- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro AMPylation/Adenylylation of Alpha-synuclein by HYPE/FICD
Published: Vol 10, Iss 18, Sep 20, 2020 DOI: 10.21769/BioProtoc.3760 Views: 4358
Reviewed by: David PaulDeepali BhandariAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protein Turnover Dynamics Analysis With Subcellular Spatial Resolution
Lorena Alamillo [...] Edward Lau
Aug 5, 2025 2397 Views

Immunopeptidomics Workflow for Isolation and LC-MS/MS Analysis of MHC Class I-Bound Peptides Under Hypoxic Conditions
Hala Estephan [...] Eleni Adamopoulou
Nov 20, 2025 1851 Views

Quantitative Proteomics of Nitrosylated Proteins in Melanoma Using the Biotin-Switch Technique Combined With Tandem Mass Tag Labeling
Vipin K. Yadav [...] Sanjay Premi
Dec 5, 2025 1540 Views
Abstract
One of the major histopathological hallmarks of Parkinson’s disease are Lewy bodies (LBs) –cytoplasmic inclusions, enriched with fibrillar forms of the presynaptic protein alpha-synuclein (α-syn). Progressive deposition of α-syn into LBs is enabled by its propensity to fibrillize into insoluble aggregates. We recently described a marked reduction in α-syn fibrillation in vitro upon posttranslational modification (PTM) by the Fic (Filamentation induced by cAMP) family adenylyltransferase HYPE/FICD (Huntingtin yeast-interacting protein E/FICD). Specifically, HYPE utilizes ATP to covalently decorate key threonine residues in α-syn’s N-terminal and NAC (non-amyloid-β component) regions with AMP (adenosine monophosphate), in a PTM termed AMPylation or adenylylation. Status quo in vitro AMPylation reactions of HYPE substrates, such as α-syn, use a variety of ATP analogs, including radiolabeled α-32P-ATP or α-33P-ATP, fluorescent ATP analogs, biotinylated-ATP analogs (N6-[6-hexamethyl]-ATP-Biotin), as well as click-chemistry-based alkyl-ATP methods for gel-based detection of AMPylation. Current literature describing a step-by-step protocol of HYPE-mediated AMPylation relies on an α-33P-ATP nucleotide instead of the more commonly available α-32P-ATP. Though effective, this former procedure requires a lengthy and hazardous DMSO-PPO (dimethyl sulfoxide-polyphenyloxazole) precipitation. Thus, we provide a streamlined alternative to the α-33P-ATP-based method, which obviates the DMSO-PPO precipitation step. Described here is a detailed procedure for HYPE mediated AMPylation of α-syn using α-32P-ATP as a nucleotide source. Moreover, our use of a reusable Phosphor screen for AMPylation detection, in lieu of the standard, single-use autoradiography film, provides a faster, more sensitive and cost-effective alternative.
Keywords: AMPylationBackground
Deposition of the small (140 amino acids), intrinsically disordered protein alpha-synuclein (α-syn) into Lewy bodies (LBs) is a major hallmark of Parkinson’s disease (PD) (Weinreb et al., 1996; Spillantini et al., 1997). On its way to forming the insoluble aggregates found in LBs, monomeric α-syn adopts a β-sheet-rich, amyloid-like conformation, leading to progressive fibrillation and concomitant insolubility (Iwai et al., 1995). Multiple lines of research describe a compelling correlation between post-translational modifications (PTMs) of α-syn and its oligomeric state. Phosphorylation at Ser 129, for example, is prevalent on aggregated α-syn found within LBs (Chen and Feany, 2005). Tyrosine nitrations of α-syn have been linked to formation of stable oligomers and reduced fibril formation (Yamin et al., 2003), while O-GlcNAcylation of threonine residues reduces α-syn aggregation (Levine et al., 2019). Recently, we described a novel α-syn modification–adenylylation or AMPylation–which reduces α-syn fibrillation in vitro (Sanyal et al., 2019). Moreover, AMPylated α-syn displayed diminished disease-linked phenotypes, such as membrane permeabilization (Ysselstein et al., 2017; Sanyal et al., 2019).
We demonstrated that HYPE, the sole Fic protein in humans, carried out AMPylation of α-syn (Worby et al., 2009; Sanyal et al., 2019). Like other members of the conserved Fic domain protein family, HYPE’s AMPylation activity is intrinsically regulated by an inhibitory helix called αinh (Engel et al., 2012). Mutation of HYPE’s conserved Glu234 to Gly within the αinh renders HYPE constitutively active for its adenylyltransferase activity (Engel et al., 2012; Sanyal et al., 2015). HYPE then covalently attaches the AMP portion of an ATP co-substrate onto target hydroxyl side-chain residues (e.g., serine or threonine) (Ham et al., 2014; Sanyal et al., 2015; Truttmann et al., 2016; Preissler et al., 2017). Indeed, tandem mass spectrometric analysis of purified, recombinant α-syn mapped AMPylation sites to three threonines–T33, T54, and T75–located within regions hypothesized to be crucial for oligomerization (Sanyal et al., 2019).
Several biochemical assays monitoring Fic-mediated AMPylation have been reported (Worby et al., 2009; Mattoo et al., 2011; Yarbrough et al., 2009; Lewallen et al., 2012; Truttmann et al., 2016; Dedic et al., 2016). Most common amongst these are assays using a fluorescent (Fl-ATP) or radioactive (α-32P-ATP or α-33P-ATP) ATP analogs. While Fl-ATP has served as a useful research tool, its large, bulky fluorescent moiety is known to stunt kinetic efficiencies (Lewallen et al., 2012). Moreover, no step-by-step protocol for Fl-ATP AMPylation has been documented. A detailed procedure for α-33P-ATP, however, is available (Truttmann and Ploegh, 2017). This method entails incubation of HYPE and its protein target with radiolabelled α-33P-ATP, followed by separation via SDS-PAGE, DMSO-PPO precipitation, and then imaging of radiolabeled proteins on autoradiography film. Though effective in detecting AMPylation, this procedure suffers from additional time-consuming steps that are expensive and require hazardous chemicals (e.g., DMSO-PPO and autoradiography exposure and development). Herein, we present an optimized method for monitoring AMPylation in vitro, using α-syn as a target and α-32P-ATP as a nucleotide source. α-32P-ATP preserves signal sensitivity, while eliminating DMSO-PPO precipitation. Our protocol also replaces the expensive single-use autoradiography film with a reusable Phosphor screen, which allows more sensitive detection of radioactive phosphate-containing proteins under ambient light and at shorter exposure times. Given the broad specificities of Fic proteins and other enzymes for co-substrates including GTP, CTP, phosphocholine, and even ATP used for phosphorylation, this protocol can be adapted to assess other PTMs in vitro (Mattoo et al., 2011; Veyron et al., 2018).
Materials and Reagents
- Kimwipes (Kimberly-Clark Professional, KimTech Science Brand, catalog number: 34155 )
- Set of micropipettes, with volumes ranging from 0.5 to 1,000 μl (generic)
- Appropriately sized sterile micropipette tips (generic)
- 2 ml pipette bulbs (VWR, catalog number: 82024-554 )
- Pasteur pipette (VWR, catalog number: 14672-380 )
- Glass thermometer, 20-100 °C range (generic)
- 1.5 ml tubes (Eppendorf Safe-lock microcentrifuge tubes, catalog number: 0 22363204 )
- Gel loading pipet tips (Fisher Scientific, Fisher Brand, catalog number: 02-707-181 )
- MTC Stop-Pop locking tubes clips (MTC Bio, catalog number: C2086 )
- Bacterially purified HYPE102-456 (WT, E234G, and E234G/H363A) (Sanyal et al., 2019)
- Bacterially purified full-length α-syn (Sanyal et al., 2019)
- ddH2O (ELGA LabWater, PURELAB Flex 2 Dispenser, model number: PF2XXXXM1 )
- HEPES (Sigma-Aldrich, Sigma-Millipore, catalog number: H4034 )
- Manganese (II) chloride tetrahydrate (Sigma-Aldrich, Sigma Millipore, catalog number: M3634 )
- 12.1 N Hydrochloric acid (Fisher Scientific, Fisher Chemical, catalog number: A144-212 )
- 50% (w/w) Sodium hydroxide solution (Fisher Scientific, Fisher Chemical, catalog number: SS254-4 )
- Tris base (Fisher Scientific, Fisher Bioreagents, catalog number: L-15863 )
- Sodium chloride (Fisher Scientific, Fisher Chemical, catalog number: L-11620 )
- Glycerol (Fisher Scientific, Fisher Bioreagents, catalog number: L-13751 )
- α-32P-ATP 3000Ci/mmol 10mCi/ml, 250 µCi (Perkin Elmer, BLU003H250UC)
- 4-20% Precast SDS-PAGE gel (Bio-Rad, Mini-PROTEAN TGX Gels, catalog number: 456-1091 )
- Protein ladder (Bio-Rad, Precision Plus Protein Dual Color Standards, catalog number: 1610374TGX )
- 2-Mercaptoethanol (Sigma-Aldrich, Sigma Life Sciences, catalog number: M3148 )
- Sodium dodecyl sulfate (Sigma-Aldrich, Sigma Life Sciences, catalog number: L4390 )
- Bromophenol Blue sodium salt, ACS (Alfa Aesar, catalog number:32639)
- Methanol (Fisher Scientific, Fisher Chemical, catalog number: L-21063 )
- Acetic acid, glacial (Fisher Scientific, Fisher Chemical, catalog number: A38C-212 )
- Glycine (Fisher Scientific, Fisher Bioreagents, catalog number: L-15696 )
- Coomassie Brilliant Blue R-250 (Amresco, catalog number: 0472-25G )
- Phosphor screen (General Electric, Kodak Phosphor Storage Screen, catalog number: 00 146931 )
- Protein Storage Buffer (see Recipes)
- 10x AMPylation Buffer (see Recipes)
- 4x SDS-PAGE Loading Buffer (see Recipes)
- 1x SDS-PAGE Running Buffer (see Recipes)
- Coomassie Stain (see Recipes)
- Destain (see Recipes)
Equipment
- Plexiglass or Lucite shield, 0.5 inches thick (Nalgene Acrylic Benchtop Beta Radiation Shield, Thermo Fisher, catalog number: 6700-1812 )
- Geiger counter (Model 3 Survey Meter, Ludlum Measurements, model: 44-7)
- ddH2O dispenser (PURELAB Flex 2 Dispenser, ELGA LabWater, model: PF2XXXXM1 )
- Heatblock (Analog Heatblock, VWR, catalog number: 12621-108 )
- pH meter (Seven Easy, Mettler Toledo, catalog number: 1227396066 )
- Centrifuge (Eppendorf, model: Centrifuge 5417C , catalog number: 00 23210 )
- Power supply ( PowerPac 300 /DCode, Bio-Rad, model: PowerPac 300 )
- PAGE system (Mini-PROTEAN Tetra Cell, 4-Gel System, Bio-Rad, catalog number: 1658004 )
- Fluorescent Image Analyzer (Typhoon FLA 9500, General Electric, model: Typhoon FLA 9000 )
- Phosphor screen eraser ( FLA Image Eraser , General Electric, model: FLA Image Eraser )
- Rocker (VWR, Rocking Platform, model: 200 )
- Image scanner (Epson, Epson Perfection V700 Photo)
Software
- Typhoon FLA 9500 (General Electric)
Procedure
- AMPylation reaction
Note: This protocol requires use of radioactive α-32P-ATP. As such, all radioactive materials or materials in contact with them must be handled, stored, and disposed of in accordance with your institution’s radioactive safety guidelines. Radioactivity must be handled behind a radiation shield at all times. A Geiger counter should be used to constantly monitor work environment for radioactive contamination. Appropriate personal protective equipment should be worn at all times.- Pipette calculated volume of ddH2O into designated 1.5 ml tubes.
- Pipette calculated volume of 10x AMPylation Buffer into designated 1.5 ml tubes.
Note: Components of 10x AMPylation Buffer may be modified as needed to accommodate differing experimental conditions, such as enzymes, substrates, etc. - Pipette 2 μg of purified, recombinant HYPE enzyme to designated 1.5 ml tubes.
Notes:- Reactions were performed at a final volume of 20 µl. The actual reaction amounts may be modified so long as a ≥ 10:1 molar substrate:enzyme ratio is maintained, and a detectable amount of each protein (i.e., ≥ 0.1 μg) is loaded onto the gel.
- All proteins are purified as previously described (Sanyal et al., 2019) and come dissolved in protein storage buffer.
- It is advisable to prepare twice the amount of required sample in case the gel needs to be rerun.
- Pipette 20 μg of purified, recombinant α-syn (or other substrate) into designated 1.5 ml tubes.
Note: When performing kinetic experiments or comparing different substrates side-by-side, molar concentrations should be used in order to ensure that the relative numbers of molecules in the reaction are consistent. - Pipette 2.5 μCi α-32P-ATP into designated 1.5 ml tubes.
Notes:- Reaction start time begins with α-32P-ATP addition. As α-32P-ATP is the only radioactive component, the other sample ingredients may be added at a standard lab bench, and samples can then be transported to a designated radioactive area for α-32P-ATP addition.
- Alternative concentrations of α-32P-ATP may be used depending on the catalytic efficiencies of the test enzyme and/or the freshness of the nucleotide. These concentrations should be determined empirically. AMPylation reactions should ideally occur within the first half-life (14.3 days) of α-32P-ATP.
- Incubate the samples in a heat block set to 30 °C for 30 min.
Notes:- Be sure to place the heat block behind a radiation shield.
- Different AMPylases may require different reaction temperatures or durations for optimal enzymatic activity.
- Samples may be snap-centrifuged for 5 s at 2,000 x g in order to consolidate any sample components not at the bottom of the tube.
- Quench reaction with an appropriate volume of 4x SDS Loading Buffer.
- Heat samples for 10 min at 95 °C on a heat block.
Note: Ensure that tube lids are firmly closed to avoid lids popping open during heating. A weighted sample tray may be placed flatly over tube lids to prevent opening. Alternatively, MTC Stop-Pop locking tubes clips (MTC Bio, Inc.) could be used. - Snap centrifuge samples for 5 s at 2,000 x g.
- SDS-PAGE separation
- Carefully remove well comb and green seal tape from precast gel cassette.
- Assemble gel cassette and tank according to manufacturers’ instructions.
Note: Fresh 1x Running Buffer should be used inside of the gel cassette assembly, although used 1x Running Buffer may be used to fill the tank. - Using gel loading pipette tips, carefully load 2-4 μl of protein ladder into the first lane.
- Using gel loading pipette tips, carefully load 10-30 μl of each sample into lanes adjacent to the first lane.
- Securely close tank lid.
- Run gel at constant 200 V for 45 min. Alternatively, better band resolution can be obtained by first running the gel at 80 V for ~30 min (until the samples clear the stacking layer), then ramping up to 120 V for ~1 h (until the dye front reaches the reference line on the gel cassette).
Note: Ensure proper current flow via production of bubbles in the running buffer and migration of the dye front towards the bottom of the gel.
- Phosphor imaging
- Carefully remove gel using a cassette lever.
Note: The gel is radioactive at this point and should be handled and disposed of in accordance with your institution’s regulations/guidelines for radioactive materials. - Place gel in between a clean, plastic cover sheet. Smooth out any air bubbles in the gel.
- Place cover sheet with gel on top of the imaging side (white) of a clean Phosphor screen.
Notes:- Phosphor screen should be erased with the Phosphor screen eraser after each use to prevent signal contamination.
- Avoid touching the imaging side of the Phosphor screen, as this can damage the screen.
- Carefully place Phosphor screen and gel inside of the foam phosphor screen storage container.
- Expose the screen for 3 h at room temperature.
Note: Duration of Phosphor screen exposure is dependent on the AMPylation reaction parameters and must therefore be determined empirically. - Place the Phosphor screen imaging-face down in an imager.
Note: Ensure imager screen is clear of debris before imaging, as these may mask sample signal. Kimwipes and ddH2O should be used to clean imager screen as needed. - Open the Typhoon FLA 9500 software.
- Under “Mode Settings”, select the “Phosphorimaging” tab.
- Under “Reader Settings,” go to the “Method” dropdown tab and select “[Phosphor]” option.
Note: The “[Phosphor]” option will be colored red and correspond to a wavelength of 673 nm. - Under “Reader Settings,” go to “PMT” (Photo-multiplier Tube) and select a value between 250-1,000 V.
Note: A PMT of 500 V is usually a good starting point to gauge band intensity; the higher the PMT, the stronger the signal. - Under “Reader Settings,” go to “Area” and select the grid area corresponding to where the gel is placed on the screen. Alternatively, the entire screen may be imaged, but this takes additional time.
- Under “Reader Settings” go to “Pixel Size” and Select “50 μm” pixel size.
Note: Lower or higher resolutions may be selected based on specific image quality needs. - Select “Scan” to scan image.
- Select “Save As” to save image as a Tiff file in the appropriate folder.
- Carefully remove gel using a cassette lever.
- Coomassie staining
- Once exposure of the Phosphor screen to the gel is complete, incubate the gel in Coomassie Stain with a rocker set to low to medium speed for 10 min.
Note: The amount of stain used will vary depending on the shape and dimensions of the container used for incubation but should be enough to fully immerse the gel. - Pour off Coomassie Stain in its original container for reuse.
- Incubate gel in Destain on rocker for 30 min or until sufficient contrast between protein bands and gel background is established.
- Discard Destain in appropriate waste container.
- Scan gel on generic document scanner between cover sheet, or with phone camera or scanning application.
Notes:- Coomassie staining is done to control for protein loading, and should yield bands that validate experimental amounts of protein (see Figure 1, bottom).
- If reactions are set up using molar ratios instead of mass-based concentrations, the apparent band sizes may vary with the differing molecular weights of the proteins.
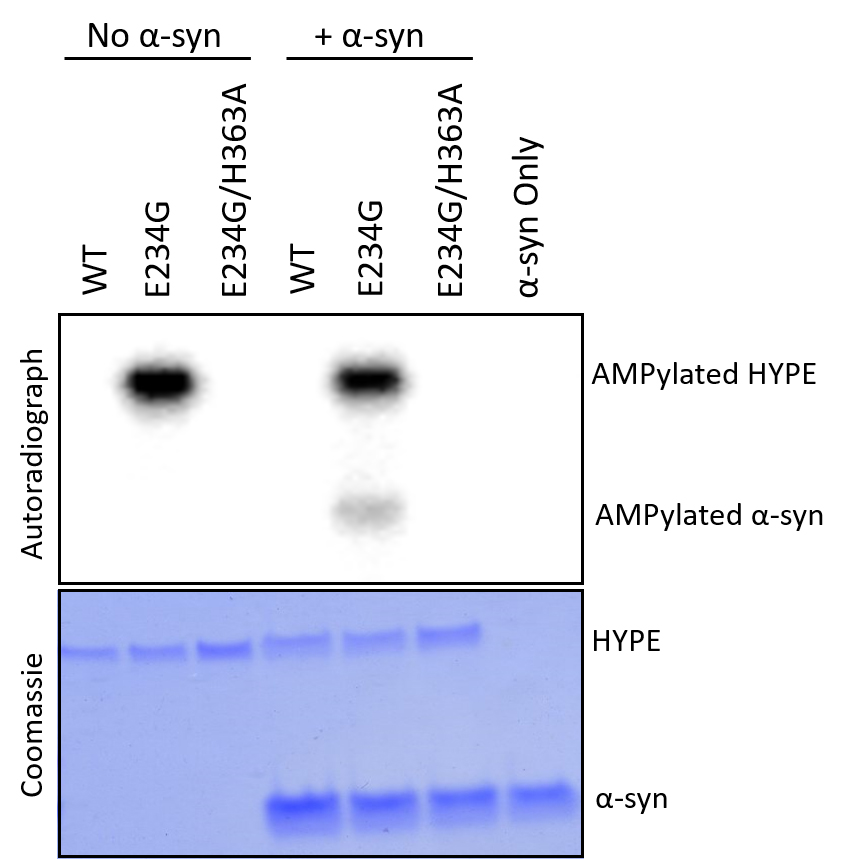
Figure 1. HYPE AMPylates α-syn in vitro. AMPylation competent E234G-HYPE robustly AMPylates α-syn and itself (auto-AMPylation) with α-32P-AMP (top). Intrinsically autoinhibited WT-HYPE and a catalytically dead E234G/H363A mutant show no auto-AMPylation or substrate AMPylation (top). 1 μg of enzyme and 10 μg of substrate from the AMPylation reaction were loaded on the gel (bottom). Phosphor screen was exposed for 3 h (top).
- Once exposure of the Phosphor screen to the gel is complete, incubate the gel in Coomassie Stain with a rocker set to low to medium speed for 10 min.
Recipes
Note: Dissolve the following solutions in ddH2O. Always add solvent before addition of other components. No pH adjustments are required unless explicitly stated. Adjust pH to appropriate value with 12.1 N HCl or 50% NaOH with pipette and rubber suction bulb. Diluted acid or base may also be prepared to avoid overshooting target pH.
- Protein Storage Buffer
50 mM Tris base
300 mM NaCl
10% (v/v) glycerol
Adjust to pH 7.5 - 10x AMPylation Buffer
50 mM HEPES
10 mM MnCl2
Adjust to pH 7.5 - 4x SDS-PAGE Loading Buffer
2.86 M BME
250 mM Tris, pH 6.8
10% (w/v) SDS
0.008% (v/v) bromophenol blue
40% (v/v) glycerol - 1x SDS-PAGE Running Buffer
0.3% Tris base
1.4% glycine
0.1% (w/v) SDS - Coomassie Stain
50% (v/v) methanol
10% (v/v) acetic acid
0.1% Coomassie Brilliant Blue - Destain
50% (v/v) methanol
10% (v/v) acetic acid
Acknowledgments
This publication was made possible with partial support from the Indiana CTSI TL1 Predoctoral Fellowship (Grant # UL1TR002529 [A. Shekhar, PI], 5/18/2018-4/30/2023, and Grant # TL1TR002531 [T. Hurley, PI], 5/18/2018-4/30/2023, from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award) to A.C.; and Eli Lilly-Stark Neuroscience Research Institute-CTSI predoctoral fellowship to S.D.; a grant from the Branfman Family Foundation to J.C.R.; and the National Institute of General Medical Sciences of the National Institute of Health (R01GM10092), an Indiana Clinical and Translational Research Grant (CTSI-106564), and a Purdue Institute for Inflammation, Immunology, and Infectious Disease Core Start Grant (PI4D-209263) to S.M. The authors would also like to thank members of the Rochet and Mattoo laboratories for their helpful discussions.
Competing interests
The authors declare no conflicting or competing interests.
References
- Chen, L. and Feany, M. B. ( 200 5). Alpha-synuclein phosphorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson disease. Nat Neurosci 8(5): 657-663.
- Dedic, E., Alsarraf, H., Welner, D. H., Ostergaard, O., Klychnikov, O. I., Hensbergen, P. J., Corver, J., van Leeuwen, H. C. and Jorgensen, R. (2016). A Novel Fic (Filamentation Induced by cAMP) protein from clostridium difficile reveals an inhibitory motif-independent adenylylation/AMPylation mechanism. J Biol Chem 291(25): 13286-13300.
- Engel, P., Goepfert, A., Stanger, F. V., Harms, A., Schmidt, A., Schirmer, T. and Dehio, C. (2012). Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins. Nature 482(7383): 107-110.
- Ham, H., Woolery, A. R., Tracy, C., Stenesen, D., Kramer, H. and Orth, K. (2014). Unfolded protein response-regulated Drosophila Fic (dFic) protein reversibly AMPylates BiP chaperone during endoplasmic reticulum homeostasis. J Biol Chem 289(52): 36059-36069.
- Iwai, A., Masliah, E., Yoshimoto, M., Ge, N., Flanagan, L., de Silva, H. A., Kittel, A. and Saitoh, T. (1995). The precursor protein of non-A beta component of Alzheimer's disease amyloid is a presynaptic protein of the central nervous system. Neuron 14(2): 467-475.
- Levine, P. M., Galesic, A., Balana, A. T., Mahul-Mellier, A.-L., Navarro, M. X., De Leon, C. A., Lashuel, H. A. and Pratt, M. R. (2019). α-Synuclein O-GlcNAcylation alters aggregation and toxicity, revealing certain residues as potential inhibitors of Parkinson’s disease. Proc Natl Acad Sci U S A 116(5): 1511-1519.
- Lewallen, D. M., Steckler, C. J., Knuckley, B., Chalmers, M. J. and Thompson, P. R. (2012). Probing adenylation: using a fluorescently labelled ATP probe to directly label and immunoprecipitate VopS substrates. Mol Biosyst 8(6): 1701-1706.
- Mattoo, S., Durrant, E., Chen, M. J., Xiao, J., Lazar, C. S., Manning, G., Dixon, J. E. and Worby, C. A. (2011). Comparative analysis of Histophilus somni immunoglobulin-binding protein A (IbpA) with other fic domain-containing enzymes reveals differences in substrate and nucleotide specificities. J Biol Chem 286(37): 32834-32842.
- Preissler, S., Rato, C., Perera, L., Saudek, V. and Ron, D. (2017). FICD acts bifunctionally to AMPylate and de-AMPylate the endoplasmic reticulum chaperone BiP. Nat Struct Mol Biol 24(1): 23-29.
- Sanyal, A., Chen, A. J., Nakayasu, E. S., Lazar, C. S., Zbornik, E. A., Worby, C. A., Koller, A. and Mattoo, S. (2015). A novel link between Fic (filamentation induced by cAMP)-mediated adenylylation/AMPylation and the unfolded protein response. J Biol Chem 290(13): 8482-8499.
- Sanyal, A., Dutta, S., Camara, A., Chandran, A., Koller, A., Watson, B. G., Sengupta, R., Ysselstein, D., Montenegro, P., Cannon, J., Rochet, J. C. and Mattoo, S. (2019). Alpha-synuclein is a target of fic-mediated adenylylation/AMPylation: possible implications for parkinson's disease. J Mol Biol 431(12): 2266-2282.
- Spillantini, M. G., Schmidt, M. L., Lee, V. M., Trojanowski, J. Q., Jakes, R. and Goedert, M. (1997). Alpha-synuclein in Lewy bodies. Nature 388(6645): 839-840.
- Truttmann, M. C., Cruz, V. E., Guo, X., Engert, C., Schwartz, T. U. and Ploegh, H. L. (2016). The Caenorhabditis elegans protein FIC-1 is an AMPylase that covalently modifies heat-shock 70 family proteins, translation elongation factors and histones. PLoS Genet 12(5): e1006023.
- Truttmann, M. C. and Ploegh, H. L. (2017). In vitro AMPylation assays using purified, recombinant proteins. Bio-protocol 7(14): e2416.
- Veyron, S., Peyroche, G. and Cherfils, J. (2018). FIC proteins: from bacteria to humans and back again. Pathog Dis 76(2).
- Weinreb, P. H., Zhen, W., Poon, A. W., Conway, K. A. and Lansbury, P. T., Jr. (1996). NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry 35(43): 13709-13715.
- Worby, C. A., Mattoo, S., Kruger, R. P., Corbeil, L. B., Koller, A., Mendez, J. C., Zekarias, B., Lazar, C. and Dixon, J. E. ( 200 9). The fic domain: regulation of cell signaling by adenylylation. Mol Cell 34(1): 93-103.
- Yamin, G., Uversky, V. N. and Fink, A. L. ( 200 3). Nitration inhibits fibrillation of human alpha-synuclein in vitro by formation of soluble oligomers. FEBS Lett 542(1-3): 147-152.
- Yarbrough, M. L., Li, Y., Kinch, L. N., Grishin, N. V., Ball, H. L. and Orth, K. ( 200 9). AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323(5911): 269-272.
- Ysselstein, D., Dehay, B., Costantino, I. M., McCabe, G. P., Frosch, M. P., George, J. M., Bezard, E. and Rochet, J. C. (2017). Endosulfine-alpha inhibits membrane-induced alpha-synuclein aggregation and protects against alpha-synuclein neurotoxicity. Acta Neuropathol Commun 5(1): 3.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Camara, A., Sanyal, A., Dutta, S., Rochet, J. and Mattoo, S. (2020). In vitro AMPylation/Adenylylation of Alpha-synuclein by HYPE/FICD. Bio-protocol 10(18): e3760. DOI: 10.21769/BioProtoc.3760.
Category
Neuroscience > Cellular mechanisms > Protein isolation
Biochemistry > Protein > Quantification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









