- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fluidigm Based Single-cell Gene Expression Library Preparation from Patient-derived Small Intestinal Organoids
Published: Vol 10, Iss 19, Oct 5, 2020 DOI: 10.21769/BioProtoc.3775 Views: 4625
Reviewed by: Tomohiro MizutaniShinya SugimotoJiamin Chen

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Preparing Chamber Slides With Pressed Collagen for Live Imaging Monolayers of Primary Human Intestinal Stem Cells
Joseph Burclaff and Scott T. Magness
Nov 20, 2024 2021 Views

In Vivo Retroviral Transduction of Cardiac Myofibroblasts Using Intramyocardial Injection Immediately Post-myocardial Infarction
Satsuki Ono [...] Michio Nakaya
Nov 20, 2025 1555 Views

A Highly Efficient siRNA Transfection Method in Primary Cultured Cortical Neurons
Xiaorong Wang [...] Zhaolong Zhang
Jan 20, 2026 266 Views
Abstract
In this protocol, we describe our methods to isolate crypts from patients' biopsy samples and to culture human intestinal stem cells as it’s called “organoid.” Beyond that, we describe how to dissociate organoids cells into single cells for single-cell analysis as a further application. This protocol should provide investigators sufficient tools to generate human organoids from biopsy samples and to accomplish a stable in-vitro assay system.
Keywords: OrganoidBackground
The intestinal epithelium is a multifunctional tissue that orchestrates homeostasis and forms a physical barrier. Each intestinal epithelial cells (IECs) arising from intestinal stem cells (ISCs) renew this epithelium every 4-5 days (Crosnier et al., 2006). ISCs are located at the bottom of the crypts and express specific markers as previously reported by various papers (Muñozet al., 2012; Clevers, 2013). Studies suggested that malfunctions of proper renewals of stem cells are related to intestinal disorders, and understandings of ISCs dynamic may elucidate the pathogenesis of various disorders including Inflammatory Bowel Disease (IBD) (Okamoto et al., 2016).
However, the studies of intestinal stem cell properties had been challenging due to the lack of efficient models that recapitulates physiological intestinal epithelial layers. The epic introduction of “organoid" has overcome various obstacles (Sato et al., 2009 and 2011). Organoids can be established from a single ISC in vitro, and faithfully retain the physiological and pathological features of their tissue of origin (Middendorp et al., 2014). Organoids have been used to dissect underlying pathologic changes in various gastrointestinal disease (Fatehullah et al., 2016; Noben et al., 2017) and has shown potentials to reflect complexed mechanisms of organs.
Also, recent advances in molecular biology techniques allow us to study single-cell modalities (Stuart and Satija, 2019). These techniques developed to have insights into each single cell diversities yet had known to be homogenous populations. Studies have shown that a heterogeneous group of cells share these ISC properties, and constitute a hierarchy within the ISC population (Smith et al., 2016). Furthermore, organoids can be one of the ideal tools that are consist of mostly stem cells and transit-amplifying cells under the undifferentiated culture. In the previous report, a single-cell analysis displayed this heterogeneity among the mouse small intestinal stem cells (Li et al., 2014). Combining organoid culture technique and single-cell analysis has the potential to open a new horizon toward the understandings of the dynamics of human intestinal stem cells. In this protocol, we describe in detail the work-flow of human intestinal organoids establishment and dissociation into single cells for further various applications. Compared to those protocols using scRNA-seq (Biton et al., 2014), the present protocol using multiplex PCR enables acquiring single cell profiles in a low-cost, short-time basis, while the number of cells and genes will be limited by the capacity of the microfluid chip format.
Materials and Reagents
- Organoid culture
- 24-well Tissue Culture Plate (Corning, Falcon, catalog Number: 353226 )
- 50 ml and 15 ml Conical centrifuge tubes (Corning, Falcon, catalog numbers: 352096 , 352070 )
- 15 ml STEMFULLTM low cell adhesion tube (Sumitomo Bakelite Co., catalog number: MS-90150Z )
- 1.5 ml Eppendorf tube (Eppendorf Safe-Lock Tubes, catalog number: 00 30121023 )
- 70 μm Cell strainer (Corning, Falcon, catalog number: 352350 )
- Patient intestinal biopsy samples obtained by endoscopies
- Phosphate Buffered Saline (PBS) (Sigma, catalog number: D8537-500ML ) (stored at 4 °C)
- Matrigel (Corning, catalog number: 356231 ) (stored at 4 °C)
- Cell recovery solution (Corning, Thermo Fisher Scientific, catalog number: 354253 ) (stored at 4 °C)
- Advanced-DMEM (Thermo Fisher Scientific, catalog number: 12491015 ) (stored at 4 °C)
- Penicillin/Streptomycin (Nacalai Tesque, catalog number: 26253-84 ) (stored at 4 °C)
- 1 mol/l-HEPES Buffer Solution (Nacalai Tesque, catalog number: 17557-94 ) (stored at 4 °C)
- GlutaMAXTM-I (100x) (Gibco, Thermo Fisher Scientific, catalog number: 35050-061 ) (stored at 4 °C)
- 0.5 M EDTA ((Thermo Fisher Scientific, catalog number: AM9260G )
- Trypan blue solution (Thermo Fisher Scientific, catalog number: 15250061 )
- N-acetylcysteine (Sigma, catalog number: A9165-5G ) (stored at -20 °C, 1 M,10 ml aliquot)
- Gastrin I (Sigma, catalog number: 3006 ) (stored at -20 °C, 100 µl, 1,000 μl aliquot)
- N2 supplement (R&D Systems, catalog number: AR009 ) (stored at -20 °C,100x, 1 vial)
- B27 supplement (R&D Systems, catalog number: AR008 ) (stored at -20 °C, 50x, 1 vial)
- Recombinant mouse EGF (R&D Systems, catalog number: 2028-EG-200 )
(stored at -20 °C, 20 μg/ml, aliquot 1,000 μl) - Recombinant mouse Noggin (R&D Systems, catalog number: 1967-NG-025/CF ) (stored at -20 °C, 20 μg/ml, aliquot 250 μl)
- Recombinant human R-spondin-1 (R&D Systems, catalog number: 4645-RS ) (stored at -20 °C, 100 μg/ml, aliquot 500 μl)
- Recombinant mouse Wnt-3a (R&D Systems, catalog number: 1324-WN-010/CF ) (stored at -20 °C, 10 μg/ml, aliquot 1,000 μl)
- Nicotinamide (R&D Systems, catalog number: 4106 ) (stored at -20 °C, 1 ml, aliquot 1,000 μl)
Note: Stored in the RT shelf as powder. Nic 6.1 g + ddH2O 50 ml, aliquot 10 ml each into 15 ml Falcon tubes. When you thaw one tube, aliquot 1.5 ml eppentubes and store. - A83-01 (Sigma-Aldrich, catalog number: 2939 ) (stored at -20 °C, 10 mM, 100 μl aliquot)
Note: Protect from light. Aliquot 10 mM dilution into Eppendolf tubes and label them with x20, further dilute 10 mM Eppendolf tube with DMSO and aliquot 100 μl each. - SB202190 (Sigma-Aldrich, catalog number: 1264 ) (stored at 4 °C, 150 μl aliquot)
Note: SB202190 10 mg + DMSO 3,018 μl. Aliquot 150 μl each and freeze at -20 until use. - Y-27632 (R&D Systems, catalog number: 1254 ) (protect from light, stored at -20 °C, 10 mM, 150 μl aliquot)
- Human intestinal basal culture medium (see Recipes)
- Human small intestinal organoid growth media (see Recipes)
- Single-cell analysis
- C1 preamp IFC (10-17 μm, Fluidigm, catalog number: 100-5480 )
- 48 x 48 IFC chip (Fluidigm, catalog number: BMK-M-48.48 )
- TrypLE Select (Thermo Fisher Scientific, catalog number: 12563011 ) (stored at 4 °C)
- C1 Single-Cell Reagent Kit for Preamp (including Module 1, Module 2: Fluidigm, catalog number; 100-5319 ) (stored at -20 °C)
- Single Cell-to-CT kit (Thermo Fisher Scientific, catalog number: 4458236 ) (stored at -20 °C)
- LIVE/DEAD viability/cytotoxicity kit (Thermo Fisher Scientific, catalog number: L3224 ) (stored at -20 °C)
- Pooled primers (see Recipes)
- Lysis final mix (see Recipes)
- RT final mix (see Recipes)
- PreAmp final mix (see Recipes)
- LIVE/DEAD cell staining (see Recipes)
- Biomark HD
Equipment
- Pipettes (5 ml, 10 ml, 25 ml, 50 ml), micro-pipettes (10 μl, 20 μl, 200 μl, 1,000 μl)
- Multi-channel pipette (Mettler Toledo, RAININ, model: E8-20 XLS+, catalog number: 17013798 )
- Centrifuge
- Vortex mixer
- Hemacytometer (Burker-Turk, Fujirika Co., A114)
- 37 °C, 5% CO2 cell culture incubator
- Fluorescence microscope (Keyence, model: BZ-X700 ,)
- C1 single cell auto prep system (Fluidigm, San Francisco, CA, USA)
- C1 single cell auto prep array IFCs (MX) (Fluidigm, San Francisco, CA, USA)
- Biomark HD system (Fluidigm, San Francisco, CA, USA)
Software
- Singular Analysis Toolset Software v3.5.2 (Fluidigm, San Francisco, CA, USA)
- R software
- The Partek Genomic Suite (Version 6.6-6.16.0812, Partek, Chesterfield, MO, USA)
Procedure
Part I: Crypt isolation and epithelial organoid culture
- Procedure to isolate and culture small intestinal organoids from patient-derived biopsy samples
- Small intestinal enteroscopic biopsy samples are obtained from patients undergoing evaluation for diseases such as small intestinal tumors, occult bleeding, or Crohn’s disease. Up to 8 biopsies from each patient are taken from a region approximately 100 cm proximal to the ileocecal valve. The Ethics Committee approval and written informed consent should be obtained from each patient.
- Using a 50 ml Falcon tube, wash the fragments with cold PBS until the supernatant is clear.
- Incubate the samples in 15 mM EDTA/PBS solution (rotating) at 4 °C for 30 min (10 ml PBS + 300 µl 0.5 M EDTA).
- After removing the EDTA buffer, add 10 ml of PBS and voltex for 2 min to isolate intestinal crypts (Figure 1).
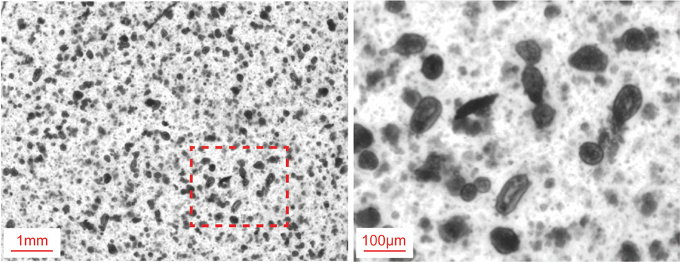
Figure 1. Freshly embedded of small intestinal crypts from patients. Scale bars = 1 mm (left) and 100 μm (right). - Confirm intact crypt isolation by observation under a microscope (see Notes).
- After settling down the specimen in the tube, collect the supernatant and transfer to a 50 ml Falcon tube filtering through a 70 μm filter.
- Repeat the Step A5 of Part I about 2-3 times till an approximately 30 ml volume of crypts containing solution is collected.
- Centrifuge the tube at 300 x g for 3 min to pellet.
- Resuspend the cell pellet with10 ml of Advanced-DMEM/F12 and transfer to one STEMFULLTM tube.
- Centrifuge the tube at 300 x g for 3 min to pellet.
- After examining the pellet under a microscope, carefully aspirate the supernatant and add the desired amount of Matrigel (30-50 µl per well) to embed the crypts (20-30 crypts per well).
- After gently pipetting within the Matrigel, gently apply 30 µl of Matrigel on the 24-well plate. Incubate 30-60 min to allow Matrigel to polymerize.
- Add the 500 µl of human intestinal growth medium to each well. Add 10 µM of Y-27632 to the culture medium for the initial 3 days.
- Procedure to maintain and passage small intestinal organoids
- After isolation, change the small intestinal growth medium every two days.
- After 13 days of isolation, you will be able to see the growth of organoids, and they start to form crypt-like structures called "budding" (Figure 2). In order to subculture, remove the growth media and add 800 µl of cold cell recovery solution to the well. Using cell recovery solution, scrape off the Matrigel containing organoids by 1,000 µl pipette.
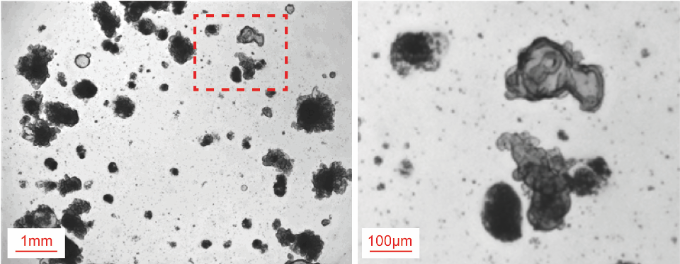
Figure 2. Establishment of small intestinal organoids from patients. Scale bars = 1 mm (left) and 100 μm (right). - Transfer the resuspension to a 15 ml STEMFULLTM tube. Using a P100 pipette, pipette up and down 50-100 times vigorously to mechanically disassociate the organoids into smaller fragments. After examining the fragments under the microscope, add 9 ml of cold Advanced-DMEM/F12 to the mixture.
- Centrifuge the cells at 300 x g for 3 min.
- Carefully aspirate the supernatant and resuspend the pellet in Matrigel. Usually, split at a 1:5-6 ratio. On the around day 13, established organoids start budding.
Part II: Single cell-level gene expression analysis
- Single-cell dissociations of cultured organoid cells
- Remove the entire volume of the growing medium from each well (use 2-3 wells to obtain a sufficient number of cells).
- Using the same method as its described in passing cultured organoids, collect the organoids in a 15 ml STEMFULLTM tube, pipette up and down 50-100 times vigorously to mechanically disassociate the organoids into smaller fragments. Add 9 ml of cold Advanced-DMEM/F12 to the mixture.
- Centrifuge at 300 x g for 3 min and aspirate the supernatant.
- Add 5 ml of TrypLE select to the pellet and vortex for 10 min at 600 rpm (add Hoechst 10 μl).
- After 10 min, shake the tube vigorously for 10 times to further dissociate them into single cells.
- Centrifuge at 500 x g for 3 min and confirm the palette.
- Resuspend with 1,000 μl of Advanced-DMEM/F12 and transfer them to a 1.5 ml Eppendorf tube and count the live cell number by using conventional Trypan Blue staining (optimized cell concentration: 3 x 105 cells/ml, expected cell viability > 70%).
- Take out the desired amount of cells containing solution and centrifuge at 500 x g for 3 min and aspirate the supernatant.
- Resuspend with 1,000 μl of PBS and proceed to C1 protocol.
- Loading single cells into the chip (C1 pre-amplification process)
- Prepare single-cell suspension for C1 loading by mixing previously prepared cell suspension (A9) and C1 Cell suspension reagent while thawing other reagents.
Components Volume (μl) Cells 166-250K/ml 30 μl C1 Cell Suspension Reagent (Fluidigm) 20 μl Total 50 μl - C1 chip priming
- Turn on the C1 machine, place a new C1 single-cell auto prep IFC and apply each reagent as shown in the below (Figure 3).
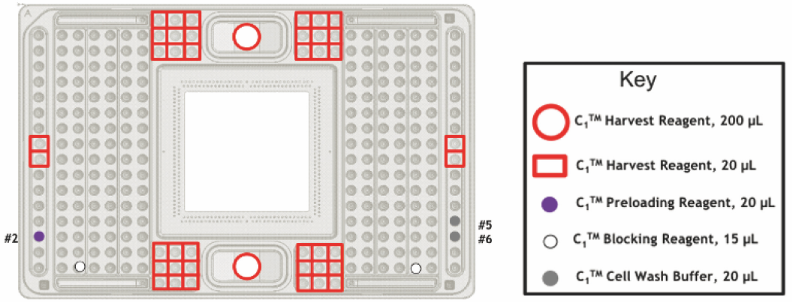
Figure 3. Application of a plate priming (Fluidigm C1 to Taqman Primers protocol, PN100-6117) - After loading each reagent, place the place on the stage and select Prime program and run (it takes 10min).
Important: After the priming, apply the sample within 1 h to prevent the priming reagents from drying out.
- Turn on the C1 machine, place a new C1 single-cell auto prep IFC and apply each reagent as shown in the below (Figure 3).
- While priming the plate, prepare Lysis Final Mix, Reverse Transcription (RT) Final Mix, and PreAmp Final Mix and keep them on ice (Recipes 4-6).
- Cell application and staining
- After the priming, remove the blocking solution as shown below (Figure 4).
- Pipet prepared cell mix for 10 times and apply 20 μl to the blue coded well.
- Prepare the C1 LIVE/DEAD (see Recipes) and apply 20 μl to the pink coded well.
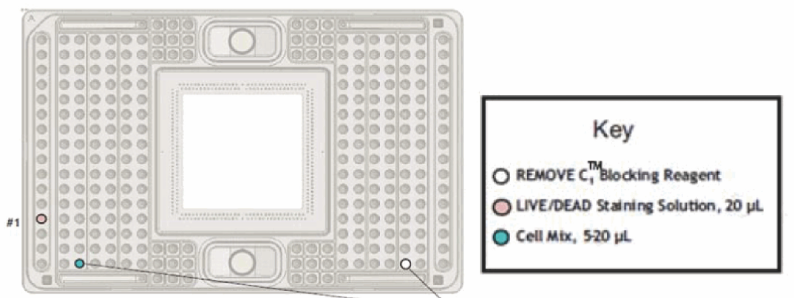
Figure 4. Cell application and staining to a C-1 plate (Fluidigm C1 to Taqman Primers protocol, PN100-6117) - Set the plate on the stage, select Cell Load & Stain program and run (this process takes 60-80 min)
- After the program is over, take out the plate and confirm the single-cell capture (Figure 5) and cell viabilities of 96 wells under a microscope (BX-X700, Keyence, Osaka, Japan).
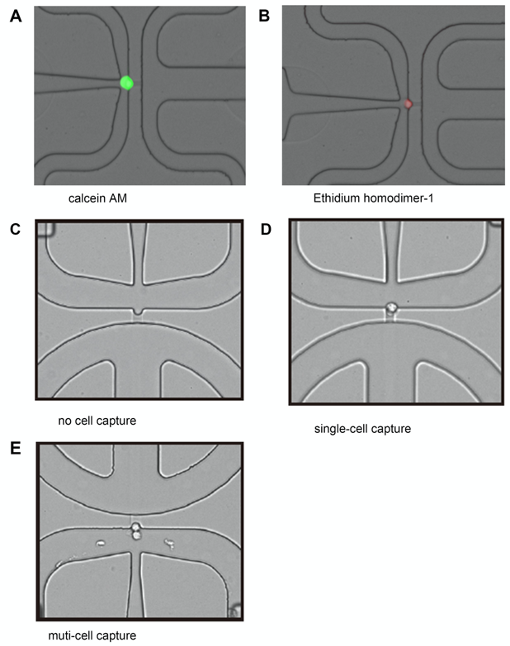
Figure 5. Examples of a viable single-cell capture positively stained with calcein AM and a dead cell stained with Ethidium homodimer-1. Scale bar = 100 μm.
- After confirming the cells, apply each solution as shown in the below (Figure 6) (Recipes 4-6)
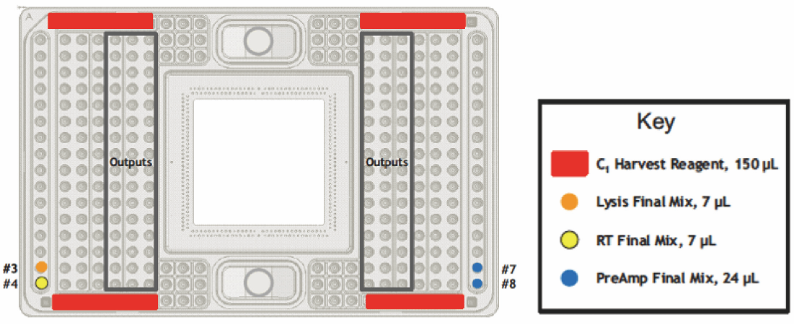
Figure 6. Application of pre-amplification solutions onto a C-1 plate (Fluidigm C1 to Taqman Primers protocol, PN100-6117) - Set the plate on the stage and select the PreAmp program and set the estimated end time (it is an over-night process).
- It is important to collect the sample within an hour of completion; otherwise, the samples will be evaporated.
- Prepare a new 96-well plate and aliquot 25 μl of C1 DNA Dilution reagent into each well of the labeled Diluted Harvest Plate
- Harvest the Pre-Amplified products showed in the below and mix with each well with DNA dilutions using an 8-channel pipette (each well has approximately 3.6 μl) (Figure 7).
- Store the harvested plate in -20 °C.
Components Volume (μl) C1 DNA Dilution Reagent (Fluidigm) 25 C1 harvest amplicons 3 Total 28 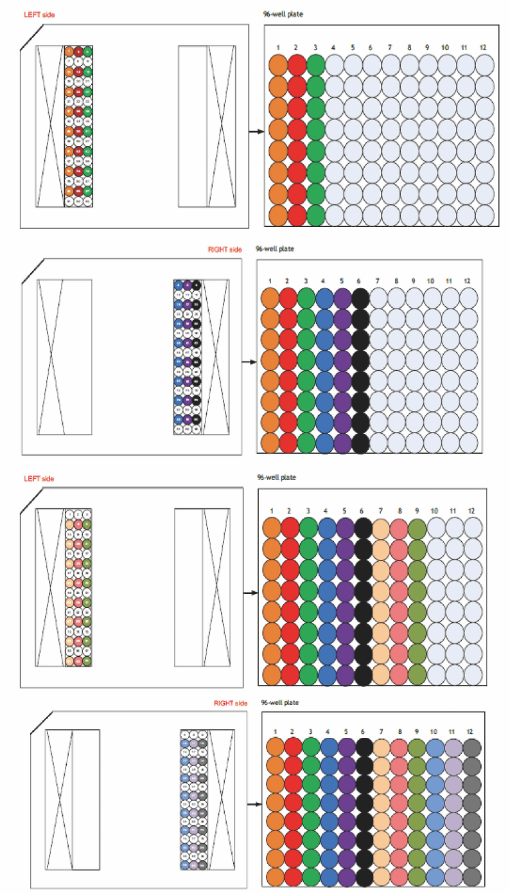
Figure 7. Pipetting steps of pre-amplified products from a C1-plate to a 96-wells plate (Fluidigm C1 to Taqman Primers protocol, PN100-6117)
- Prepare single-cell suspension for C1 loading by mixing previously prepared cell suspension (A9) and C1 Cell suspension reagent while thawing other reagents.
- Biomark HD workflow
Further amplification and data acquirement of the multiplex quantitative PCR analysis will be performed using the Biomark 48.48. Dynamic array IFC and Biomark HD system (Fluidigm, San Francisco, CA, USA). Dynamic array IFC system operates nanofluidic IFC circuits that automatically combines harvested samples with sets of gene assays.- Preparing
- Prepare both “10x assay mix” and “Sample mix” as they are shown below.
TaqMan primers 10x assay mix
Store at -20°CComponents Volume per Inlet (μl) Volume per Islet with Overage (μl) Volume for 48.48 (μl) Volume for 96.96 (μl) 2x Assay Loading Reagent 2.5 6.0 X 60 = 360 X 120 = 600 20x TaqMan Gene Expression Assay 2.5 6.0 Total volume 5.0 6.0 300.0 600.0 Final concentration at 10x Primers: 9 μM Probe: 2.5 μM
Sample MixComponents Volume per Inlet (μl) Volume per Islet with Overage (μl) Volume for 48.48 (μl) Volume for 96.96 (μl) 2x Mater Mix 2.5 3.0 X 60 = 180.0 X 120 = 36.0 20x GE Sample Loading Reagent 0.25 0.3 X 60 = 18.0 X 120 = 36.0 cDNA 2.25 2.7 Total 5.0 6.0 198 396.0
- Prepare both “10x assay mix” and “Sample mix” as they are shown below.
- Priming IFC and sample loading
Make sure to use IFC Controller MX for 48.48 Dynamic Array IFC, and IFC Controller HX for 96.96 Dynamic Array IFC.- Inject control line fluid into each accumulator on the IFC.
- Remove and discard the protective film from the bottom of the IFC.
- Place the IFC into the appropriate controller, then run the appropriate “Prime” script to prime the control line fluid into the IFC. Each IFC type corresponds to a distinct script number, and the plate ID number will be identified by the controller (Priming takes up to 30 min).
Important: Please make sure to run “Load” within 1 h after completing the priming as the plate will dry out. - Remove the IFC from the controller, and pipette samples and assay solutions into the inlets on the IFC (5 μl each).
- Return the IFC to the controller.
- Run the appropriate “Load Mix” script. Air pressure forces samples and gene assay solutions into the IFC where they mix.
- Remove the loaded IFC from the controller.
- Place the IFC into the Biomark HD instrument, making sure the A1 corner on the IFC aligns with the A1 on the instrument tray.
Important: Make sure to run Biomark HD within 1 h after completing the “Load Mix” script as the plate will dry out.
- Running Biomark HD
- Turn on the Biomark HD and the computer.
- Click the icon named “Biomark Data Collection”.
- Wait until the application shows “Ready” when the temperature of the CCD camera reaches to -5 °C.
- Click “Start a New Run” of the default desktop. Confirm that the “Data Collection” screen appears in response.
- Place the pre-loaded IFC on the tray and click “Load”. The tray retracts and the system scans the barcode and identifies the IFC.
- Click “Next”. Browse and select a file that you desire to save.
- Select the Gene Expression application. Select the Passive Reference (ROX). Select Probe and select “Next”.
- Select Browse and Protocol file. Start the Run.
- Preparing
Data analysis
Data acquired from multiplex single-cell gene expression analysis were processed using the Singular Analysis Toolset Software v3.5.2 (Fluidigm, San Francisco, CA, USA) and the Partek Genomic Suite (Version 6.6-6.16.0812, Partek, Chesterfield, MO, USA) by following standard workflows.
Notes
Crypts that can be harvested from endoscopic biopsy samples are very limited in number. After step A4, take out 1ml of the crypt suspension and spread it onto a 24-well plate to observe under a microscope and confirm existence of viable crypts.
Recipes
- Human intestinal basal medium
Components Stock Con. Final con. /500 ml medium Advanced-DMEM/F12 1x 500 ml GlutaMAX-1 200 mM 2 mM 5 ml HEPES 1 M 10 mM 5 ml Penicillin/Streptomycin 10,000 U/ml 100 units/ml 5 ml N-acetylcysteine 500 mM 1 mM 1 ml Gastrin 100 μM 10 mM 50 μl N2 supplement 100x 1x 5 ml B27 supplement 50x 1x 10 ml - Human small intestinal organoid growth media
Components Stock Cons Final Cons Unit ml 1 well 2 wells Basal Medium- - - ml 0.25 0.5 m EGF 20 μg/ml 50 ng/ml μl 1.25 2.5 m Noggin 20 μg/ml 100 ng/ml μl 2.5 5 m R-spondin-1 100 μg/ml 1 μg/ml μl 5 10 m Wnt-3A 10 μg/ml 300 ng/ml μl 15 30 μl 1 M 10 mM μl 5 10 A83-01 500 μM 500 nM μl 0.5 1 SB202190 10 mM 10 μM μl 0.5 1 - Pooled primers
Components Volume (μl) 1 μl each primer pair (100 μM each) 1.0 (x 93 = 93 μl) Optional RNA Spike primers 1.0 (x 3 = 3 μl) C1 DNA Dilution Reagent 104.0 Total 200.0 Can save up to 6 months (-20 °C) - Lysis final mix
Components Volume (μl) C1 DNA Dilution Reagent (Fluidigm) 0.90 Single-Cell Lysis Solution (Thermo Fisher Scientific) 12.75 C1 Lysis Plus Reagent (Fluidigm) 4.35 Total 18.0 - RT final mix
Components Volume (μl) Single-Cell VOLO RT Mix (Thermo Fisher Scientific) 5.84 Single-Cell SuperScript RT (Thermo Fisher Scientific) 3.62 Stop solution (Thermo Fisher Scientific) 1.94 C1 Loading Reagent (Fluidigm) 0.60 Total 12.00 - PreAmp final mix
Components Volume (μl) Single-Cell PreAmp Mix (Thermo Fisher Scientific) 12.0 C1 Loading Reagent (Fluidigm) 3.0 DNA-Free Water (DPEC) 30.0 Pooled Primers (500 nM) 15.0 Total 60.0 - LIVE/DEAD cell staining
Components Volume (μl) C1 Cell Wash Buffer (Fluidigm) 1250.0 Ethidium homodimer-1 (LIVE/DEAD kit, Thermo Fisher Scientific) 2.5 Calcein AM (LIVE/DEAD kit, Thermo Fisher Scientific) 0.625 Total 1253.125
Acknowledgments
This protocol was adapted from Sato et al. (2009) and Suzuki et al. (2018) (Fluidigm, C1 microRNA PreAmp protocol). This work was supported by MEXT/JSPS KAKENHI (18K15774, 18K15743, 19H03634, 19K17484); the Research Center Network Program for Realization of Regenerative Medicine from AMED (18bm03041h0006, 18bk0104008h0001, 19bm0304001h0007, 19bk0104008h0002, 19bm0404055h000).
Competing interests
The authors declare that they have no conflict of interest.
Ethics
The Ethics Committees of Tokyo Medical & Dental University and Yokohama Municipal Hospital approved our study (M2000-2093 and M2000-1176); and written informed consent forms were obtained from each patient.
References
- Biton, M., Haber, A. L., Rogel, N., Burgin, G., Beyaz, S., Schnell, A., Ashenberg, O., Su, C. W., Smillie, C., Shekhar, K., Chen, Z., Wu, C., Ordovas-Montanes, J., Alvarez, D., Herbst, R. H., Zhang, M., Tirosh, I., Dionne, D., Nguyen, L. T., Xifaras, M.E., Shalek, A. K., von Andrian, U. H., Graham, D. B., Rozenblatt-Rosen, O., Shi, H. N., Kuchroo, V., Yilmaz, O. H., Regev, A. and Xavier, R. J. (2018). T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell 175(5):1307-1320.
- Clevers, H. (2013). The intestinal crypt, a prototype stem cell compartment. Cell 154(2): 274-284.
- Crosnier, C., Stamataki, D. and Lewis, J. (2006). Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat Rev Genet 7(5): 349-359.
- Fatehullah, A., Tan, S. H. and Barker, N. (2016). Organoids as an in vitro model of human development and disease. Nat Cell Biol 18(3): 246-254.
- Li, N., Yousefi, M., Nakauka-Ddamba, A., Jain, R., Tobias, J., Epstein, J. A., Jensen, S. T. and Lengner, C. J. (2014). Single-cell analysis of proxy reporter allele-marked epithelial cells establishes intestinal stem cell hierarchy. Stem Cell Reports 3(5): 876-891.
- Middendorp, S., Schneeberger, K., Wiegerinck, C. L., Mokry, M., Akkerman, R. D., van Wijngaarden, S., Clevers, H. and Nieuwenhuis, E. E. (2014). Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells 32(5): 1083-1091.
- Muñoz, J., Stange, D. E., Schepers, A. G., van de Wetering, M., Koo, B. K., Itzkovitz, S., Volckmann, R., Kung, K. S., Koster, J., Radulescu, S., Myant, K., Versteeg, R., Sansom, O. J., van Es, J. H., Barker, N., van Oudenaarden, A., Mohammed, S., Heck, A. J. and Clevers, H. (2012). The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4' cell markers. EMBO J 31(14): 3079-3091.
- Noben, M., Vanhove, W., Arnauts, K., Santo Ramalho, A., Van Assche, G., Vermeire, S., Verfaillie, C. and Ferrante, M. (2017). Human intestinal epithelium in a dish: Current models for research into gastrointestinal pathophysiology. United European Gastroenterol J 5(8): 1073-1081.
- Okamoto, R. and Watanabe, M. (2016). Role of epithelial cells in the pathogenesis and treatment of inflammatory bowel disease. J Gastroenterol 51(1): 11-21.
- Sato, T., Stange, D. E., Ferrante, M., Vries, R. G., Van Es, J. H., Van den Brink, S., Van Houdt, W. J., Pronk, A., Van Gorp, J., Siersema, P. D. and Clevers, H. (2011). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141(5): 1762-1772.
- Sato, T., Vries, R. G., Snippert, H. J., van de Wetering, M., Barker, N., Stange, D. E., van Es, J. H., Abo, A., Kujala, P., Peters, P. J. and Clevers, H. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459(7244): 262-265.
- Smith, N. R., Gallagher, A. C. and Wong, M. H. (2016). Defining a stem cell hierarchy in the intestine: markers, caveats and controversies. J Physiol 594(17): 4781-4790.
- Stuart, T. and Satija, R. (2019). Integrative single-cell analysis. Nat Rev Genet 20(5): 257-272.
- Suzuki, K., Murano, T., Shimizu, H., Ito, G., Nakata, T., Fujii, S., Ishibashi, F., Kawamoto, A., Anzai, S., Kuno, R., Kuwabara, K., Takahashi, J., Hama, M., Nagata, S., Hiraguri, Y., Takenaka, K., Yui, S., Tsuchiya, K., Nakamura, N., Ohtsuka, K., Watanabe, M., Okamoto, R. (2018). Single cell analysis of Crohn's disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties. J Gastroenterol 53: 1035-1047.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Suzuki, K. and Okamoto, R. (2020). Fluidigm Based Single-cell Gene Expression Library Preparation from Patient-derived Small Intestinal Organoids. Bio-protocol 10(19): e3775. DOI: 10.21769/BioProtoc.3775.
Category
Stem Cell > Adult stem cell > Intestinal stem cell
Cell Biology > Cell-based analysis > Gene expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








