the 100 µm section as assessed by following each process from the cell body and ensuring none exit the section. To achieve this aim to image cells in the middle of the 100 µm section.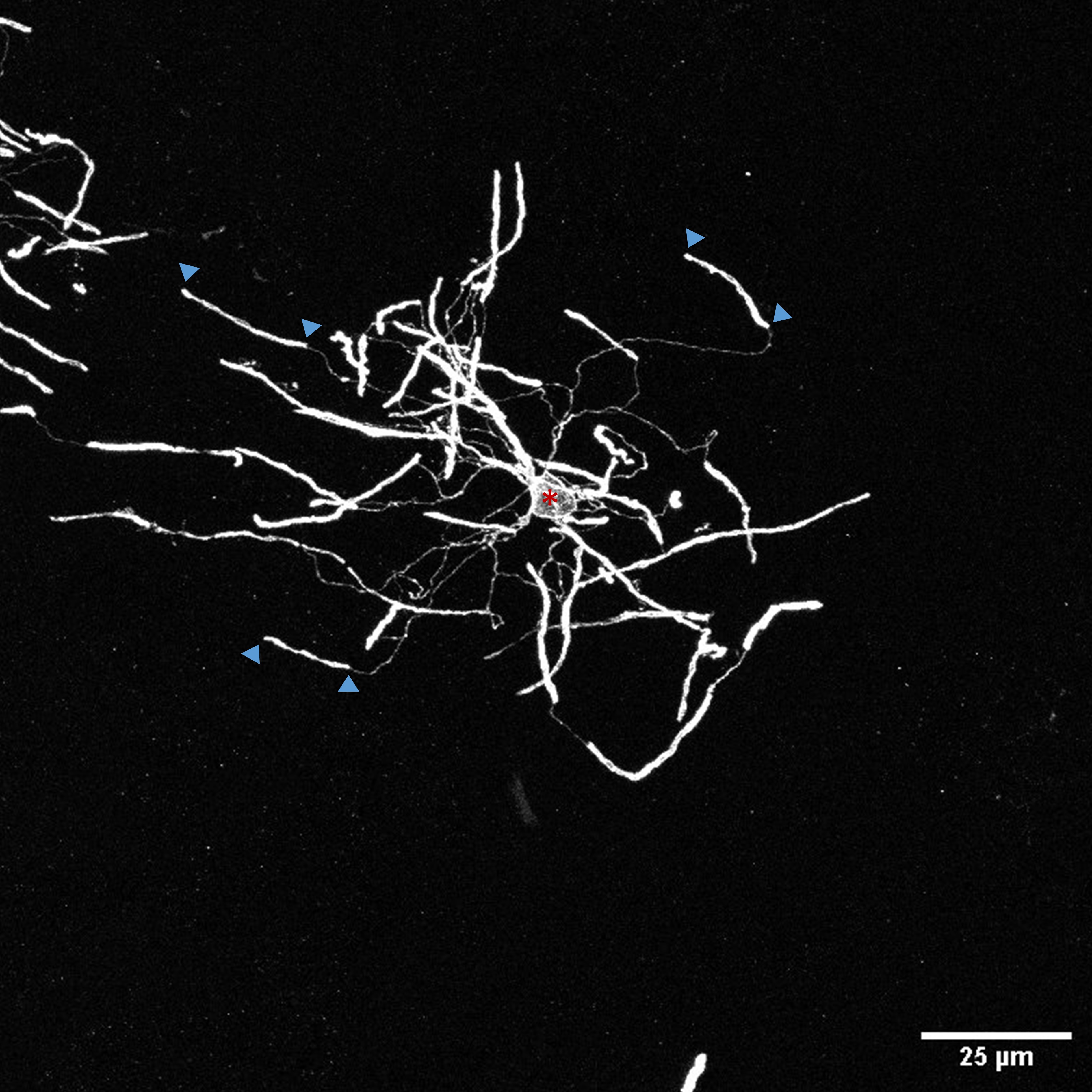
Figure 3. Thin CNPase positive processes connect cell body (asterisk) to myelin sheaths (arrowheads depicting start and end of example sheaths). - To measure a myelin sheath in 3-dimensions click at the start of a myelin sheath. The start of a sheath can be defined where bright sheath signal begins but not the end of the section or image. Move through the z-stack and follow the sheath. Click along the sheath and accept the path (y) if the sheath has traced correctly or cancel (n) if incorrect. At the end of a sheath select finish path (f). Ends of sheaths can be defined where signal ends but not the end of the section or image.
Note: Ends of myelin sheaths can also be confirmed with Caspr staining, see materials and methods for a Caspr antibody. - The length in microns of completed sheaths are recorded in the “All Paths” box.
- Return to the oligodendrocyte cell body and trace another process to a different sheath and measure as above.
- When all sheaths from one cell have been measured, select “File” from the tracing window, then “Export as CSV…”
- A Microsoft Excel file will be created containing the lengths of myelin sheaths measured.
Note: In wild type mice we have observed oligodendrocytes generate an average of 50 myelin sheaths per cell with a range from 25-70. Myelin sheaths length can range from 5 µm up to over 150 µm.
- Using a confocal microscope at 63x magnification and a z step size of 0.5 µm, image individual CNPase positive oligodendrocytes in the sparsely myelinated layers II-III of the mouse cortex (Figure 2).
Data analysis
- From the Excel file generated the lengths of myelin sheaths measured can be plotted and analyzed in various forms (e.g., Swire et al., 2019; Vasistha et al., 2019). For example, sheath lengths can be binned and a frequency distribution generated.
- By counting the number of sheaths measured from one individual oligodendrocyte the number of myelin sheaths generated per cell can be plotted.
- When data from suitable numbers of oligodendrocytes and cells have been analyzed, relevant statistical tests can be performed to identify changes to myelin sheaths number or length.
Recipes
- 2% Low gelling temperature agarose in H2O
2 g Low gelling temperature agarose
100 ml distilled H2O - Antigen retrieval buffer (10 mM citrate, 0.05% Tween, pH 6)
2.94 g Tri-sodium citrate
500 μl Tween-20
1 L H2O - Blocking buffer (10% goat serum, 0.25% triton, in PBS)
1 ml goat serum
25 μl Triton X-100
8.975 ml PBS
Acknowledgments
We would like to thank past and present members of the ffrench-Constant, Lyons, Williams and Miron labs for technical assistance and helpful discussions. This work was supported by a MS Society Research Grant PhD studentship (Grant Reference 950) and a Wellcome Trust Senior Investigator Award to CffC. Protocol adapted from Swire et al. (2019).
Competing interests
No conflicting interests.
Ethics
Animal experimentation: Animal husbandry and experiments were performed under UK Home Office project licenses issued under the Animals (Scientific Procedures) Act, under project license 70/8748. All animal experiments were reviewed, revised and approved by the University of Edinburgh Bioresearch & Veterinary Services team.
References
- Chong, S. Y., Rosenberg, S. S., Fancy, S. P., Zhao, C., Shen, Y. A., Hahn, A. T., McGee, A. W., Xu, X., Zheng, B., Zhang, L. I., Rowitch, D. H., Franklin, R. J., Lu, Q. R. and Chan, J. R. (2012). Neurite outgrowth inhibitor Nogo-A establishes spatial segregation and extent of oligodendrocyte myelination. Proc Natl Acad Sci U S A 109(4): 1299-1304.
- Hill, R. A., Li, A. M. and Grutzendler, J. (2018). Lifelong cortical myelin plasticity and age-related degeneration in the live mammalian brain. Nat Neurosci 21(5): 683-695.
- Hughes, E. G., Orthmann-Murphy, J. L., Langseth, A. J. and Bergles, D. E. (2018). Myelin remodeling through experience-dependent oligodendrogenesis in the adult somatosensory cortex. Nat Neurosci 21(5): 696-706.
- Huxley, A. F. and Stampfli, R. (1949). Evidence for saltatory conduction in peripheral myelinated nerve fibres. J Physiol 108(3): 315-339.
- Longair, M. H., Baker, D. A. and Armstrong, J. D. (2011). Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27(17): 2453-2454.
- Osanai, Y., Shimizu, T., Mori, T., Yoshimura, Y., Hatanaka, N., Nambu, A., Kimori, Y., Koyama, S., Kobayashi, K. and Ikenaka, K. (2017). Rabies virus-mediated oligodendrocyte labeling reveals a single oligodendrocyte myelinates axons from distinct brain regions. Glia 65(1): 93-105.
- Pajevic, S., Basser, P. J. and Fields, R. D. (2014). Role of myelin plasticity in oscillations and synchrony of neuronal activity. Neuroscience 276: 135-147.
- Perez-Cerda, F., Sanchez-Gomez, M. V. and Matute, C. (2015). Pio del Rio Hortega and the discovery of the oligodendrocytes. Front Neuroanat 9: 92.
- Rivers, L. E., Young, K. M., Rizzi, M., Jamen, F., Psachoulia, K., Wade, A., Kessaris, N. and Richardson, W. D. (2008). PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat Neurosci 11(12): 1392-1401.
- Rushton, W. A. (1951). A theory of the effects of fibre size in medullated nerve. J Physiol 115(1): 101-122.
- Swire, M., Kotelevtsev, Y., Webb, D. J., Lyons, D. A. and Ffrench-Constant, C. (2019). Endothelin signalling mediates experience-dependent myelination in the CNS. Elife 8: 49493.
- Vasistha, N. A., Johnstone, M., Barton, S. K., Mayerl, S. E., Thangaraj Selvaraj, B., Thomson, P. A., Dando, O., Grunewald, E., Alloza, C., Bastin, M. E., Livesey, M. R., Economides, K., Magnani, D., Makedonopolou, P., Burr, K., Story, D. J., Blackwood, D. H. R., Wyllie, D. J. A., McIntosh, A. M., Millar, J. K., Ffrench-Constant, C., Hardingham, G. E., Lawrie, S. M. and Chandran, S. (2019). Familial t(1;11) translocation is associated with disruption of white matter structural integrity and oligodendrocyte-myelin dysfunction. Mol Psychiatry 24(11): 1641-1654.
- Young, K. M., Psachoulia, K., Tripathi, R. B., Dunn, S. J., Cossell, L., Attwell, D., Tohyama, K. and Richardson, W. D. (2013). Oligodendrocyte dynamics in the healthy adult CNS: evidence for myelin remodeling. Neuron 77(5): 873-885.
- Waxman, S. G. (1997). Axon-glia interactions: building a smart nerve fiber. Curr Biol 7(7): R406-410.
Article Information
Copyright
Swire and ffrench-Constant. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Swire, M. and ffrench-Constant, C. (2020). Staining and Quantitative Analysis of Myelinating Oligodendrocytes in the Mouse Grey Matter. Bio-protocol 10(20): e3792. DOI: 10.21769/BioProtoc.3792.
- Swire, M., Kotelevtsev, Y., Webb, D. J., Lyons, D. A. and Ffrench-Constant, C. (2019). Endothelin signalling mediates experience-dependent myelination in the CNS. Elife 8: 49493.
Category
Neuroscience > Development > Immunofluorescence
Cell Biology > Tissue analysis > Tissue staining
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









