- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Evaluation of Viable Cells in Pseudomonas aeruginosa Biofilms by Colony Count and Live/Dead Staining
Published: Vol 10, Iss 18, Sep 20, 2020 DOI: 10.21769/BioProtoc.3762 Views: 6806
Reviewed by: Juan Facundo Rodriguez AyalaAlba BlesaChao Jiang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Inexpensive Imaging Platform to Record and Quantitate Bacterial Swarming
Weijie Chen [...] Jay X. Tang
Sep 20, 2021 3636 Views

Purification of the Bacterial Amyloid “Curli” from Salmonella enterica Serovar Typhimurium and Detection of Curli from Infected Host Tissues
Murugesan Sivaranjani [...] Aaron P. White
May 20, 2022 3233 Views

A Guideline for Assessment and Characterization of Bacterial Biofilm Formation in the Presence of Inhibitory Compounds
Bassam A. Elgamoudi and Victoria Korolik
Nov 5, 2023 3104 Views
Abstract
Pseudomonas aeruginosa is a human pathogen capable to form robust biofilms. P. aeruginosa biofilms represent a serious problem because of the adverse effects on human health and industry, from sanitary and economic points of view. Typical strategies to break down biofilms have been long used, such as the use of disinfectants or antibiotics, but also, according to their high resistance to standard antimicrobial approaches, alternative strategies employing photocatalysis or control of biofilm formation by modifying surfaces, have been proposed. Colony forming units (cfu) counting and live/dead staining, two classic techniques used for biofilm quantification, are detailed in this work. Both methods assess cell viability, a key factor to analyze the microbial susceptibility to given treatment, then, they represent a good approach for evaluation of an antibiofilm strategy.
Keywords: Pseudomonas aeruginosaBackground
Bacterial biofilms, complex structures attached to surfaces, are matrices composed by proteins, DNA, polysaccharides and water networks, in which cells are embedded (Costerton et al., 1995). Pseudomonas aeruginosa is a versatile bacterium that can be found in terrestrial and aquatic environments, or as human pathogen, either as free cells or as cells in robust biofilms. P. aeruginosa biofilms represent a serious problem because of the adverse effects on human health and industry (Nickel et al., 1985; Gibson et al., 1999; Willcox et al., 2001; Ramsey and Wozniak, 2005; Rajasekar et al., 2010; Mulcahy et al., 2014) and their high resistance to antibacterial agents (Mah and O’Toole, 2001; Mah et al., 2003). Because of their resistance and robustness, P. aeruginosa biofilms represent a model for biofilm studies (Ciofu and Tolker-Nielsen, 2019). The development of P. aeruginosa biofilms is regulated by a complex genetic program; in addition, biofilm formation is modified by environmental factors (O’Toole et al., 2000; Di Bonaventura et al., 2007; Ben Said et al., 2011; Gambino and Cappitelli, 2016; Pezzoni et al., 2018).
Factors related to the high resistance of biofilms include impaired diffusion of antibacterial compounds, reduced sensitivity due to slow growth rate of cells in biofilms, emergence of resistant bacterial phenotypes, presence of antioxidant products in the biofilm matrix, among others (Pezzoni et al., 2014; Hall and Mah, 2017). Strategies employed to combat biofilms include chemical and physical treatments such as application of disinfectants, antibiotics and ultrasound (Bridier et al., 2011; Wu et al., 2014; Gnanadhas et al., 2015). The high resistance of biofilm cells to commonly used disinfectants and the risk to human health and the environment by the use of increasing bactericide doses prompted to redirect research to safer strategies, such as the use of photocatalytic techniques, proposed as inexpensive, safe and effective (Dalrymple et al., 2010; Gamage McEvoy and Zhang, 2010; Pezzoni et al., 2020). On the other hand, since adhesion of microorganisms to surfaces depends on the surface topography and roughness, preventing biofilm formation by tuning surfaces at nanoscale level is a major opportunity in the development of antibiofilm strategies (Katsikogianni and Missirlis, 2004; Pezzoni et al., 2017).
In order to deepen in the understanding of biofilm properties and/or evaluate the effectiveness of antibacterial treatments, several techniques have been employed to evaluate the amount of live bacteria in biofilms (Welch et al., 2012). Among them, two known methods are the counting of the number of colony forming units (cfu) and the live/dead staining (Merritt et al., 2005; Smith and Hunter, 2008; Tsvetanova, 2020). Both methods provide valuable but different information. In the cfu method, a viable cell is one capable to form a colony. On the other hand, the live/dead staining evaluates membrane integrity. The live/dead staining method employs the fluorescent stains SYTO 9 and Propidium iodide (PI), and viability is evaluated by fluorescence microscopy. PI is a red dye that only enters cells with permeabilized cytoplasmic membrane, while SYTO 9 is a green dye that stains all types of cells. According to this criterion, green cells (intact membrane) are live and red cells (disrupted membrane) are dead. While the colony count method requires biofilm disruption and a subsequent step of cell culture to visualize individual colonies, the live/dead method can be applied on entire biofilms and allows us to evaluate the morphological aspects of the biofilm. Since the two methods are based on different criteria, the data obtained from them will not be identical but they will follow the same tendency upon a given antibacterial treatment (Berney et al., 2006; Bosshard et al., 2010; Pezzoni et al., 2014 and 2020). In this work, we detailed how to apply both methods to P. aeruginosa biofilms.
Materials and Reagents
- 14 ml sterile Borosilicatetubes (Pyrex®, catalog number:SLW1622/09M))
- 1.5 ml sterile Eppendorf centrifuge tubes (Eppendorf, catalog number: 022364111)
- Sterile pipette tips 1-200, 100-1,000 (Corning®, catalog numbers: S4860, S9032)
- Sterile Petri dish glass plates (40 mm and 80 mm diameter) (Brand®, catalog numbers: BR455701, BR455732)
- Sterile cotton plugs (Nunn Finer, catalog number: 562)
- Glass coupons (approximately 15 mm x 15mm x 1mm)
Obtained from glass slides cut with an emery stone. To prepare the coupons, the slides are sliced gently with successive passes of the emery stone. You can wet the area to facilitate the cut. Finally, the glass slide is wrapped with a towel and the final cut is made by pressing hard on the marked area. - Glass slides (YEGREN®, catalog number: 7101)
- Emery stone (Value-Tec, catalog number 52-003094)
- Tupper container (20 cm x 30 cm x 6 cm)
- Rake shaped glass spreaders (Chemglass Life Science, catalog number: CLS-1350-01)
- Aluminum foil (Reynolds Wrap Heavy Duty Aluminum Foil, catalog number: 625)
- Paper towel (WypAll* X 60 Jumbo Roll, KCWW, Kimberly-Clark, catalog number: 30218593)
- Inoculating loop (DeconTM, catalog number: MP 19025)
- P. aeruginosa strain PAO1 (B.H. Holloway)
Pseudomonas aeruginosa PAO1 is maintained in glycerol stocks in liquid nitrogen. Stocks are prepared by mixing by inversion 130 µl of sterile glycerol and 770 µl of an overnight bacterial culture grown in LB medium. Bacteria are streaked with a sterile inoculating loop from glycerol stocks onto solid LB plates and grow overnight at 37 °C in an incubation stove. Single colonies are used as inoculums. - Glycerol (Sigma-Aldrich, catalog number: G5516-500ML)
- Distilled water (G-BIOSCIENCES, catalog number: 786-1713)
- Etanol 96% (EMSURE® Reag, catalog number: 159010)
- Inmersion oil (Leica, standar and type “F”)
- Tryptone (OXOID, catalog number: LP0042)
- Yeast extract (Merck, catalog number: 103753)
- Granulated agar (Difco, catalog number: 214530)
- Sodium Chloride (NaCl) (Biopack, catalog number: 1646.08)
- SYTOTM 9 Green Fluorescent Nucleic Acid Stain (Invitrogen, catalog number: S34854)
- Propidium Iodide (Invitrogen, catalog number: P1304MP)
- LB medium (see Recipes)
- LB agar solid medium (see Recipes)
- 4 M NaCl solution(see Recipes)
- Saline solution (see Recipes)
- 3.5 μM SYTO 9 and 20 μM PI solution (see Recipes)
Equipment
- 125 ml sterile Erlenmeyers flasks (Duran®, catalog number: 2121628)
- 2-20 µl, 20-100 µl,100-1,000 µl Kartell pluripet micropipettes (Kartell LABWARE, catalog numbers: 13000, 13210, 13220) and 1-10 ml Acura® manual micropipette (Socorex Swiss, catalog number: 825/835)
- Sterile 50, 100 and 1,000 ml borosilicate measuring cylinders (VILABO, catalog numbers: 3501114, 3501115, 3501118)
- Sterile 100 ml glass beakers (Brand®, catalog number: BR91224)
- Spatula (Arnaldochapini®, catalog number: 1929)
- Stainless steel dissecting tweezer (Fisherbrand, catalog number: 12-000-132)
- Conventional incubator shaker (New Brunswick Scientific Co., INC, model: G25)
- UV-Vis Spectrophotometer (Biotraza, model: 752)
- Autoclave (Hirayama HICLAVETM, model: HVE-50)
- Hot air oven sterilizer (DalvoIntrumentos, model: OHR/T)
- Vortex (Velp, model: ZV3, 201251076)
- Epifluorescence microscope (Olympus, model: BX51)
- Incubation stove (Precision, Scientific Group, model 4, catalog number: 31483)
- Bunsen burner (EiscoTM, catalog number: CH0091B)
Software
- ImageJ software (Rasband, 1997)
Procedure
The steps of biofilm formation and biofilm evaluation are summarized in Figure 1.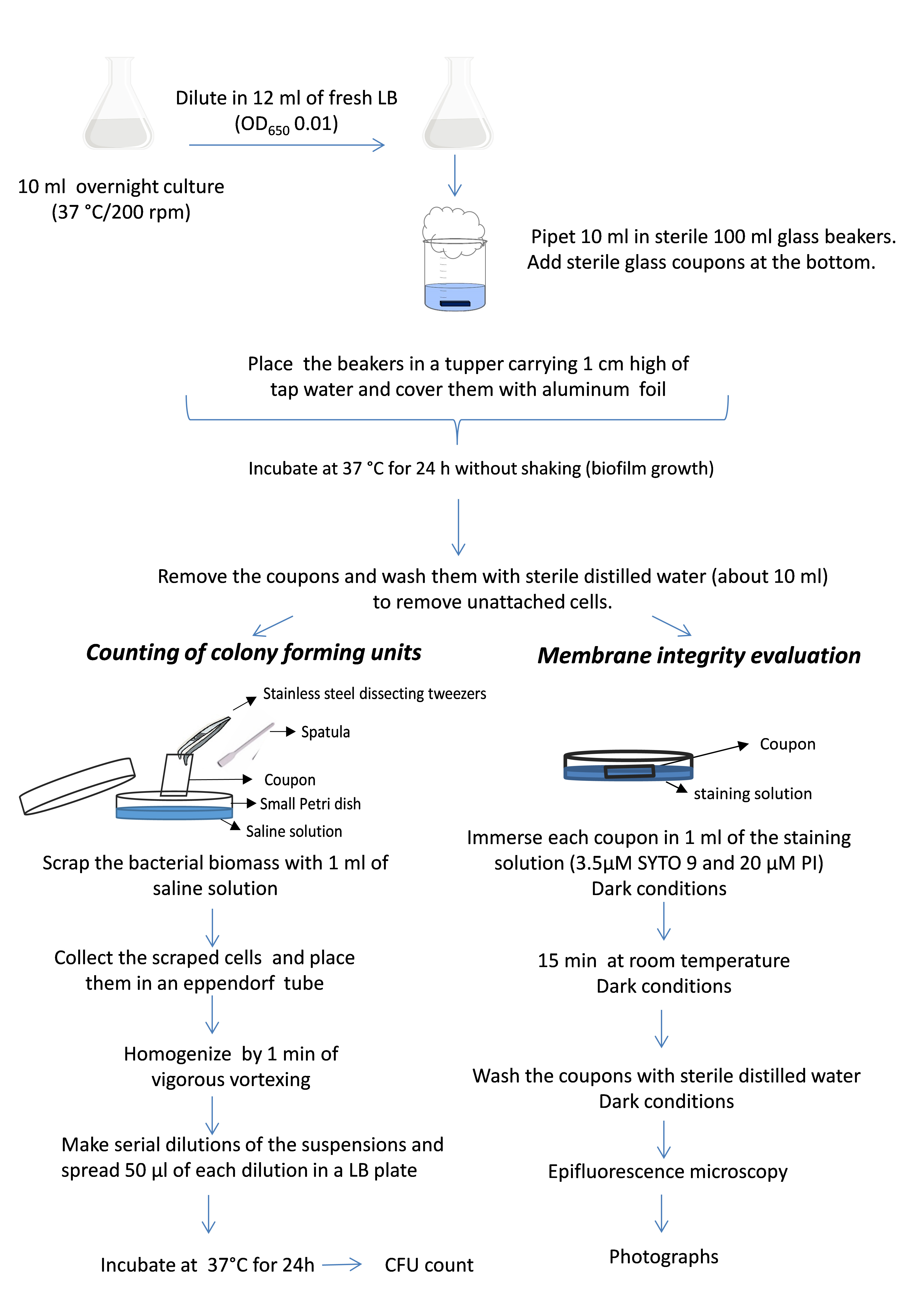
Figure 1. Schematic diagram of the experimental procedure
- Biofilm formation
- Pipet 10 ml of LB medium in sterile 125 ml Erlenmeyer flasks. Then pick, using an inoculation loop, PAO1 colonies from an LB media plate, and shake the loop until the fragment is visibly floating in the liquid LB medium. Grow overnight at 37 °C with shaking (200 rpm) in an incubator shaker. Keep the Bunsen burner on during the entire procedure. Work slowly, carefully, and at all times within this sterile area created by the Bunsen burner.
- Dilute this overnight culture (desired OD650 2.5-3) in 12 ml of fresh LB medium in a new sterile 125 ml Erlenmeyer flask to achieve a final OD650 of about 0.01. Keep the Bunsen burner on during the entire procedure. Work slowly, carefully, and at all times within this sterile area created by the Bunsen burner.
- Pipet 10 ml of these suspensions in sterile100 ml glass beakers. By using a sterile stainless steel dissecting tweezer, place a sterile glass coupon horizontally at the bottom of each beaker to allow biofilm formation on it. Cap the beaker with a cotton plug (Figure 2).
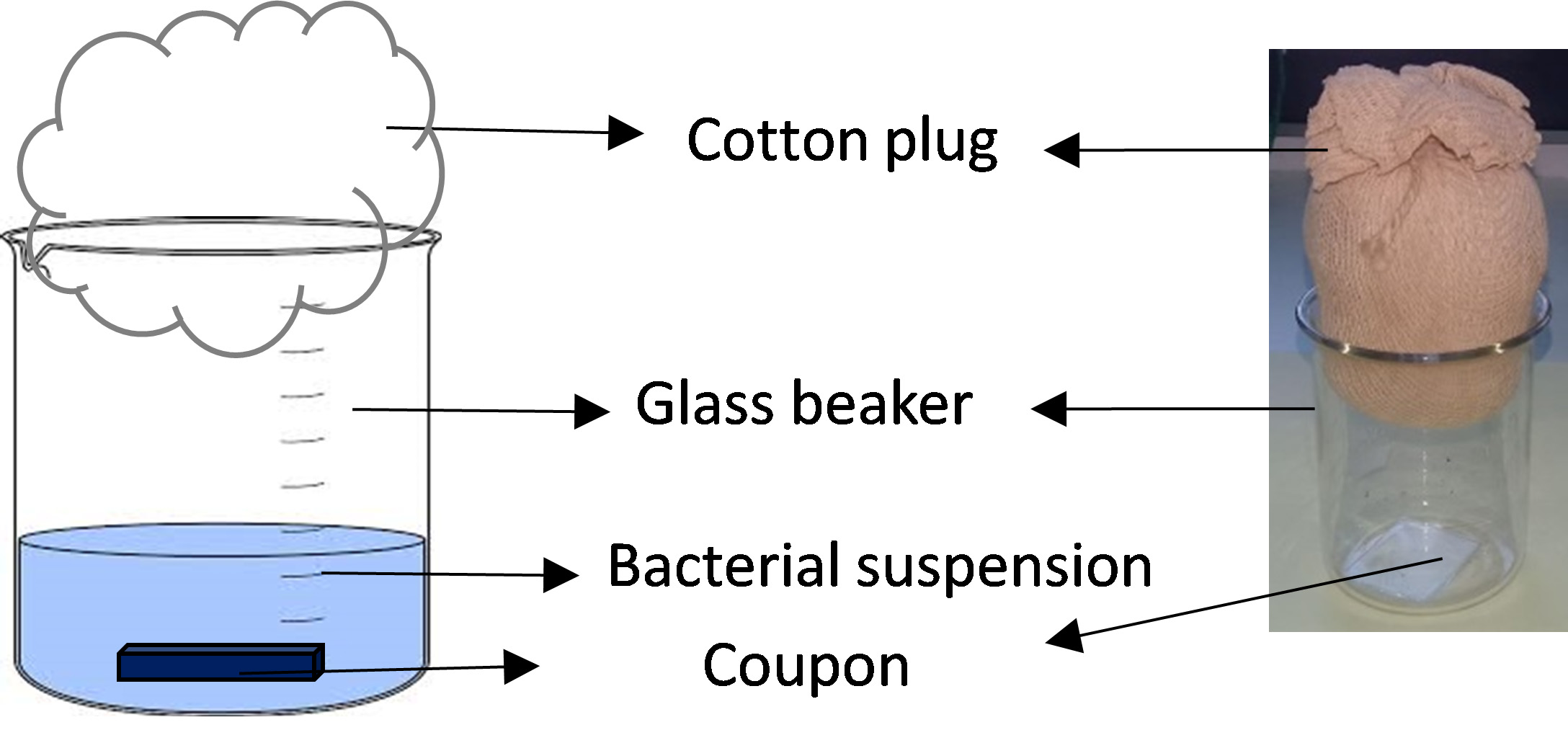
Figure 2. Biofilm formation device - Place the glass beakers in a tupper carrying 1 cm high of tap water and cover them with aluminum foil to create a humid chamber.
- Incubate at 37 °C in an incubation stove for 24 h without shaking.
- Biofilm evaluation
Counting of colony forming units (cfu)- Prepare fresh LB agar plates by placing 25 ml of solid LB medium in 80 mm Petri dishes. After solidification, allow them to dry open and upside down in a stove for at least 30 min at 55 °C.
- Remove the coupons from the beakers with a sterile stainless steel dissecting tweezer.
- Wash the coupons carefully by letting sterile distilled water (about 10 ml) drop down gently on them to remove unattached cells.
- Hold the coupons vertically with the tweezer and scrap for 1 min the bacterial biomass from them with a sterile spatula in a 40 mm Petri plates containing 1 ml of saline solution (Figure 3).
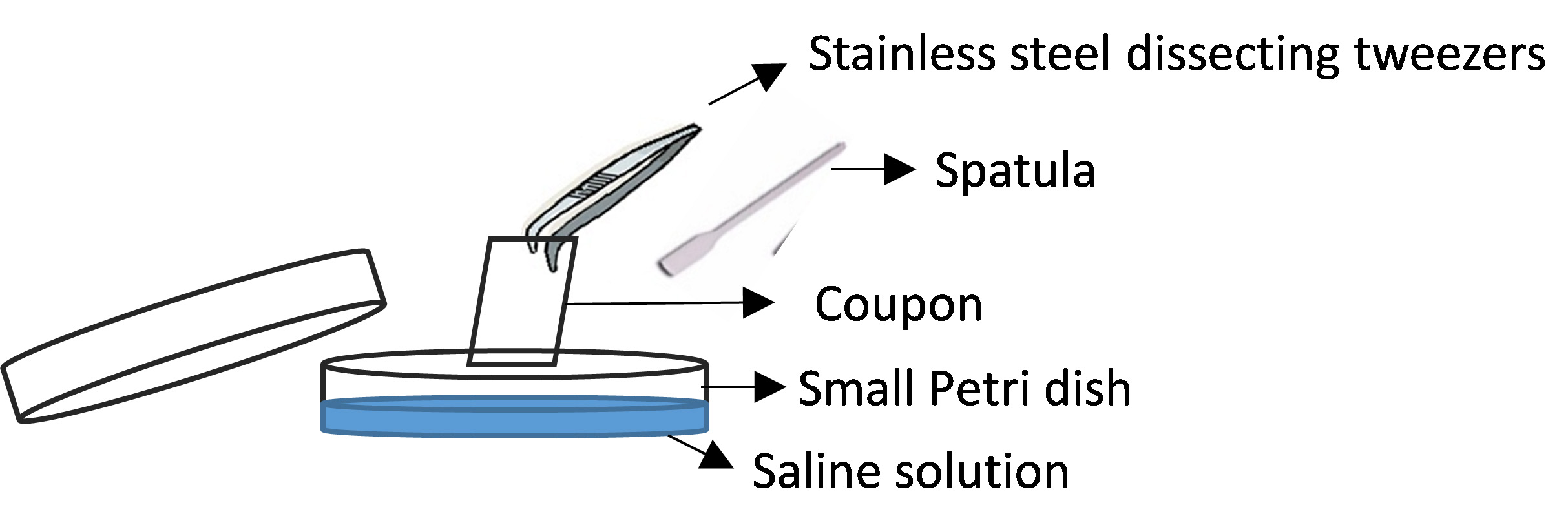
Figure 3. Coupon scraping - Wash the coupons by pipetting the saline solution in the small Petri dish to remove all attached bacteria.
- Place the bacterial suspensions in 1.5 ml sterile Eppendorf tubes.
- Homogenize by 1 min of vigorous vortexing (2,400 rpm).
- Make serial dilutions of the suspensions. To do this, add 0.5 ml of the initial sample to a sterile borosilicate glass tube carrying 4.5 ml of saline solution and mix by vortexing (10-1 dilution). Then, take 0.5 ml of 10-1 dilution and pass it to a new tube carrying 4.5 ml of saline solution to make the next dilution (10-2 dilution) and so on until 10-5 dilution.
- Spread 50 µl of each dilution in a sterile dried LB plate by using a sterile rake shaped spreader.
- Incubate the plates at 37 °C for 24 h in an incubation stove.
- Count the colonies formed onto the LB agar in the Petri plates. Only plates containing about 20-200 colonies must be taken into account.
- Determine the number of the viable cells in cfu cm-2. First, the total cfu in a coupon is calculated according the following formula: no. of colonies count in the plate x dilution factor x total volume of biofilm biomass (1 ml)/volume plated (0.05 ml). This value must be divided by the total surface of the coupon to obtain the number of cfu per cm2. At least three coupons are needed for statistical analysis.
Membrane integrity evaluation- Remove the coupons by taking them with a pair of stainless steel dissecting tweezer as for cfu counting procedure.
- Wash the coupons carefully by letting sterile distilled water (about 10 ml) drop down gently on them to remove unattached cells.
- Immerse each coupon in 1 ml of a solution containing 3.5 μM SYTO 9 and 20 μM PI in distilled water placed in a small Petri plate. This procedure must be performed in dark conditions.
- Incubate for 15 min at room temperature in the dark.
- Wash the coupons with sterile distilled water to remove the staining solution. To do this, dip the coupon in a 100 ml glass beaker carrying 20 ml of distilled water for a few seconds and then let dry at room temperature on a paper towel. This procedure must be performed in dark conditions.
- Observe the coupon under an epifluorescence microscope with 100x objective lens and immersion oil.
- Take at least 6 representative photographs per slide.
- Use the ImageJ Program (ImageJ software [Rasband, 1997]) to calculate the percentage of red areas in the images:
- Open the image with the ImageJ Program.
- Select the red areas using any of the drawing/selection tools (i.e., rectangle, circle, polygon or freeform).
- Then, from the Analyze menu, select Set measurements and look of the area value.
- Repeat the procedure with all the red areas.
- Calculate the percentage of red areas based on the total area of the image.
Notes:- After washing the slides, the stained biofilms could be stored under dark conditions at room temperature for at least 24 h to observe them by epifluorescence microscopy.
- Avoid all contact with SYTO9 and PI; they need to be handled carefully and with gloves. The generation of waste should be avoided or minimized whenever possible. All waste must be discarded as “special waste” under the safety standards of hazardous waste.
- The staining and microscope procedures do not need to maintain sterility.
Data analysis
As example for the described methodologies, results of the effect of different photocatalytic titania surfaces on P. aeruginosa biofilms are shown (Pezzoni et al., 2020). Surfaces were obtained through sol-gel and evaporation-induced self-assembly, by combining titania and surfactants under controlled conditions. They were: non-mesoporous (NM) and mesoporous titania surfaces with different pore sizes, which were achieved based on the use of surfactants Brij-58 (MB) and Pluronics-F127 (MF). In addition, two structural forms of titania were assayed: amorphous and anatase. Biofilms were grown on these surfaces and submitted to ultraviolet-A (UVA) radiation to promote the photocatalytic killing. The biofilms were exposed for 180 min at a fluence rate of 20 W m-2 (total dose 216 Kj m-2).
Note: Data presented in Figures 4 and 5 were reported in: Pezzoni, M., Catalano P. N., Delgado, D. C., Pizarro, R. A., Bellino, M. G. and Costa, C. S. (2020). Antibiofilm effect of mesoporous titania coatings on Pseudomonas aeruginosa biofilms. J Photochem Photobiol B 203:111762.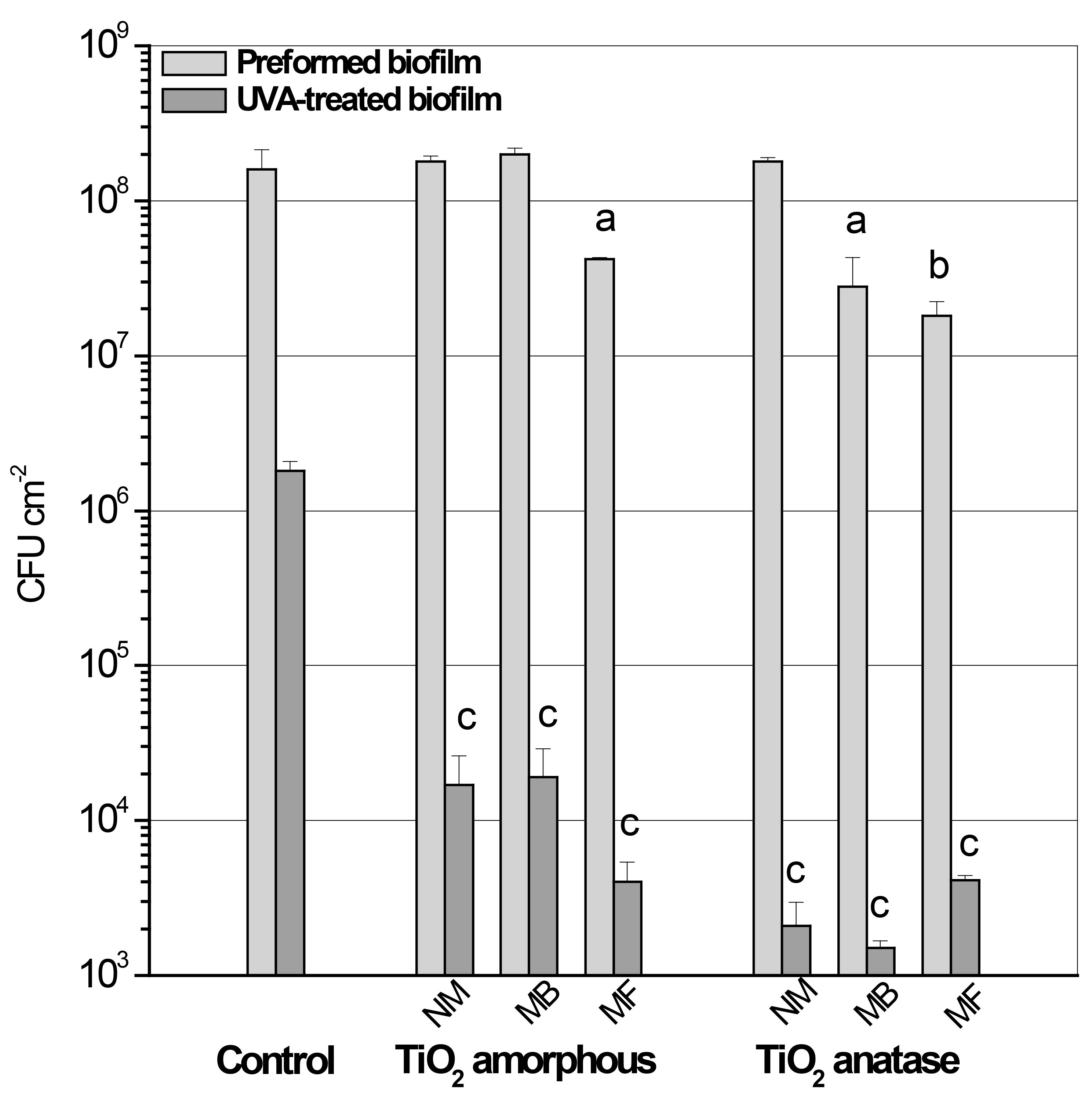
Figure 4. Counting of cfu showing: i) the effect of different titania surfaces on biofilm formation (pre-formed biofilm), and ii) the effect of photocatalytic treatment (UVA-treated biofilm) on these biofilms. 24 h biofilms (Preformed biofilm) were obtained on Control (non-TiO2) or titania coated surfaces (amorphous NM, MB or MF and anatase NM, MB or MF) and exposed to UVA at a fluence rate of 20 W m−2 (UVA-treated biofilm). Appropriate dilutions of the bacterial biomass were plated for cfu count before and after UVA exposure to determine efficiency of biofilm formation and cell survival. Error bars represent the standard deviations of a least three independent experiments. a (P < 0.05) and b (P < 0.005) represent significant difference between preformed biofilms grown on titania and the control surface before UVA exposure. c (P < 0.0005) represents significant difference between biofilms grown on each titania surface and the control surface after UVA exposure.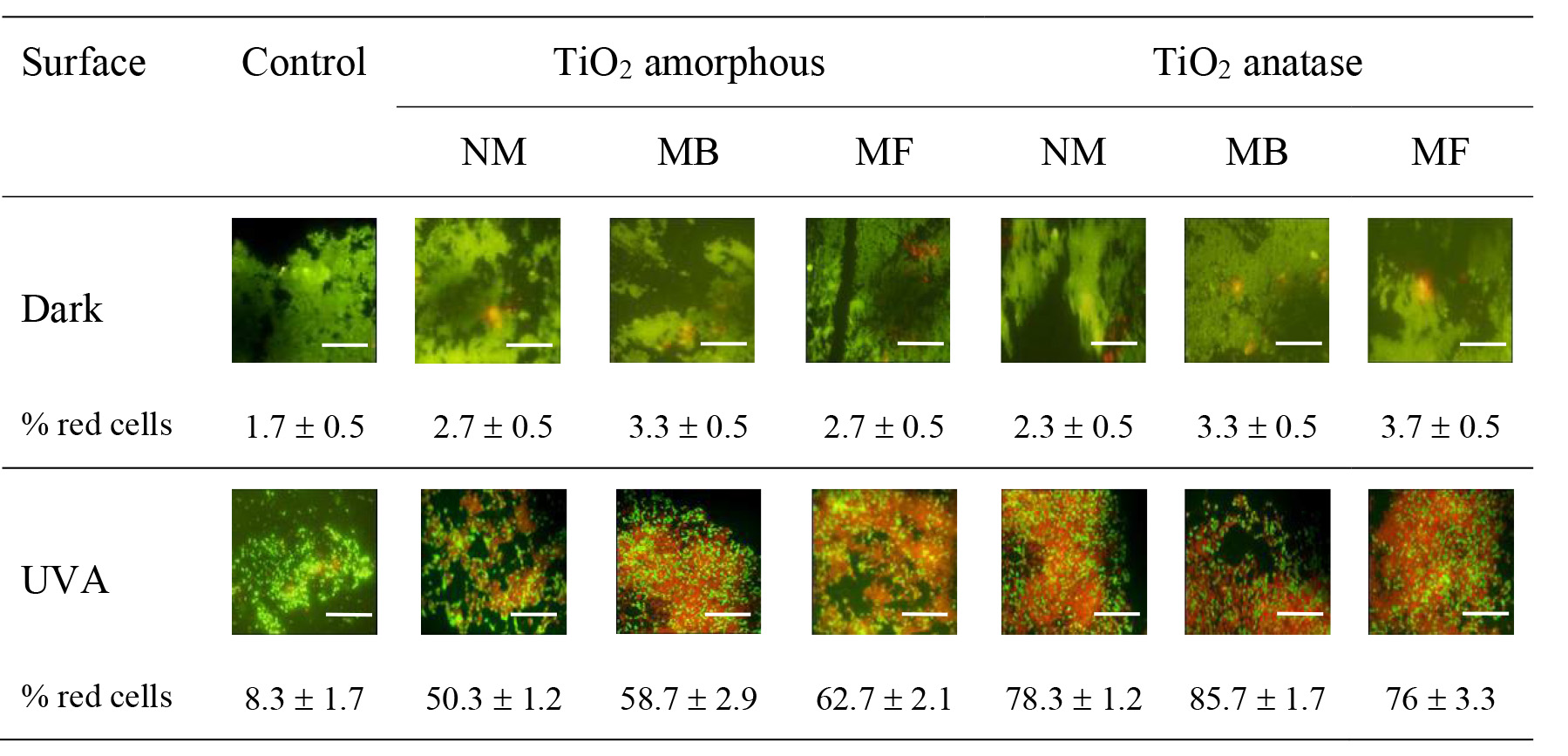
Figure 5. Evaluation of membrane integrity as indicator of the effect of photocatalytic treatment on biofilms obtained on different surfaces. Biofilms grown on Control (no TiO2) or titania coated surfaces (amorphous NM, MB or MF and anatase NM, MB or MF) were maintained in the dark or exposed to UVA at fluence rate of 20 W m−2 for 180 min. Representative epifluorescence images of biofilms stained with the stains SYTO 9 and PI are shown. The experiments were repeated at least three times. Each value is the mean of three independent tests. The scale bars represent 150 nm.
Recipes
- LB medium
- Dissolve 10 g tryptone, 5 g yeast extract, 5 g NaCl
- Bring the volume up to 1,000 ml in distilled water
- Autoclave at 1atm for 20 min
- LB solid medium
- Dissolve 10 g tryptone, 5 g yeast extract, 5 g NaCl, 15 g agar
- Bring the volume up to 1,000 ml in distilled water
- Autoclave at 1atm for 20 min
- 4 M NaCl
- Dissolve 46.7 g NaCl in 200 ml of distilled water
- Autoclave at 1atm for 20 min
- Saline solution
Mix 7.5 ml sterile 4 M NaCl in 300 ml of sterile distilled water - 3.5 μM SYTO 9 and 20 µM PI solution
0.7 μl of 5 mM SYTO 9 (stock solution; stored at -20 °C protected from the light)
26 μl of 0.77 mM PI (stock solution; stored at 4 °C protected from the light)
1,000 µl of distilled water
Prepare fresh for every experiment
The solution can be kept in the dark at room temperature during the experiment
Acknowledgments
The excellent technical assistance of Ms. P. Pereyra Schuth is gratefully acknowledged. Financial support for this research was received from the Comisión Nacional de Energía Atómica (Argentina). M.P. is investigator at Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina).
The counting of colony forming units procedure is based on a previously protocol of Merritt, Kadouri and O'Toole (2005). The live/dead staining method for membrane integrity evaluation is a modification of a protocol published by Smith and Hunter (2008).
Competing interests
The authors declare no conflict of interest.
References
- Ben Said, M., Khefacha, S., Maalej, L., Daly, I. and Hassen, A. (2011). AJMR 525: 4353-4358.
- Berney, M., Weilenmann, H. U. and Egli, T. (2006). Flow-cytometric study of vital cellular functions in Escherichia coli during solar disinfection (SODIS). Microbiology 152(Pt 6): 1719-1729.
- Bosshard, F., Bucheli, M., Meur, Y. and Egli, T. (2010). The respiratory chain is the cell's Achilles' heel during UVA inactivation in Escherichia coli. Microbiology 156(Pt 7): 2006-2015.
- Bridier, A., Briandet, R., Thomas, V. and Dubois-Brissonnet, F. (2011). Resistance of bacterial biofilms to disinfectants: a review. Biofouling 27(9): 1017-1032.
- Ciofu, O. and Tolker-Nielsen, T. (2019). Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents-How P. aeruginosa Can Escape Antibiotics. Front Microbiol 10: 913.
- Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R. and Lappin-Scott, H. M. (1995). Microbial biofilms. Annu Rev Microbiol 49: 711-745.
- Dalrymple, O. K., Stefanakos, E., Trotz, M. A. and Goswami, D. Y. (2010). A review of the mechanisms and modeling of photocatalytic disinfection. Applied Catalysis B Environmental 98(1): 27-38.
- Di Bonaventura, G., Stepanović, S., Picciani, C., Pompilio, A. and Piccolomini, R. (2007). Effect of environmental factors on biofilm formation by clinical Stenotrophomonas maltophilia isolates. Folia Microbiol (Praha) 52(1): 86-90.
- Gamage McEvoy, J. and Zhang, Z. (2010). Applications of Photocatalytic Disinfection. Int J Photoenergy 2010: 764870.
- Gambino, M. and Cappitelli, F. (2016). Mini-review: Biofilm responses to oxidative stress. Biofouling 32(2): 167-178.
- Gibson, H., Taylor, J. H., Hall, K. E. and Holah, J. T. (1999). Effectiveness of cleaning techniques used in the food industry in terms of the removal of bacterial biofilms. J Appl Microbiol 87(1): 41-48.
- Gnanadhas, D. P., Elango, M., Janardhanraj, S., Srinandan, C. S., Datey, A., Strugnell, R. A., Gopalan, J. and Chakravortty, D. (2015). Successful treatment of biofilm infections using shock waves combined with antibiotic therapy. Sci Rep 5(1): 17440.
- Hall, C. W. and Mah, T. F. (2017). Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev 41(3): 276-301.
- Katsikogianni, M. and Missirlis, Y. F. (2004). Concise review of mechanisms of bacterial adhesion to biomaterials and of techniques used in estimating bacteria-material interactions. Eur Cell Mater 8: 37-57.
- Mah, T. F., Pitts, B., Pellock, B., Walker, G. C., Stewart, P. S. and O'Toole, G. A. (2003). A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426(6964): 306-310.
- Mah, T. F. and O'Toole, G. A. (2001). Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9(1): 34-39.
- Merritt, J. H., Kadouri, D. E. and O'Toole, G. A. (2005). Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1: Unit 1B 1.
- Mulcahy, L. R., Isabella, V. M. and Lewis, K. (2014). Pseudomonas aeruginosa biofilms in disease. Microb Ecol 68(1): 1-12.
- Nickel, J. C., Wright, J. B., Ruseska, I., Marrie, T. J., Whitfield, C. and Costerton, J. W. (1985). Antibiotic resistance of Pseudomonas aeruginosa colonizing a urinary catheter in vitro. Eur J Clin Microbiol 4(2): 213-218.
- O'Toole, G., Kaplan, H. B. and Kolter, R. (2000). Biofilm formation as microbial development. Annu Rev Microbiol 54: 49-79.
- Pezzoni, M., Catalano, P. N., Delgado, D. C., Pizarro, R. A., Bellino, M. G. and Costa, C. S. (2020). Antibiofilm effect of mesoporous titania coatings on Pseudomonas aeruginosa biofilms. J Photochem Photobiol B 203: 111762.
- Pezzoni, M., Catalano, P. N., Pizarro, R. A., Desimone, M. F., Soler-Illia, G., Bellino, M. G. and Costa, C. S. (2017). Antibiofilm effect of supramolecularly templated mesoporous silica coatings. Mater Sci Eng C Mater Biol Appl 77: 1044-1049.
- Pezzoni, M., Pizarro, R. A. and Costa, C. S. (2014). Protective role of extracellular catalase (KatA) against UVA radiation in Pseudomonas aeruginosa biofilms. J Photochem Photobiol B Biol 131: 53-64.
- Pezzoni, M., Pizarro, R. A. and Costa, C. S. (2018). Exposure to low doses of UVA increases biofilm formation in Pseudomonas aeruginosa. Biofouling 34(6): 673-684.
- Rajasekar, A., Anandkumar, B., Maruthamuthu, S., Ting, Y. P. and Rahman, P. K. (2010). Characterization of corrosive bacterial consortia isolated from petroleum-product-transporting pipelines. Appl Microbiol Biotechnol 85(4): 1175-1188.
- Ramsey, D. M. and Wozniak, D. J. (2005). Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol 56(2): 309-322.
- Rasband, W. S. (1997). ImageJ. U.S. NationalInstitutesofHealth, Bethesda, Maryland, A.
- Smith, K. and Hunter, I. S. (2008). Efficacy of common hospital biocides with biofilms of multi-drug resistant clinical isolates. J Med Microbiol 57(Pt 8): 966-973.
- Tsvetanova, Z. (2020). Quantification of the bacterial community of drinking water-associated biofilms under different flow velocities and changing chlorination regimes. Appl Water Sci 10:3.
- Welch, K., Cai, Y. and Strømme, M. (2012). A method for quantitative determination of biofilm viability. J Funct Biomater 3(2): 418-431.
- Willcox, M. D., Harmis, N., Cowell, Williams, T. and Holden. (2001). Bacterial interactions with contact lenses; effects of lens material, lens wear and microbial physiology. Biomaterials 22(24): 3235-3247.
- Wu, H., Moser, C., Wang, H. Z., Hoiby, N. and Song, Z. J. (2015). Strategies for combating bacterial biofilm infections. Int J Oral Sci 7(1): 1-7.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Pezzoni, M., Pizarro, R. A. and Costa, C. S. (2020). Evaluation of Viable Cells in Pseudomonas aeruginosa Biofilms by Colony Count and Live/Dead Staining. Bio-protocol 10(18): e3762. DOI: 10.21769/BioProtoc.3762.
Category
Microbiology > Microbial biofilm > Biofilm culture
Cell Biology > Cell viability > Cell proliferation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









