- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Method to Efficiently Cryopreserve Mammalian Cells on Paper Platforms
(*contributed equally to this work) Published: Vol 10, Iss 18, Sep 20, 2020 DOI: 10.21769/BioProtoc.3764 Views: 4784
Reviewed by: Alessandro DidonnaPreeti SharmaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protocol for 3D Bioprinting a Co-culture Skin Model Using a Natural Fibrin-Based Bioink as an Infection Model
Giselle Y. Díaz [...] Stephanie M. Willerth
Jul 20, 2025 3761 Views

A Simplified 3D-Plasma Culture Method for Generating Minimally Manipulated Autologous Equine Muscle-Derived Progenitor Cells
Hélène Graide [...] Didier Serteyn
Dec 5, 2025 1251 Views

Simple and Rapid Model to Generate Differentiated Endometrial Floating Organoids
Adriana Bajetto [...] Tullio Florio
Feb 5, 2026 96 Views
Abstract
This protocol describes a simple method to cryopreserve mammalian cells within filter papers as an alternative to conventional slow-freezing approach. The method involves treating paper fibers with fibronectin, using low concentrations of the cryoprotectant dimethyl sulfoxide (DMSO), and slow freezing cells to -80 °C at a 1 °C min-1 rate. In our method, the biocompatibility, large surface area, 3D porosity and fiber flexibility of the paper, in combination with the fibronectin treatment, yield recovery of cells comparable to conventional approaches, with no additional fine-tuning to freezing and thawing procedures. We expect that the paper-based cryopreservation method will bring several advantages to the field of preserving mammalian cells, including accommodation of a higher number of cells within a unit volume and no cell loss after release. The method requires a minimal storage space, where paper platforms with large areas can be rolled and/or folded and stored in stocks, and allows for efficient transportation/distribution of cells in an on-demand manner. Moreover, an additional feature of this method includes the formation and cryopreservation of cellular spheroids and 3D cell cultures.
Keywords: PaperBackground
Successful preservation, long-term storage, maintenance and distribution of mammalian cells are important research areas that are still under intense scientific investigation. In particular, the timely and steady supply of frozen cells is pertinent in tissue engineering research such as cell culture, drug development and testing, and regenerative and biotherapeutic medicine.
Current conventional cell cryopreservation protocols include slow and rapid freezing and vitrification (Pegg, 2002; Baust et al., 2009). In these approaches, cryoprotectant agents at various concentrations are added to the cell suspension, followed by cooling down the medium at temperature rates as low as 1 °C min-1 (slow freezing) to as high as 120 °C min-1 (rapid freezing), or by placing samples directly in -195 °C liquid nitrogen tanks (vitrification). As a result, with the protective role of cryoprotectants, the cell damage or death is minimized during freezing (Karlsson and Toner, 1996; Asghar et al., 2014; Jang et al., 2017). However, these approaches possess limitations in terms of requiring large storage spaces to house the cells in thousands to millions of small cryotubes and cryobags (Heidemann et al., 2010; Massie et al., 2014). The distribution of cells then faces challenges in terms of loss, misidentification, and logistics management (Tomlinson, 2005).
Recent approaches described the use of various engineered porous scaffolds for cryopreservation of cell tissue constructs. Examples include the use of corn starch-polycaprolactone fiber meshes (Costa et al., 2012), electrospun-polyurethane nanofiber sheets (Batnyam et al., 2017), alginate-gelatin cryogel sponges (Katsen-Globa et al., 2014), and reticulated polyvinyl formal resins (Miyoshi et al., 2010). The results of these studies proved that the biocompatibility, mechanical support, and 3D porosity of scaffolds provide a suitable balance in creating a protective microenvironment for the cells during their cryopreservation. However, with these scaffolds, substrates need to be repeatedly manufactured (i.e., engineered) for their use in relevance to cryopreservation of cells, which brings a heavy dose of hurdle to the field.
Remarkably, paper-based platforms have emerged as an attractive alternative for tissue engineering development, and especially for 3D cell culture. With added features such as cost-effectiveness and tunable fiber surface characteristics, these ready-to-use scaffolds offer remarkable applicability for large-scale, multilayer biological testing (Derda et al., 2009; Mosadegh et al., 2014). As a result, various cellular applications utilizing paper platforms have been intensively investigated (Ng et al., 2017; Wu et al., 2018; Rosqvist et al., 2020). Yet, despite its outstanding potential as a substrate for 3D cell culture and molecular sampling, paper platform has never been utilized to its full potential in directly cryopreserving cells, until our recent work (Alnemari et al., 2020). Instead, it was used as a vitrification container (2D paper substrate) to enhance cryopreservation of mouse embryos (Paul et al., 2018), bovine oocytes (Kim et al., 2012), and bovine blastocysts (Lee et al., 2013). FTA cards, on the other hand, were used for the collection, storage (at room temperature or at +4 °C, -20 °C and -80 °C), transportation, and molecular analysis of nucleic acids (Santos, 2018).
In this protocol, we give step-by-step explanations (Figure 1) on how to 3D preserve and release mammalian cells using our developed paper-based cryopreservation method (Alnemari et al., 2020). The technique starts with cutting the filter paper into small strips (e.g., 3 × 3 cm2). Then, paper fibers are treated with fibronectin to enhance the post-thawing cell release. This is trailed by suspending cells in serum medium containing low concentration dimethyl sulfoxide (DMSO). Following, cell suspension is pipetted onto fibronectin-treated papers. Immediately after cells penetrate within the paper’s 3D porous matrix, papers are rolled and placed in standard cryotubes, slow freezed to -80 °C at a 1 °C min-1 rate, and kept in -195 °C liquid nitrogen for long-term storage. The cells can then be thawed and, depending on the need, either released from the paper to expand as typical 2D culture in flasks or kept inside the paper to grow as 3D cultures and spheroids.
In the developed method, cells are ubiquitous in the 3D porous environment of the paper, where paper fibers provide a natural protective and supportive environment during their cryopreservation. As a result, after their freeze, thawed cells are efficiently released from paper with high viability rates by gently shaking the paper. Here, any paper type, with pores suitable for cells to penetrate within, can be used by simply optimizing the fibronectin concentration for the effective release of cells. The paper also provides a versatile environment for the remaining cells within the paper to grow as aggregates (spheroids) and, as well, successfully enables the formation (by using Matrigel matrix) and cryopreservation of 3D cell cultures (Alnemari et al., 2020). The paper-based cryopreservation offers space-saving and efficient cell transportation/distribution solutions in a cost-effective, fast and easy-to-manage manner, since large paper sheets can be rolled and/or folded to fit standard cryotubes (or other contianers) and stored in stocks and cut into small pieces without the need to thaw the entire platform.
Figure 1. Paper-based cell cryopreservation method. A. An outline of stages involved in the paper-based cryopreservation method. After the thaw, cells can be either released from paper by gentle shaking and expand as 2D culture in flasks or 3D cultured in vitro as needed. B. Micrographs visualize the way a 3 × 3 cm2 paper strip is treated and placed in a cryotube after rolling.
Materials and Reagents
The materials and reagents presented below are for paper-based cryopreservation of cervical HeLa cell lines. For cryopreservation of breast MCF-7, prostate PC3 and lymphocyte JKT cell lines, see the section “Notes” for specific details.
Required
- Sterile 10 ml serological pipettes (Costar Stripette, catalog number: 4488 )
- Sterile 15 ml centrifuge tubes (ThermoFisher, catalog number: 339650 )
- Sterile 1-20 μl, 200 μl, 1,000 μl pipette tips (ThermoFisher, catalog numbers: 10380792 , 10619331 , 10390792 )
- Sterile T75 tissue culture flasks (ThermoFisher, catalog number: 156499 )
- Sterile 1 ml cryogenic tubes (cryotubes) (Sigma-Aldrich, catalog number: CLS430487 )
- Cryo-safe vial storage boxes (cryoboxes) (Sigma-Aldrich, catalog number: Z756776 )
- Mr. Frosty freezing container (ThermoFisher, catalog number: 5100-0001 )
- Sterile 35 mm x 10 mm Petri dishes (Corning, catalog number: 430165 )
- Sterile 60 mm x 10 mm Petri dishes (Falcon, catalog number: 351007 )
- Whatman Grade 114 cellulose filter papers (Sigma-Aldrich, catalog number: 1114-185 )
- Warm water (37 °C)
- Liquid nitrogen (-196 °C)
- Dulbecco’s phosphate buffered saline (DPBS)-10x (Sigma-Aldrich, catalog number: 59331C )
- Fibronectin human plasma (Sigma-Aldrich, catalog number: F0895 )
- Human cervical HeLa cancer cell line (ATCC, catalog numbers: CCL-2 )
- Dulbecco's modified Eagle’s medium (DMEM) (Gibco, catalog number: 11965092 )
- Fetal bovine serum (FBS) (Sigma-Aldrich, catalog number: F7524 )
- Penicillin-streptomycin (Pen-Strep) solution (Sigma-Aldrich, catalog number: P4333 )
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D2650 )
- TrypLE express enzyme (Gibco, catalog number: 12604021 )
- Corning Matrigel matrix (Sigma-Aldrich, catalog number: DLW356231 )
- Cell counting chamber slides (ThermoFisher, catalog number: C10228 )
- Trypan blue exclusion assay (Optional, Sigma-Aldrich, catalog number: T6146 )
- Invitrogen Live/Dead assay kit for mammalian cells, containing green calcein-AM and red ethidium homodimer-1 fluorescent dyes (ThermoFisher, catalog number: L3224 )
- Complete Roswell Park Memorial Institute (RPMI) medium (Gibco, catalog number: 11875093 )
Equipment
Required
- All-purpose scissors with size of 18 cm
- Ruler of length 15 cm
- High precision tweezers (Dumont, catalog number: 5627 )
- Timer (Sunnex, catalog number: 360594 )
- Microbalance (Mettler Toledo, catalog number: ME54 )
- Autoclave (Runyes, catalog number: SEA 18L-DIG-USB )
- Centrifuge (Eppendorf, catalog number: 5810 )
- Laboratory water bath (Witeg, catalog number: WITEG 20002 )
- Humidifying cell incubator (New Brunswick, catalog number: Galaxy 48R )
- Class II laminar air flow hood (NuAire, catalog number: NU-437S )
- -80 °C freezer (Arctiko, catalog number: ULUF 65 )
- Liquid nitrogen tank (Arpege, catalog number: 40 )
Optional
- Countess II FL automated cell counter (ThermoFisher, catalog number: AMQAF1000 )
- Olympus FV1000 inverted confocal microscope (Olympus America)
Software
- (Optional) Imaris 9.2 image analysis software (Oxford Instruments)
Procedure
- Preparation of paper (see Video 1)
- Autoclave the paper and cut it, using sterile scissors, into ~3 x 3 cm2 strips under the hood (Papers can be cut to other sizes and shapes as well, provided that the pipetted cell suspension saturates the paper. See Notes for details).
- Place the paper strips in sterile 60 mm x 10 mm Petri dishes (one strip per dish) and pipette 100 µl DPBS containing 10 µg/ml fibronectin concentration under the hood. After adding the fibronectin solution, incubate the paper fibers for 20 min at room temperature. To avoid water evaporation, make a humidifying chamber by using wet tissues inside Petri dishes and cover their lid (see Notes for the evaporation rate).
- Remove the excess fibronectin by rinsing the paper strips twice with 100 µl DPBS for about 5 s.
- Keep the fibronectin-treated paper strips in the humidifying chamber until the cells are prepared and loaded.Video 1. Steps involved in preparation of paper strips for use in cryopreserving mammalian cells. Paper strips of any type, size and shape can be used provided that the paper pores are suitable for cells to penetrate within and the fibronectin is saturating the paper.
- Preparation of cells
- Culture ~106 cells in T-75 flasks using 10 ml complete DMEM (see Recipes) and place them in a humidifying incubator at 37 °C and 5% CO2.
- Passage cells for a maximum of 15 to 24 passages.
- Dissociate the cells from the flasks when they are ~80% confluent using 2 ml TrypLE for 5 min. Transfer the cells to 15 ml centrifuge tubes to centrifuge at 300 x g for 5 min at 37 °C.
- Resuspend the cell pellet (~107 cells) in 1 ml room temperature DPBS and apply trypan blue exclusion assay to count the number of live and dead cells using cell counting chamber slides and automated cell counter.
- Centrifuge cells at 300 x g for 5 min at 37 °C and resuspend the cell pellet (~107 cells) in 300 µl DMEM-based freezing medium (see Recipes).
- Cryopreservation of cells (see Video 2)
- Transfer fibronectin-coated paper strips to sterile 35 mm x 10 mm Petri dishes (one strip per dish) and pipette onto them 300 µL of cell suspensions (~107 cells/ml per cm2, see Notes).
- Roll the paper strips with tweezers within 1 min to prevent evaporation of cell suspension and place them immediately in cryotubes. Then, place the cryotubes in Mr. Frosty freezing container. To control the rate of freezing, fill the container with 100% isopropyl alcohol to the indicated line.
- Slow freeze the cells in Mr. Frosty container for overnight to -80 °C at a -1 °C min-1 rate.
- Transfer the cells to cryoboxes for long-term storage in liquid nitrogen at -196 °C.Video 2. Cell loading into paper strips. Roll the paper strips and place them in standard cryotubes within one minute to prevent evaporation of the suspension medium.
- Thawing cells on-demand
- Take the cryotubes from liquid nitrogen and thaw them in water bath at 37 °C for 30 s.
- Cells can be cryopreserved in larger sheets. In this case, cut a small portion of the sheet using sterilized scissors, followed by thaw. Place the unused sheet back in liquid nitrogen. Verify the cell viability on sheets after each sample retrieval cycle.
- Releasing cells from paper
- Remove the cell-loaded paper strips from the cryotubes under the hood and place them in 15 ml centrifuge tubes containing 10 ml of complete DMEM.
- Manually shake centrifuge tubes for about 20 s to release the cells from paper.
- Remove the paper strips from the tubes using sterile tweezers.
- Centrifuge the cell suspension in the tubes at 300 x g for 5 min at 37 °C to pellet the cells.
- Wash the cell pellet three times with 1 ml room temperature DPBS.
- Resuspend the cells in complete DMEM in T75 flasks and place the flasks in humidifying incubator at 37 °C and 5% CO2 to expand them as 2D culture (Figure 2).
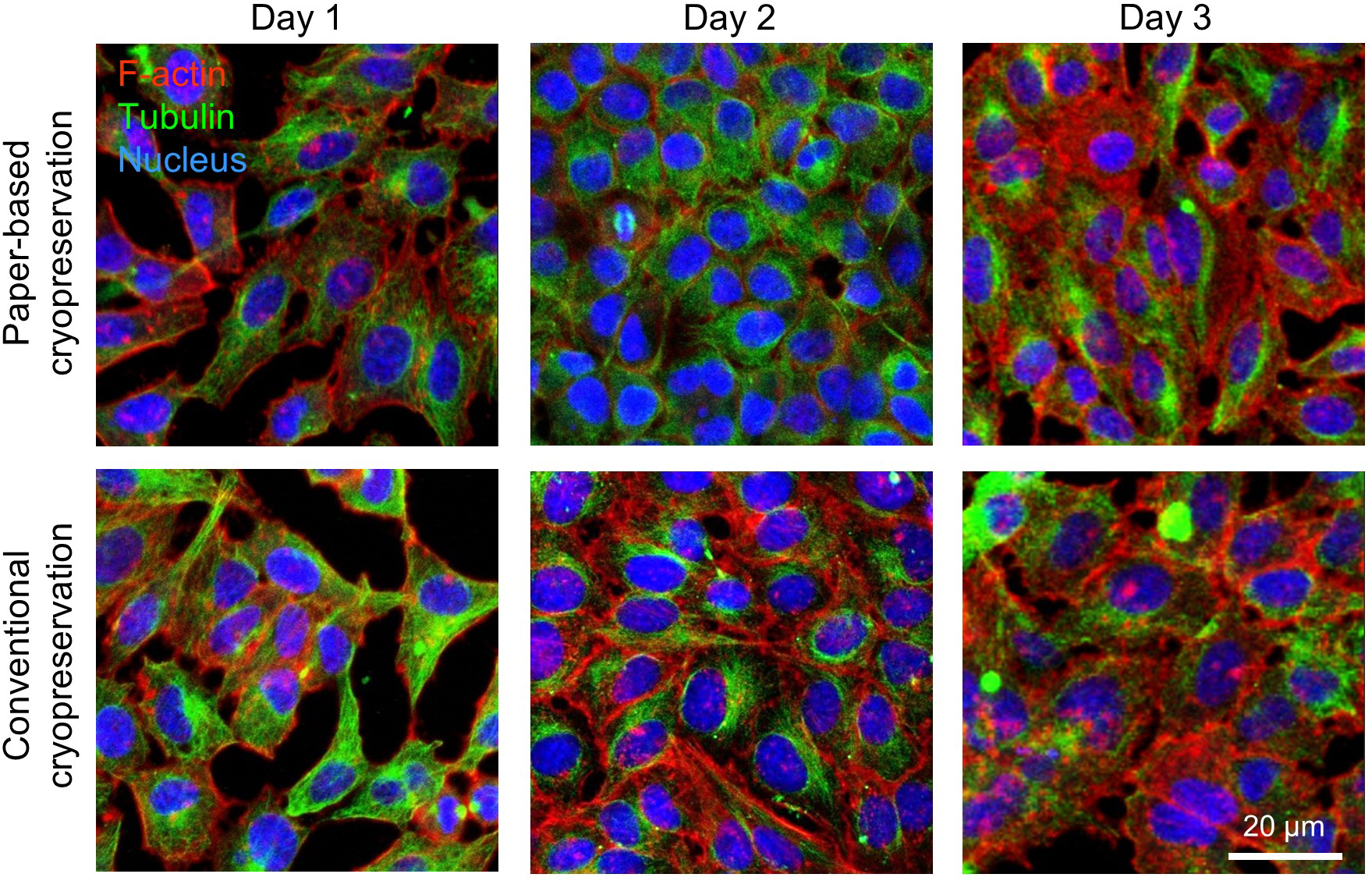
Figure 2. Confocal microscope images show cells expanded as 2D culture after paper-based and conventional (control) cryopreservation. Following their proliferation for 3 days, the spread of actin (red) and tubulin (green) and the presence of nucleus (blue) confirmed no morphological abnormalities or differences between paper-retrieved and freshly cultured HeLa cells. - (Optional) Apply trypan blue exclusion assay to directly count the number of released live and dead cells using cell counting chamber slides and automated cell counter.
- (Optional) Utilize the cells that still remain on the paper (Figure 3) for further in vitro 3D culture growth and spheroid formations (Alnemari et al., 2020) or any other cellular application. This is achieved by placing the paper, with remaining cells, in sterile Petri dishes and allowing cells to 3D expand inside humidifying incubator at 37 °C and 5% CO2. Note that this step has been extended in our recent study (Alnemari et al., 2018) to form arrays of 3D cell cultures on paper platforms, and currently we are advancing its applicability to cryopreserving these arrays.
- Cryopreservation of 3D cell cultures
- Add cells (~107 cells/ml concentration) to 100% Matrigel at 4 °C (placed on ice) and subsequently pipette the cell suspensions in Matrigel onto the autoclaved paper strips placed in sterile Petri dishes (~107 cells/ml per cm2).
- Leave paper strips for 10 min at room temperature for the Matrigel to solidify.
- Submerge paper strips in complete DMEM for 7 days in humidifying incubator at 37 °C and 5% CO2 to allow the proliferation of cells within the paper.
- Roll the paper strips and place them in standard cryotubes containing DMEM-based freezing medium. Place the cryotubes in cryoboxes.
- Slow freeze the proliferated cells for overnight to -80 °C at a 1 °C min-1 rate.
- Transfer the cells to liquid nitrogen at -196 °C for long-term storage.
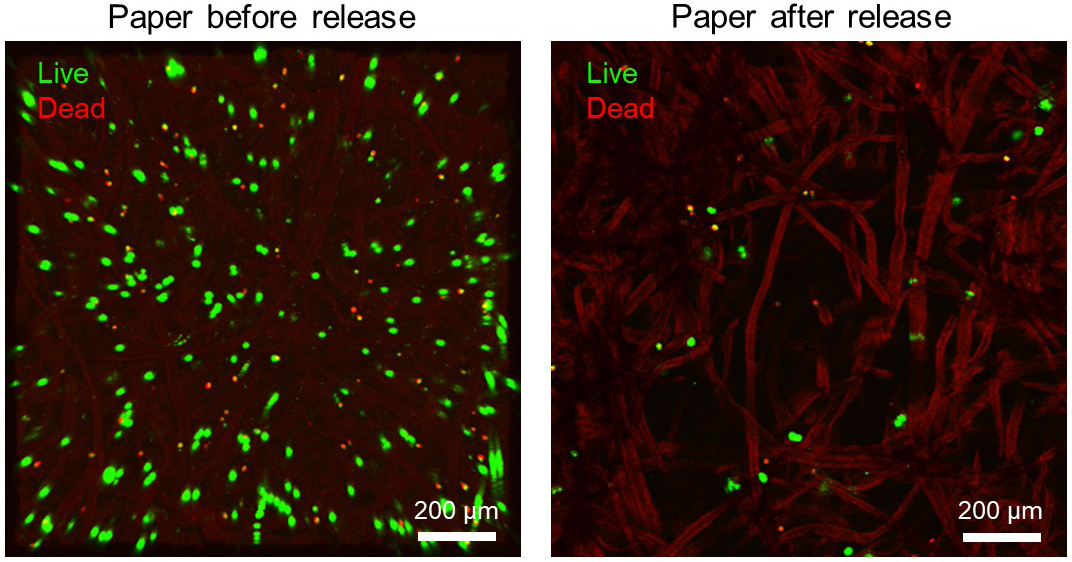
Figure 3. Confocal microscope images show the viable cell release from paper after thaw. Following their load onto 10 µg/ml fibronectin-treated papers and subsequent freeze and thaw, the relative distribution of live (green) and dead (red) HeLa cells within the paper platform visually demonstrate the effective release of viable cells from paper.
- Thawing and providing 3D cell cultures on-demand
- Take the cryotubes from liquid nitrogen and thaw them in water bath at 37 °C for 30 s.
- In case the cell culture is cryopreserved in larger sheets; cut a small portion of the sheet, followed by thaw. Place the unused sheet back in liquid nitrogen.
- Submerge the paper strips with remaining cells in complete DMEM in sterile Petri dishes for additional cell proliferation in humidifying incubator at 37 °C and 5% CO2.
- (Optional) Use Live/Dead assay to visually assess (as control) the distribution of live/dead cells within the papers (Figure 4). This is a useful step especially when working with 3D cell cultures (see Recipes for procedure details).
Data analysis
The Live/Dead analysis was carried by imaging cells with Olympus FV1000 inverted confocal laser scanning microscope (Olympus Corporation) using green (488 nm), and red (612 nm) excitation wavelengths and post-processing images with Imaris software. 10× air objective lens was used to image up to 160 µm deep in paper. The z-stack imaging was performed in 5 µm increments and their 3D projections were created using Imaris software.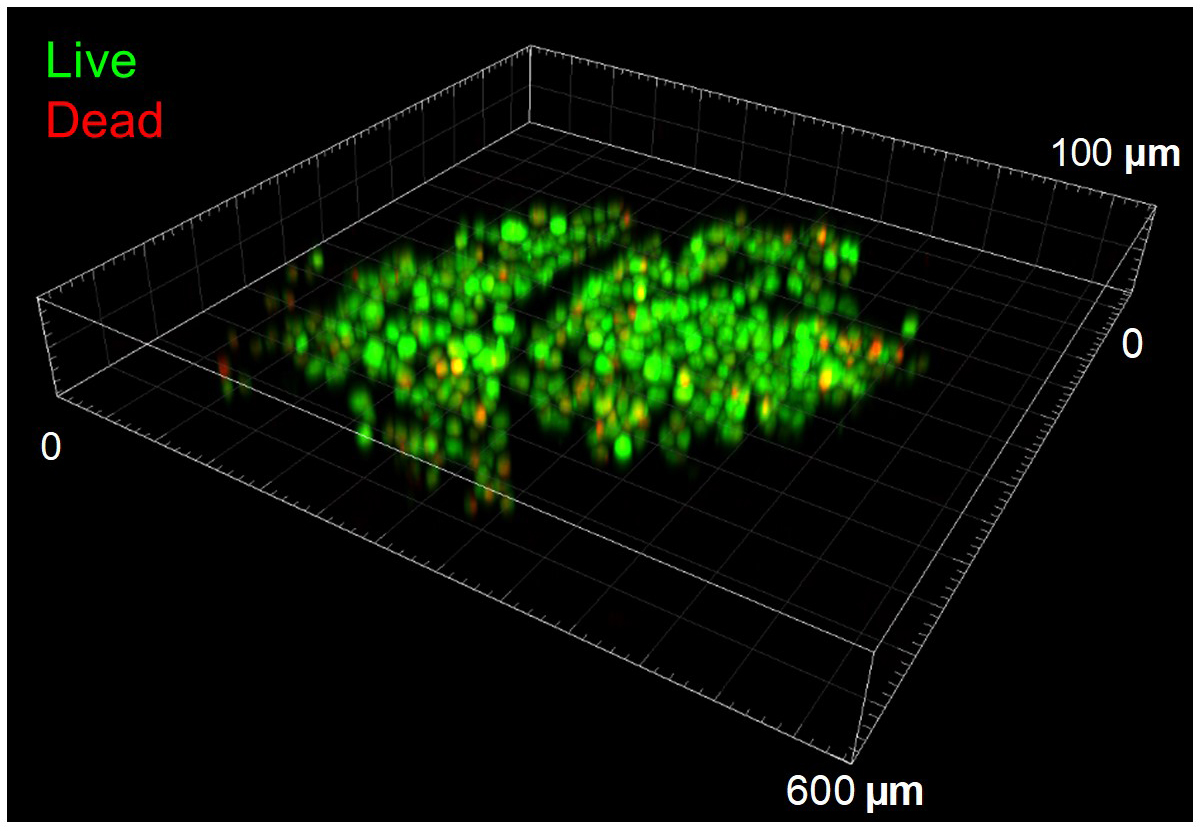
Figure 4. Confocal microscope image shows the survival of cells in paper-based 3D culture after thaw. Following 7 days of 3D paper-based HeLa cell culture, freezing for overnight to -80 °C at a 1 °C min-1 rate, and finally thawing; experiments showed integrity and survival of viable cells.
Notes
- Breast MCF-7 cancer cell line, prostate PC3 cancer cell line, and lymphocyte JKT cell line (ATCC, catalog numbers: CRL-3435, CRL-1435, and TIB-152, respectively) can also be used for paper-based cryopreservation. See Alnemari et al. (2020) for comparison. Below are details of procedures for preparing the cells. The rest of the cryopreservation and thawing procedures are the same when HeLa cells are used.
- Use complete DMEM for MCF-7 cells and complete RPMI medium for PC3 and JKT cells, both supplemented with 10% (vol/vol) FBS and 1% (vol/vol) Pen-Strep.
- Dissociate PC3 and MCF-7 cells from the flasks when they are ~80% confluent using TrypLE and centrifuge at 300 x g for 5 min. Collect suspended JKT cells and centrifuge at 200 x g for 7 min.
- Resuspend the cell pellets (~107 cells) in complete freezing medium with 10% (vol/vol) DMSO for PC3 and MCF-7 cells, and with 5% (vol/vol) DMSO for JKT cells.
- The change in the water mass within the paper at room temperature (water evaporation rate) is 0.002 g min-1 per 9 cm2 paper strips.
- 300 µl cell suspension (107 cells/ml concentration) is just enough to saturate the 3 × 3 × 0.02 cm3 paper volume with cells.
Recipes
- Complete medium
To prepare complete cell culture medium for HeLa cells, supplement the DMEM with 10% (vol/vol) FBS and 1% (vol/vol) Pen-Strep - Freezing medium
To prepare freezing medium for HeLa cells, add 10% (vol/vol) DMSO to complete DMEM - Live/Dead assay
- Dilute 2.5 µl of calcein-AM and 10 µl ethidium homodimer-1 in 5 ml room temperature DPBS
- Pipette 100 µl of working solutions onto paper stripes and incubate for 30 min in the dark
- Place the paper on coverslip and seal it with another coverslip using mounting medium
- Observe the cells under microscope for the integrity, survival, and viability
Acknowledgments
The work was financially supported by NYU Abu Dhabi (NYUAD). We acknowledge the technical support from Core Technology Platforms at NYU Abu Dhabi. This protocol was revised from the procedures described in Alnemari et al. (2020).
Competing interests
The authors declare no competing interest.
References
- Alnemari, R., Sukumar, P., Deliorman, M. and Qasaimeh, M. A. (2020). Paper-based cell cryopreservation. Adv Biosyst 4: 1900203.
- Alnemari, R., Sukumar, P., Deliorman, M. and Qasaimeh, M. A. (2018). Paper based microarrays for 3D tumor spheroid modeling. The 22nd International Conference on Miniaturized Systems for Chemistry and Life Sciences (µTAS 2018), Kaohsiung, Taiwan, 11-15 November, pp. 1484-1486.
- Asghar, W., El Assal, R., Shafiee, H., Anchan, R. M. and Demirci, U. (2014). Preserving human cells for regenerative, reproductive, and transfusion medicine. Biotechnol J 9: 895-903.
- Batnyam, O., Suye, S. and Fujita, S. (2017). Direct cryopreservation of adherent cells on an elastic nanofiber sheet featuring a low glass-transition temperature. RSC Adv 7: 51264-51271.
- Baust, J. G., Gao, D. Y. and Baust, J. M. (2009). Cryopreservation: An emerging paradigm change. Organogenesis 5: 90-96.
- Costa, P. F., Dias, A. F., Reis, R. L. and Gomes, M. E. (2012). Cryopreservation of cell/scaffold tissue-engineered constructs. Tissue Eng Part C Meth 18: 852-858.
- Derda, R., Laromaine, A., Mammoto, A., Tang, S. K. Y., Mammoto, T., Ingber, D. E. and Whitesides, G. M. (2009). Paper-supported 3D cell culture for tissue-based bioassays. P Natl Acad Sci USA 106, 18457-18462.
- Heidemann, R., Lunse, S., Tran, D. and Zhang, C. (2010). Characterization of cell-banking parameters for the cryopreservation of mammalian cell lines in 100-ml cryobags. Biotechnol Progr 26: 1154-1163.
- Jang, T. H., Park, S. C., Yang, J. H., Kim, J. Y., Seok, J. H., Park, U. S., Choi, C. W., Lee, S. R. and Han, J. (2017). Cryopreservation and its clinical applications. Integr Med Res 6: 12-18.
- Karlsson, J. O. M. and Toner, M. (1996). Long-term storage of tissues by cryopreservation: critical issues. Biomaterials 17: 243-256.
- Katsen-Globa, A., Meiser, I., Petrenko, Y. A., Ivanov, R. V., Lozinsky, V. I., Zimmermann, H. and Petrenko, A. Y. (2014). Towards ready-to-use 3-D scaffolds for regenerative medicine: adhesion-based cryopreservation of human mesenchymal stem cells attached and spread within alginate-gelatin cryogel scaffolds. J Mater Sci Mater Med 25: 857-871.
- Kim, Y. M., Uhm, S. J., Gupta, M. K., Yang, J. S., Lim, J. G., Das, Z. C., Heo, Y. T., Chung, H. J., Kong, I. K., Kim, N. H. et al. (2012). Successful vitrification of bovine blastocysts on paper container. Theriogenology 78: 1085-1093.
- Lee, K. H., Sun, J. C., Chuang, C. K., Guo, S. F., Tu, C. F. and Ju, J. C. (2013). An efficient and mass reproducible method for vitrifying mouse embryos on a paper in cryotubes. Cryobiology 66: 311-317.
- Massie, I., Selden, C., Hodgson, H., Fuller, B., Gibbons, S. and Morris, G. J. (2014). GMP cryopreservation of large volumes of cells for regenerative medicine: active control of the freezing process. Tissue Eng Part C Meth 20: 693-702.
- Miyoshi, H., Ehashi, T., Ohshima, N. and Jagawa, A. (2010). Cryopreservation of fibroblasts immobilized within a porous scaffold: Effects of preculture and collagen coating of scaffold on performance of three-dimensional cryopreservation. Artif Organs 34: 609-614.
- Mosadegh, B., Dabiri, B. E., Lockett, M. R., Derda, R., Campbell, P., Parker, K. K. and Whitesides, G. M. (2014). 40 54" target="_blank">Three-dimensional paper-based model for cardiac ischemia. Adv Healthc Mater 3: 1036-1043.
- Ng, K., Gao, B., Yong, K. W., Li, Y. H., Shi, M., Zhao, X., Li, Z. D., Zhang, X. H., Pingguan-Murphy, B., Yang, H. and Xu, F. (2017). Paper-based cell culture platform and its emerging biomedical applications. Mater Today 20: 32-44.
- Paul, A. K., Liang, Y. Y., Srirattana, K., Nagai, T. and Parnpai, R. (2018). Vitrification of bovine matured oocytes and blastocysts in a paper container. Anim Sci J 89: 307-315.
- Pegg, D. E. (2002). The history and principles of cryopreservation. Semin Reprod Med 20: 5-13.
- Rosqvist, E., Niemela, E., Frisk, J., Oblom, H., Koppolu, R., Abdelkader, H., Veliz, D. S., Mennillo, M., Venu, A. P., Ihalainen, P. et al. (2020). A low-cost paper-based platform for fast and reliable screening of cellular interactions with materials. J Mater Chem B 8: 1146-1156.
- Santos, G. D. (2018). FTA cards for preservation of nucleic acids for molecular assays: A review on the use of cytologic/tissue samples. Arch Pathol Lab Med 142: 308-312.
- Tomlinson, M. (2005). Managing risk associated with cryopreservation. Hum Reprod 20: 1751-1756.
- Wu, X. C., Suvarnapathaki, S., Walsh, K. and Camci-Unal, G. (2018). Paper as a scaffold for cell cultures: Teaching an old material new tricks. MRS Commun 8: 1-14.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Deliorman, M., Sukumar, P., Alnemari, R. and Qasaimeh, M. A. (2020). A Method to Efficiently Cryopreserve Mammalian Cells on Paper Platforms. Bio-protocol 10(18): e3764. DOI: 10.21769/BioProtoc.3764.
Category
Cell Biology > Cell isolation and culture > Cryopreservation
Cell Biology > Cell isolation and culture > 3D cell culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









