- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Immunoprecipitation of Acetyl-lysine and Western Blotting of Long-chain acyl-CoA Dehydrogenases and Beta-hydroxyacyl-CoA Dehydrogenase in Palmitic Acid Treated Human Renal Tubular Epithelial Cells
Published: Vol 10, Iss 18, Sep 20, 2020 DOI: 10.21769/BioProtoc.3765 Views: 5093
Reviewed by: Xiaoyi ZhengYing ShiJake J. Wen

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Experimental Protocol for the Boyden Chamber Invasion Assay With Absorbance Readout
Kathleen C. Brown [...] Piyali Dasgupta
Aug 5, 2024 2761 Views

In-Gel Activity Assay of Mammalian Mitochondrial and Cytosolic Aconitases, Surrogate Markers of Compartment-Specific Oxidative Stress and Iron Status
Wing-Hang Tong and Tracey A. Rouault
Dec 5, 2024 2200 Views

Reliable and Sensitive Detection of Carbonylated Proteins by Oxime Blot
Filip Luka Mikulić [...] Mladen Merćep
Aug 5, 2025 1239 Views
Abstract
As one of the main energy metabolism organs, kidney has been proved to have high energy requirements and are more inclined to fatty acid metabolism as the main energy source. Long-chain acyl-CoA dehydrogenases (LCAD) and beta-hydroxyacyl-CoA dehydrogenase (beta-HAD), key enzymes involved in fatty acid oxidation, has been identified as the substrate of acetyltransferase GCN5L1 and deacetylase Sirt3. Acetylation levels of LCAD and beta-HAD regulate its enzymes activity and thus affect fatty acid oxidation rate. Moreover, immunoprecipitation is a key assay for the detection of LCAD and beta-HAD acetylation levels. Here we describe a protocol of immunoprecipitation of acetyl-lysine and western blotting of LCAD and beta-HAD in palmitic acid treated HK-2 cells (human renal tubular epithelial cells). The scheme provides the readers with clear steps so that this method could be applied to detect the acetylation level of various proteins.
Keywords: ImmunoprecipitationBackground
Post-translational modifications (PTMs) enable the cell with a highly dynamic mechanism for regulation of cellular pathways (Zhao et al., 2010). Acetylation has emerged as one of the major post-translational protein modifications. Accumulating evidences indicate that acetylation rivals phosphorylation as a regulatory modification in mitochondria (Henriksen et al., 2012). Over 60% of mitochondrial proteins are acetylated, being of notice, which were involved in energy metabolism such as tricarboxylic acid (TCA) cycle, oxidative phosphorylation (OXPHOS), fatty acid oxidation and amino acid metabolism (Hirschey et al., 2010; Hebert et al., 2013). Acetylation is also considered to be one of the main adaptive mechanisms for mitochondria in sensing nutritional challenges. Under the condition of excess energy supplement, mitochondrial proteins are hyperacetylated. Insufficient energy availability leads to a decreased acetylation level of mitochondrial proteins (Schwer et al., 2009). A large number of mitochondrial metabolic enzymes are acetylated in diabetes, including the FAO enzymes LCAD and beta-HAD (Kosanam, 2014). Dharendra et al. indicated that the upregulation of acetylation status of mitochondrial fatty acid oxidation enzymes promoted fatty acid oxidation in the heart of high fat diet mice (Thapa et al., 2018). Our previous study also found that the acetylation levels of FAO enzymes LCAD and β-HAD were enhanced in HK-2 cells treated with palmitic acid (PA) (Lv et al., 2019). Protein lysine acetylation refers to covalent addition of acetyl group to lysine residue (N-lysine acetylation). At present, since there are no acetylated antibodies of LCAD and HAD, the detection of LCAD and HAD acetylation levels mainly depends on immunoprecipitation analysis which mainly concentrates the acetylated proteins by using acetyl-lysine antibody and then detects the content of acetylated LCAD and acetylated HAD by using LCAD antibody and HAD antibody respectively (Figure 1).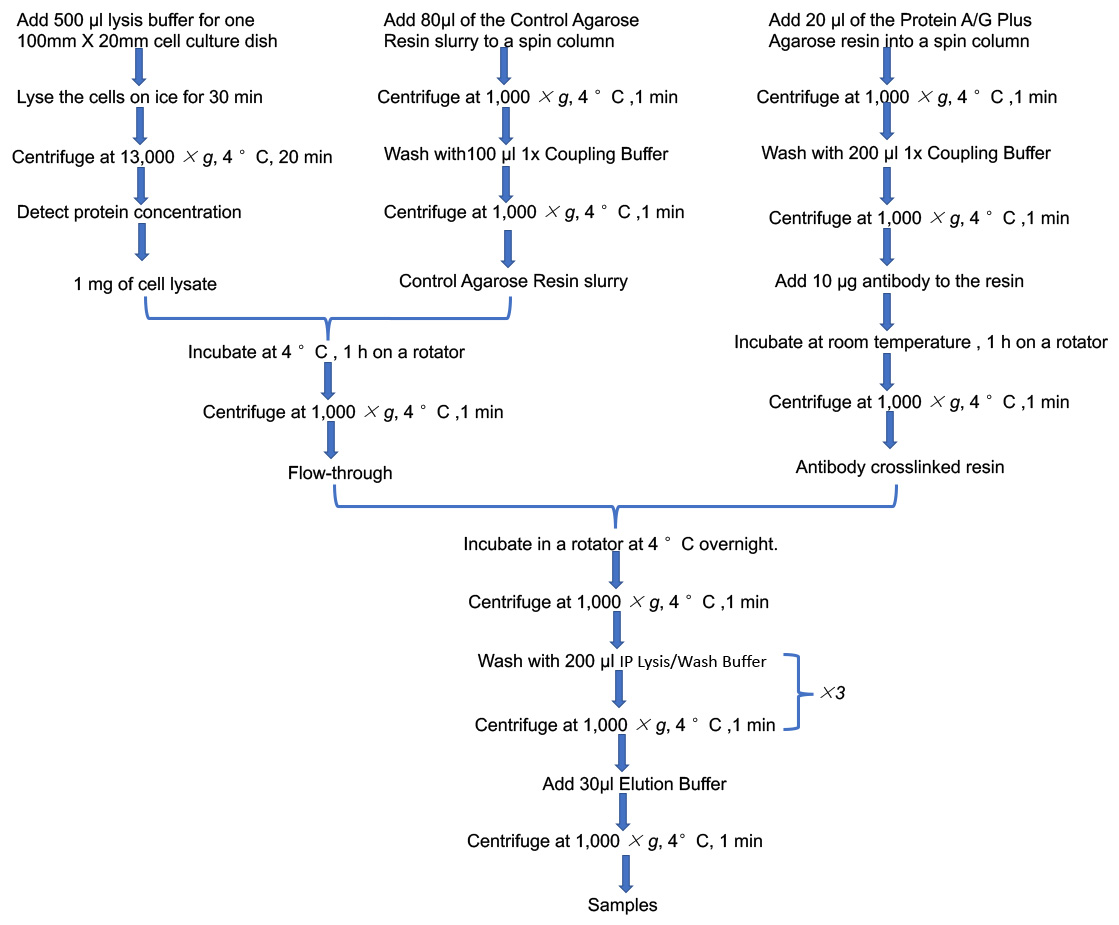
Figure 1. Flowchart of IP
Materials and Reagents
- 100 mm x 20 mm cell culture dishes (Coring, catalog number: 430167 )
- 1.5 ml microcentrifuge tube
- Disposable syringe filters (Millipore, catalog number: SLGP033RB )
- Polyvinylidene fluoride (PVDF) Transfer Membrane (Thermo Scientific, catalog number: 88518 )
- HK-2 cells (ATCC, catalog number: CRL-2190 )
- Pierce Crosslink Immunoprecipitation Kit (Thermo Scientific, catalog number: 26147 )
- Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, catalog number: 11885084 )
- Fetal bovine serum (FBS) (Gibco, catalog number: 10099141C )
- Palmitic acid (PA) (Sigma, catalog number: P5585 )
- Albumin Bovine (BSA) (Solarbio, catalog number: A8850 )
- Halt Protease and Phosphatase inhibitor cocktail (Thermo Scientific, catalog number: 78442 )
- Phosphate buffered saline (PBS) (Solarbio, catalog number: P1020 )
- Mouse-Acetyl-lysine antibody (Abcam, catalog number: ab22550 )
- Rabbit-LCAD antibody (Proteintech, catalog number: 17526-1-AP )
- Rabbit-beta-HAD antibody (Sigma, catalog number: HPA037539 )
- Rabbit-COXIV antibody (Proteintech, catalog number: 11242-1-AP )
- Secondary anti-rabbit IgG HRP-conjugated antibody
- Glycine (Solarbio, catalog number: G8200 )
- Sodium dodecyl sulfate (SDS) (Solarbio, catalog number: S8010 )
- Ammonium persulfate (APS) (Solarbio, catalog number: A8090 )
- N,N,N',N'-Tetramethylethylenediamine (TEMED) (Invitrogen, catalog number: 15524010 )
- Tween-20 (Solarbio, catalog number: T8220 )
- Tris (Solarbio, catalog number: T8060 )
- 1.5 M Tris(hydroxymethyl)aminomethane (Tris-HCl) (pH 8.8) (Solarbio, catalog number: T1010 )
- 1.0 M Tris(hydroxymethyl)aminomethane (Tris-HCl ) (pH 6.8) (Solarbio, catalog number: T1020 )
- 30% Acrylamide (Solarbio, catalog number: A1010 )
- Bicinchoninic acid (BCA) Protein Assay Kit (Beyotime, catalog number: P0012S )
- Immobilon Western Chemiluminescent HRP Substrate (Millipore, catalog number: WBKLS0050 )
- Methanol (Tianjin Kaitong, China)
- Sodium chloride (Tianjin Kaitong, China)
- 12% Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) separating gels (10 ml) (see Recipes)
- SDS-PAGE stacking gels (4 ml) (see Recipes)
- 1x TBST (Tris-HCl, NaCl, Tween 20) (see Recipes)
- 2.5% BSA (see Recipes)
- 10 mM PA (PA stock solution) (see Recipes)
- 200 μM PA (see Recipes)
Equipment
- Centrifuge (Eppendorf, catalog number: 5810R )
- P1000 pipette (Eppendorf, catalog number: 3111000858 )
- P200 pipette (Eppendorf, catalog number: 3111000840 )
- P20 pipette (Eppendorf, catalog number: 3111000823 )
- P10 pipette (Eppendorf, catalog number: 3111000815 )
- Rotator
- Benchtop rocker (HEIDOLPH, catalog number: Titramax 101 )
- Spectrophotometer
- Blot and gel imager (General Electric Company, catalog number: Amersham Imager 680 )
Procedure
- Preparation of cells
Plate cells at 100 x 20 mm cell culture dishes. Culture cells in DMEM with 10% FBS (Fujiki et al., 2019; Guillaume Canaud et al., 2019; Das et al., 2020) and 200 μM PA in a humidified atmosphere of 5% CO2 at 37 °C. - Procedure for Immunoprecipitation
- Cell Lysis
- Carefully discard culture medium from cells.
- Gently wash the cells once with 4 ml pre-chilled PBS.
- Lyse the cells with pre-chilled IP lysis/wash buffer (IP kit) containing protease and phosphatase inhibitor on ice for 10 min.
Note: 500 μl lysis buffer is used for one 100 mm x 20 mm cell culture dish. - Collect the lysate to a 1.5 ml microcentrifuge tube and continue lysing on ice for 20 min.
Note: Mix every 10 min to ensure complete lysis. - Centrifuge at 13,000 x g, 4 °C, for 20 min.
- Collect the supernatant to a new tube and discard the cellular debris.
- Detect protein concentration of cell lysate using BCA Protein Assay Kit and spectrophotometer. Prepare 1 mg protein for IP and 30 μg protein for input.
Note: The range of the STD curve of BCA assay is 50-2,000 μg/ml, and the ODs of the samples fall on the end of the STD curve.
- Pre-clear cell lysate
- Add 80 μl of the Control Agarose Resin slurry to a spin column.
Note: Gently swirl the bottle of Control Agarose Resin before use. For 1 mg of cell lysate use 80 μl of the Control Agarose Resin. The volume of the Control Agarose Resin was proportional to the mass of the cell lysate. - Centrifuge at 1,000 x g, 4 °C for 1 min to remove storage buffer.
- Add 100 μl 1x Coupling Buffer to the column, centrifuge at 1,000 x g, 4 °C for 1 min and discard the flow-through.
- Add 1 mg of cell lysate to the column containing the resin and incubate at 4 °C for 1 h on a rotator.
- Centrifuge at 1,000 x g, 4 °C for 1 min, collect the flow-through to a new tube and place it on ice for later use.
Note: The resin and column are from the IP kit.
- Add 80 μl of the Control Agarose Resin slurry to a spin column.
- Antibody bind to protein A/G plus agarose
- Add 20 μl of the Protein A/G Plus Agarose resin (IP kit) into a spin column. Place column into a centrifuge tube and centrifuge at 1,000 x g, 4 °C for 1 min. Discard the flow-through.
Note: Gently swirl the bottle of Pierce Protein A/G Plus Agarose before use. - Wash agarose resin twice with 200 μl of 1x Coupling Buffer and centrifuge at 1,000 x g, 4 °C for 1 min after each wash. Discard the flow-through.
- Add 10 μl (1 μg/μl) of mouse acetyl-lysine antibody, 5 μl of 20x Coupling Buffer and 85 μl of ultrapure water to the resin in the column. Do the same for normal mouse IgG control.
- Incubate in a rotator at room temperature for 60 min.
- Centrifuge at 1,000 x g, 4 °C for 1 min and discard the flow-through.
- Add 20 μl of the Protein A/G Plus Agarose resin (IP kit) into a spin column. Place column into a centrifuge tube and centrifuge at 1,000 x g, 4 °C for 1 min. Discard the flow-through.
- Antigen Immunoprecipitation
- Add 1 mg cell lysate into the acetyl-lysine antibody crosslinked resin in the column.
Note: The volume of protein lysates is better to between 300 μl and 600 μl. - Incubate in a rotator at 4 °C overnight.
- Centrifuge at 1,000 x g, 4 °C for 1 min and discard the flow-through.
- Add 200 μl of IP Lysis/Wash Buffer to wash the sample, centrifuge at 1,000 x g, 4 °C for 1 min and discard the flow-through. Repeat this wash once.
- Add 1 mg cell lysate into the acetyl-lysine antibody crosslinked resin in the column.
- Antigen Elution
- Add 30μl of Elution Buffer (IP kit) and incubate for 5 min at room temperature.
- Centrifuge at 1,000 x g, 4 °C for 1 min and collect the flow-through.
- Add the flow-through (Step B5b) into column and centrifuge again. Collect the flow-through (IP samples).
- Sample preparation for Western blotting analysis
- Add 4x SDS-PAGE loading buffer to samples to make 1x final solutions.
- Heated the IP samples and Input samples at 100 °C for 5 min to denature the protein.
- Cell Lysis
- Procedure for western blotting analysis
- Prepare 12% SDS-PAGE gels.
- Load 30μg protein per mini-gel well. Run the gel with 80 voltage, 30 min and then 120 voltage, 45 min.
- Transfer proteins of the gel to PVDF membranes under wet transfer condition at 100 voltage about 60 min in ice/water bath under room temperature.
- Blocking the membranes with 5% defatted milk for 1 h at room temperature.
- Wash the membranes with TBST for 5 min at room temperature.
- Incubate membranes with Anti-LCAD antibody (1:1,000), Anti-beta-HAD antibody (1:1,000) and Anti-COXIV antibody (1:5,000) which diluent in 2% defatted milk on a rocker at 4 °C overnight.
- Wash the membranes three times for 10 min with TBST at room temperature on a rocker.
- Incubation with secondary Anti-rabbit IgG HRP-conjugated antibody for 60 min at room temperature.
- Wash the membranes three times for 10 min with TBST at room temperature on a rocker.
- Visualize protein bands by enhanced chemiluminescence (ECL) (Figure 2). Exposure time is about 20 s.

Figure 2. The acetylation level of LCAD and β-HAD was increased after PA stimulation. Ac-LCAD and Ac-β-HAD was detected by immunoprecipitation and western blotting. IgG group was negative control to detect no-specific binding protein. Since LCAD and β-HAD were mitochondrial protein, the mitochondrial contents were normalized by using the internal mitochondrial reference COX IV.
Data analysis
The acetylation level of LCAD or beta-HAD was presented by ratio of acetylated LCAD to total LCAD or acetylated HAD to total HAD.
Recipes
- 12% SDS-PAGE separating gels (5 ml)
ddH2O 1.6 ml 30% Acrylamide 2 ml 1.5 M Tris (pH 8.8) 1.3 ml 10% SDS 0.05 ml 10% APS 0.05 ml TEMED 0.002 ml - SDS-PAGE stacking gels (2 ml)
ddH2O 1.4 ml 30% Acrylamide 0.33 ml 1.0 M Tris (pH 6.8) 0.25 ml 10% SDS 0.02 ml 10% APS 0.02 ml TEMED 0.002 ml - 1x TBST
Tris 0.606 g NaCl 4.4 g ddH2O constant volume 500 ml Tween-20 500 μl - 2.5% BSA
BSA 0.25 g ddH2O 10 ml - 10 mM PA (PA stock solution)
Using disposable syringe filters to sterilize the solutionPA 0.025642 g 95% ethanol 1 ml 2.5% BSA 9 ml - 200 μM PA (PA working solution)
10 mM PA 1 ml DMEM with 10% FBS 49 ml
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants No. 81770729 and 91749111) and Shandong Province Taishan Scholar Project (Grants No. tsqn 20161073). This protocol was adapted from our previous work (Lv et al., 2019).
Competing interests
The authors declare that they have no conflict of interest.
References
- Das, N. A., Carpenter, A. J., Belenchia, A., Aroor, A. R., Noda, M., Siebenlist, U., Chandrasekar, B. and DeMarco, V. G. (2020). Empagliflozin reduces high glucose-induced oxidative stress and miR-21-dependent TRAF3IP2 induction and RECK suppression, and inhibits human renal proximal tubular epithelial cell migration and epithelial-to-mesenchymal transition. Cell Signal 68: 109506.
- Fujiki, K., Inamura, H., Sugaya, T. and Matsuoka, M. (2019). Blockade of ALK4/5 signaling suppresses cadmium- and erastin-induced cell death in renal proximal tubular epithelial cells via distinct signaling mechanisms. Cell Death Differ 26(11): 2371-2385.
- Guillaume Canaud, C.R.B., Seiji Kishi, Kensei Taguchi,, and Kenji Nishimura, S.M., Adam Scott, Li-Li Hsiao, Takaharu Ichimura, Fabiola Terzi, Li Yang, Joseph V. (2019). Bonventre Cyclin G1 and TASCC regulate kidney epithelial cell G2-M arrest and fibrotic maladaptive repair. Sci Transl Med 11(476): eaav4754.
- Hebert, A. S., Dittenhafer-Reed, K. E., Yu, W., Bailey, D. J., Selen, E. S., Boersma, M. D., Carson, J. J., Tonelli, M., Balloon, A. J., Higbee, A. J., Westphall, M. S., Pagliarini, D. J., Prolla, T. A., Assadi-Porter, F., Roy, S., Denu, J. M. and Coon, J. J. (2013). Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell 49(1): 186-199.
- Henriksen, P., Wagner, S. A., Weinert, B. T., Sharma, S., Bacinskaja, G., Rehman, M., Juffer, A. H., Walther, T. C., Lisby, M. and Choudhary, C. (2012). Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol Cell Proteomics 11(11): 1510-1522.
- Hirschey, M. D., Shimazu, T., Goetzman, E., Jing, E., Schwer, B., Lombard, D. B., Grueter, C. A., Harris, C., Biddinger, S., Ilkayeva, O. R., Stevens, R. D., Li, Y., Saha, A. K., Ruderman, N. B., Bain, J. R., Newgard, C. B., Farese, R. V., Jr., Alt, F. W., Kahn, C. R. and Verdin, E. (2010). SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464(7285): 121-125.
- Kosanam, H., Thai, K., Zhang, Y., Advani, A., Connelly, K. A., Diamandis, E. P. and Gilbert, R. E. (2014). Diabetes induces lysine acetylation of intermediary metabolism enzymes in the kidney. Diabetes 63(7): 2432-2439.
- Lv, T., Hu, Y., Ma, Y., Zhen, J., Xin, W. and Wan, Q. (2019). GCN5L1 controls renal lipotoxicity through regulating acetylation of fatty acid oxidation enzymes. J Physiol Biochem 75(4): 597-606.
- Schwer, B., Eckersdorff, M., Li, Y., Silva, J. C., Fermin, D., Kurtev, M. V., Giallourakis, C., Comb, M. J., Alt, F. W. and Lombard, D. B. (2009). Calorie restriction alters mitochondrial protein acetylation. Aging Cell 8(5): 604-606.
- Thapa, D., Wu, K., Stoner, M. W., Xie, B., Zhang, M., Manning, J. R., Lu, Z., Li, J. H., Chen, Y., Gucek, M., Playford, M. P., Mehta, N. N., Harmon, D., O'Doherty, R. M., Jurczak, M. J., Sack, M. N. and Scott, I. (2018). The protein acetylase GCN5L1 modulates hepatic fatty acid oxidation activity via acetylation of the mitochondrial beta-oxidation enzyme HADHA. J Biol Chem 293(46): 17676-17684.
- Zhao, S., Xu, W., Jiang, W., Yu, W., Lin, Y., Zhang, T., Yao, J., Zhou, L., Zeng, Y., Li, H., Li, Y., Shi, J., An, W., Hancock, S. M., He, F., Qin, L., Chin, J., Yang, P., Chen, X., Lei, Q., Xiong, Y. and Guan, K. L. (2010). Regulation of cellular metabolism by protein lysine acetylation. Science 327(5968): 1000-1004.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lv, T., Zhu, S., Ma, Y., Feng, H. and Wan, Q. (2020). Immunoprecipitation of Acetyl-lysine and Western Blotting of Long-chain acyl-CoA Dehydrogenases and Beta-hydroxyacyl-CoA Dehydrogenase in Palmitic Acid Treated Human Renal Tubular Epithelial Cells. Bio-protocol 10(18): e3765. DOI: 10.21769/BioProtoc.3765.
Category
Cell Biology > Cell metabolism > Other compound
Cell Biology > Cell-based analysis > Enzymatic assay
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









