- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ratiometric Measurement of Protein Abundance after Transient Expression of a Transgene in Nicotiana benthamiana
Published: Vol 10, Iss 17, Sep 5, 2020 DOI: 10.21769/BioProtoc.3747 Views: 5982
Reviewed by: Shin‐nosuke HashidaGazala AmeenAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Agrobacterium-Mediated Transient Gene Expression Optimized for the Bioenergy Crop Camelina sativa
Pawan Kumar [...] Jean T. Greenberg
Apr 5, 2024 2114 Views

Identification of S-locus F-box Protein Sequences in Diploid Potato, Solanum okadae, via Degenerate PCR
Amar Hundare [...] Timothy P. Robbins
Jun 5, 2025 1995 Views

Quantitative Analysis of the Arabidopsis Leaf Secretory Proteome via TMT-Based Mass Spectrometry
Sakharam Waghmare [...] Rucha Karnik
Nov 20, 2025 2020 Views
Abstract
Ratiometric reporters are tools to dynamically measure the relative abundance of a protein of interest. In these systems, a target protein fused to a fluorescent or bioluminescent reporter is expressed with fixed stoichiometry to a reference protein fused to a second reporter. Both fusion proteins are encoded on a single transcript but are separated during translation by a 2A “self-cleaving” peptide. This approach enables changes in the relative abundance of a target protein to be detected sensitively, reducing variability in expression of the ratiometric reporter transgene that may occur across different tissues or transformation events. We recently developed a set of Gateway-compatible plant transformation vectors termed pRATIO that combine a variety of promoters, fluorescent and bioluminescent reporters, and 2A peptides derived from foot-and-mouth disease virus. Here, we describe in detail how to use the dual-fluorescent ratiometric reporter pRATIO3212 to examine the relative abundance of a target protein after transient expression in Nicotiana benthamiana leaves. For this example, we analyze degradation of the SUPPRESSOR OF MAX2 1 (SMAX1) protein from Arabidopsis thaliana in response to treatments with karrikins and rac-GR24. This protocol provides a simple, rapid, and readily scalable method for in vivo analysis of relative protein abundance in Agrobacterium-infiltrated Nicotiana leaf tissues.
Keywords: Ratiometric reporterBackground
Karrikins (KARs) are butenolide compounds found in smoke that stimulate seed germination and enhance seedling photomorphogenesis of Arabidopsis thaliana (Flematti, 2004; Nelson et al., 2009, 2010 and 2012). KAR responses in Arabidopsis require the α/β-hydrolase KARRIKIN INSENSITIVE2/HYPOSENSITIVE TO LIGHT (KAI2/HTL) (Sun and Ni, 2011; Waters et al., 2012). KAI2 can bind KAR1 in vitro and is thought to function as a KAR receptor (Guo et al., 2013). KAI2 works with the F-box protein MORE AXILLARY GROWTH2 (MAX2) to mediate KAR responses, likely through polyubiquitination and degradation of SUPPRESSOR OF MAX2 1 (SMAX1) (Nelson et al., 2011; Stanga et al., 2013; Waters et al., 2017). This signaling mechanism is highly similar to that of the plant hormone strigolactone (SL). In SL signaling, DWARF14 (D14)/DECREASED APICAL DOMINANCE2 (DAD2), which is an ancient paralog of KAI2, works with MAX2 to target a subset of SMAX1-LIKE proteins for degradation (Hamiaux et al., 2012; Waters et al., 2012; Jiang et al., 2013; Zhou et al., 2013; Soundappan et al., 2015; Wang et al., 2015). Interestingly, KAI2 and D14 are both responsive to rac-GR24, a commonly used racemate of synthetic SL analogs. However, they show different preferences for the two enantiomers in this mixture, only one of which has a stereochemical configuration that mimics natural SLs (Scaffidi et al., 2014; Waters et al., 2015; Flematti et al., 2016).
In addition to mediating KAR responses, there is growing evidence that the primary role of KAI2 is the recognition of an unknown endogenous signal known as KAI2 ligand (KL) (Conn and Nelson, 2015). Identification of KL will require a highly specific assay for the activation of KAI2. Recently, we found evidence that SMAX1 degradation occurs after KAR and rac-GR24 treatment, and this response requires MAX2 and KAI2 (Khosla et al., 2020b). This observation led us to develop a bioassay for SMAX1 degradation that is appealing as a direct and specific readout of KAI2 activation.
We generated a series of ratiometric reporter vectors (pRATIO) that can be used to assess protein abundance in vivo by monitoring the relative abundance of two co-expressed fluorescent or bioluminescent reporters, one of which is fused to a target protein of interest (Khosla et al., 2020a). Rather than express a pair of target and reference genes from two promoters on a single vector or two co-transformed vectors, in the pRATIO system the target and reference genes are transcribed on the same mRNA. Importantly, pRATIO vectors can normalize for differences in transformation efficiency or transgene expression across samples by encoding a 2A ribosomal skipping peptide from foot-and-mouth disease virus (FMDV) between the target and reference genes. During translation, the nascent 2A peptide blocks the ribosomal exit channel. This disrupts formation of the bond between the C-terminal Gly and Pro residues of the 2A peptide. Translation can resume on the mRNA, producing two proteins in near stoichiometric ratios (Luke and Ryan, 2018).
Here we present a detailed protocol for using the pRATIO system. As an example, we monitor KAR- and rac-GR24- induced proteolysis of Arabidopsis SMAX1 that has been transiently expressed in Nicotiana benthamiana.
Materials and Reagents
- CorningTM 96-Well Black Polystyrene Microplates (Fisher, catalog number: 07-200-590 )
- 5 ml Centrifuge tubes (VWR, catalog number: 10002-731 )
- Polystyrene culture tubes (VWR, catalog number: 60818-703 )
- 1 ml Norm-Ject Syringes (Fisher, catalog number: 1481725 )
- 50 ml Falcon tubes (Genesee Scientific, catalog number: 28-108 )
- 24-Well, Flat Bottom plates (Olympus, catalog number: 25-102 )
- 96-Well Microplates, Clear (Greiner Bio-One, catalog number: 655801 )
- Potting soil (Professional Growing Mix, sungro Horticulture) supplemented with Gnatrol WDG, Marathon (imidacloprid), and Osmocote 14-14-14 fertilizer
- Wild type Nicotiana benthamiana seeds (seeds kindly provided by Dr. Martha Orozco-Cardenas; Plant Transformation Research Center)
- Agrobacterium tumefaciens strain GV3101 strain with helper plasmid pMP90 (Koncz and Schell, 1986; kindly provided by Dr. Meng Chen, University of California, Riverside)
- pRATIO3212-SMAX1 plasmid (Khosla et al., 2020b)
- Agrobacterium tumefaciens strain GV3101 with helper plasmid pMP90 (Koncz and Schell, 1986) carrying pBIN61-p19 construct (Habibi et al., 2018)
- Antibiotics
Gentamicin Sulfate (GoldBio, catalog number: G-400-10 )
Spectinomycin Dihydrochloride Pentahydrate (GoldBio, catalog number: S-140-5 )
Kanamycin Monosulfate (GoldBio, catalog number: K-120-10 )
Rifampicin (GoldBio, catalog number: R-120-1 ) - Magnesium chloride hexahydrate (MgCl2·6H2O) (Fisher, catalog number: 14222322 )
- 2-(N-Morpholino) ethanesulfonic acid (MES) (Fisher, catalog number: BP300100 )
- Potassium hydroxide (KOH) (Fisher, catalog number: P250-500 )
- Acetosyringone (Fisher, catalog number: AC115540010 )
- Dimethyl Sulfoxide (DMSO) (Fisher, catalog number: BP231-100 )
- Luria-Bertani (LB) broth (Fisher, catalog number: BP9723-2 )
- Bacteriological agar (Sigma, catalog number: A5306-1KG )
- Solid LB plates with 1.5% (w/v) agar
- KAR1
- KAR2
- rac-GR24
- Acetone
- MgCl2 (1 M stock) (see Recipes)
- MES (0.5 M stock) (see Recipes)
- Acetosyringone (1 M stock) (see Recipes)
- Infiltration Buffer (see Recipes)
- KAR1, KAR2, rac-GR24 (50 mM stock) (see Recipes)
Notes:- KAR2 was synthesized as previously reported (Goddard-Borger et al., 2007). rac-GR24 was synthesized according to Mangus et al. and recrystallized from diethyl ether/hexanes (Mangnus et al., 1992).
- The chemicals can be purchased from several commercial suppliers: KAR1: Toronto Research Chemicals (M305480), Chiralix (CX27716); KAR2: Toronto Research Chemicals (F864800), Chiralix (CX94877); rac-GR24: Chiralix (CX23880), Strigolab (ST23b rac), PhytoTech Labs (G3324), Toronto Research Chemicals (S687590).
- 10 µM working chemical stocks (see Recipes)
- 0.02% (v/v) acetone control (see Recipes)
Equipment
- Centrifuge (Eppendorf, model: 5804 R )
- 30 °C incubator shaker (ThermoFisher, model: MaxQ 6000 R )
- CLARIOstar microplate reader (BMG Labtech)
- 4 mm Round Ticket Hole Punch
- Dumont #5 Fine Forceps (Fine Science Tools, catalog number: 11252-20 )
- 1 mm electroporation cuvette (Genesee Scientific, catalog number: 40-100 )
- MicroPulser Electroporator (Bio-Rad, catalog number: 165-2100 )
- Vacuum-driven filter system (Olympus, catalog number: 25-227 )
- 3.5-inch square plastic pots (Farrand, catalog number: K0-25SQ-G )
Software
- Prism (v8.2.0, GraphPad Software Inc.)
Procedure
- Transformation of Agrobacterium tumefaciens
- Transform pRATIO3212-SMAX1 into Agrobacterium tumefaciens strain GV3101 by electroporation method (Mersereau et al., 1990).
- Select the transformants on LB agar plates [containing 50 µg/ml Spectinomycin (expression construct), 25 µg/ml Gentamicin (helper plasmid), and 25 µg/ml Rifampicin (genomic)] at 30 °C for 2-3 days.
- Propagation of Nicotiana benthamiana
- Germinate ~50-100 seeds in a 3.5-inch square plastic pot containing potting soil at ~21-24 °C and 16 h light/8 h dark photoperiod with ~80-100 μmol/m2s light intensity. Cover the pot with a plastic cover to ensure high humidity.
- As soon as the first true leaves emerge (10-12 days), transfer the seedlings to 3.5-inch plastic pots containing potting soil (two seedlings per pot) and cover again for 4 days with a plastic cover. Remove cover and maintain in growth conditions described above.
- Water plants every second day or as required.
- At 3-3.5 weeks after transplanting, the plant has reached the optimal developmental stage for agroinfiltration.
Note: It is best to stop watering plants one day before the infiltration.
- Transient expression
- Culture A. tumefaciens carrying pRATIO3212-SMAX1 or pBIN61-p19 (hereafter, p19) in 2-3 ml liquid LB medium containing 25 µg/ml Gentamicin (helper plasmid), 25 µg/ml Rifampicin (genomic), 50 µg/ml Spectinomycin (pRATIO3212-SMAX1), 50 µg/ml Kanamycin (p19); grow overnight at 30 °C incubator, 200-220 rpm.
Notes:- The presence of p19 partially blocks RNA silencing and thus establishes a higher and more uniform transgene expression.
- For each transformed strain, prepare glycerol stocks by mixing 0.5 ml of culture with 0.5 ml of 50% glycerol. Store at -80 °C.
- Dilute the starter culture 1:1,000 in 15 ml LB + antibiotics in a 50 ml Falcon tube. Grow overnight at 30 °C incubator, 200 -220 rpm.
Note: Perform this step in the evening. The culture volume can be scaled up depending on the number of transformations intended. However, we recommend keeping the culture volume less than 30 ml in a 50 ml Falcon tube for proper aeration. - Pellet cells at 3,900 x g, 10 min at room temperature, remove supernatant, add 10 ml of infiltration media (see Recipes) and resuspend.
- Repeat Step C3, resuspend the pelleted cells in ~3-4 ml of infiltration media.
Note: These washes remove traces of antibiotics that can kill the leaf tissue after infiltration. - Measure absorbance of the culture at 600 nm. Add 100 µl Agrobacterium culture to a clear 96-well plate and measure the optical density at 600 nm (OD600) with a microplate reader.
Notes:- Blank the plate reader with a well containing infiltration media and use blank subtracted OD600 readings for further calculations.
- Alternatively, OD600 can be measured in a 1 ml plastic cuvette using a spectrophotometer.
- Determine the titer of culture required for infiltration and calculate the dilution factor for a final volume of 4.0-5.0 ml of culture in infiltration media, depending on the number of transformations intended. For example, if you require a final OD600 of 0.6 and culture reads 1.5, then you will need initial volume = (final OD600 x final volume)/initial OD600. Therefore, the required initial volume = (0.6 x 4 ml)/1.5 = 1.6 ml.
Note: In this protocol, SMAX1 and p19 were diluted to OD600 of 0.6 and 0.4, respectively, in the final infiltration volume of 4.0 ml. However, the optimum ratio might be different for other proteins. The final OD600 should not exceed 1.2 as this induces tissue necrosis, while OD600 of 0.3 and lower on the other hand drastically reduces transient expression efficiencies (Kim et al., 2009) (see Note 1). - Vortex and incubate the cultures for 4 h at room temperature (without shaking).
- Choose the proper leaves for infiltration. Select the healthiest looking plants and avoid leaves that are torn or otherwise damaged. The ideal leaves are young leaves that are ~3-5 cm wide. Use 2-3 leaves per plant and at least 3 leaves (from independent plants) per construct.
Notes:- The developmental stage of the Nicotiana benthamiana plants is important for the success of this method. Healthy plants should have at least two large leaves, which may be the third or fourth leaf from the apical meristem.
- Avoid using small leaves as they are harder to infiltrate and display reduced transformation rates.
- Plants should not have flowered at the time of infiltration.
- Swirl the solution in the tube before drawing it up into the syringe, in order to evenly suspend the Agrobacterium. Press the nozzle of a 1 ml syringe without a needle against the abaxial (lower) surface of the leaf and hold a gloved finger on the other side and inject slowly. Try to be as gentle as possible when injecting, to avoid damaging the leaf. The infiltrated area appears as a dark, water-soaked region. If the infiltrated region stops expanding without pushing too hard, infiltrate again in a different area of the leaf. After infiltration, pat dry the leaf surface with a Kimwipe and mark the edge of the infiltrated circle with a Sharpie permanent marker. Mark the leaf by attaching a small tag to the petiole.
Notes:- (Optional) Before infiltration, a small incision can be made at the site of infiltration using a sterile razor. Make sure that the wounds do not perforate the leaf, which could decrease the efficiency of the infiltration.
- We usually do not infiltrate more than one construct per leaf.
- It is not necessary to infiltrate the total leaf space. We typically infiltrate 2-3 spots in a single leaf.
- If you are infiltrating leaves with different constructs on the same plant, you should clean your gloves with 70% ethanol (or wipe them with Kimwipe) in between infiltrations and ensure that the leaves are not touching each other to prevent cross-contamination.
- We suggest using leaves transformed with p19 only (untransformed leaves are less preferable) as negative controls.
- Return infiltrated N. benthamiana plants to the growth room, water them, and incubate for 3 days under normal growth conditions.
- Culture A. tumefaciens carrying pRATIO3212-SMAX1 or pBIN61-p19 (hereafter, p19) in 2-3 ml liquid LB medium containing 25 µg/ml Gentamicin (helper plasmid), 25 µg/ml Rifampicin (genomic), 50 µg/ml Spectinomycin (pRATIO3212-SMAX1), 50 µg/ml Kanamycin (p19); grow overnight at 30 °C incubator, 200-220 rpm.
- Degradation Assay
- Before setting up treatment plates, it is necessary to verify the expression of the reporter proteins.
- Prepare a black 96-well microplate (low autofluorescence) and add 300 µl dH2O in the wells you are going to use. Excise a single leaf disc from each transformed (and p19 only or untransformed) leaf using a hole punch and place them in a microplate abaxial (i.e., the leaf underside) side up. Read the plate as per section E to identify those leaves that show a good ratio of transformed over p19 (or untransformed) signal, and proceed with these samples (see Notes 2 and 3).
Note: This step is critical to identify leaves suitable for the degradation assay (Steps D3-D6). - Prepare a 24-well treatment plate for sample harvesting and treatments (Figure 1). Label wells and add 1 ml of freshly prepared 10 µM KAR1, KAR2, rac-GR24, or 0.02% (v/v) acetone control (see Recipe 9) to each well.
- Excise 5 or 6 leaf discs per leaf per treatment from within the infiltrated region/circle and float them (abaxial side down) on chemical treatments. Place no more than 4 discs in each well.
Notes:- It is critical to minimize mechanical damage to the leaf during handling (cleanly punch the disc edges and try to avoid the forcep damage). Exclude major veins from the excised discs.
- For each construct, we recommend using at least 3 different leaves from independent plants. Also, include multiple negative control leaf discs per treatment.
- For each leaf, punch out leaf discs at random from lamina on each side of the midrib. When the desired number of leaf discs are collected, they are immediately and randomly transferred to the treatment plates to prevent them from drying. Alternatively, place the discs in distilled water until ready to be transferred.
- For each construct, randomly allocate discs from the same leaf to different treatments rather than allocating discs from one leaf for one treatment and another leaf to a different treatment.
- Cover the plate and incubate at room temperature 12-16 h.
Note: The plate should be carefully placed in the lab drawer (in the dark) and not allowed to remain on the bench top. - Set up a black 96-well microplate with 300 µl dH2O in each well to be used. Transfer one leaf disc per well (abaxial side up) for measurement.
Notes:- Discard leaf discs that appear translucent (water-logged) or had submerged after overnight treatment.
- It is necessary to include negative controls (p19 only or untransformed) on each plate, as these need to be measured with the same gain to perform the background subtraction correctly.
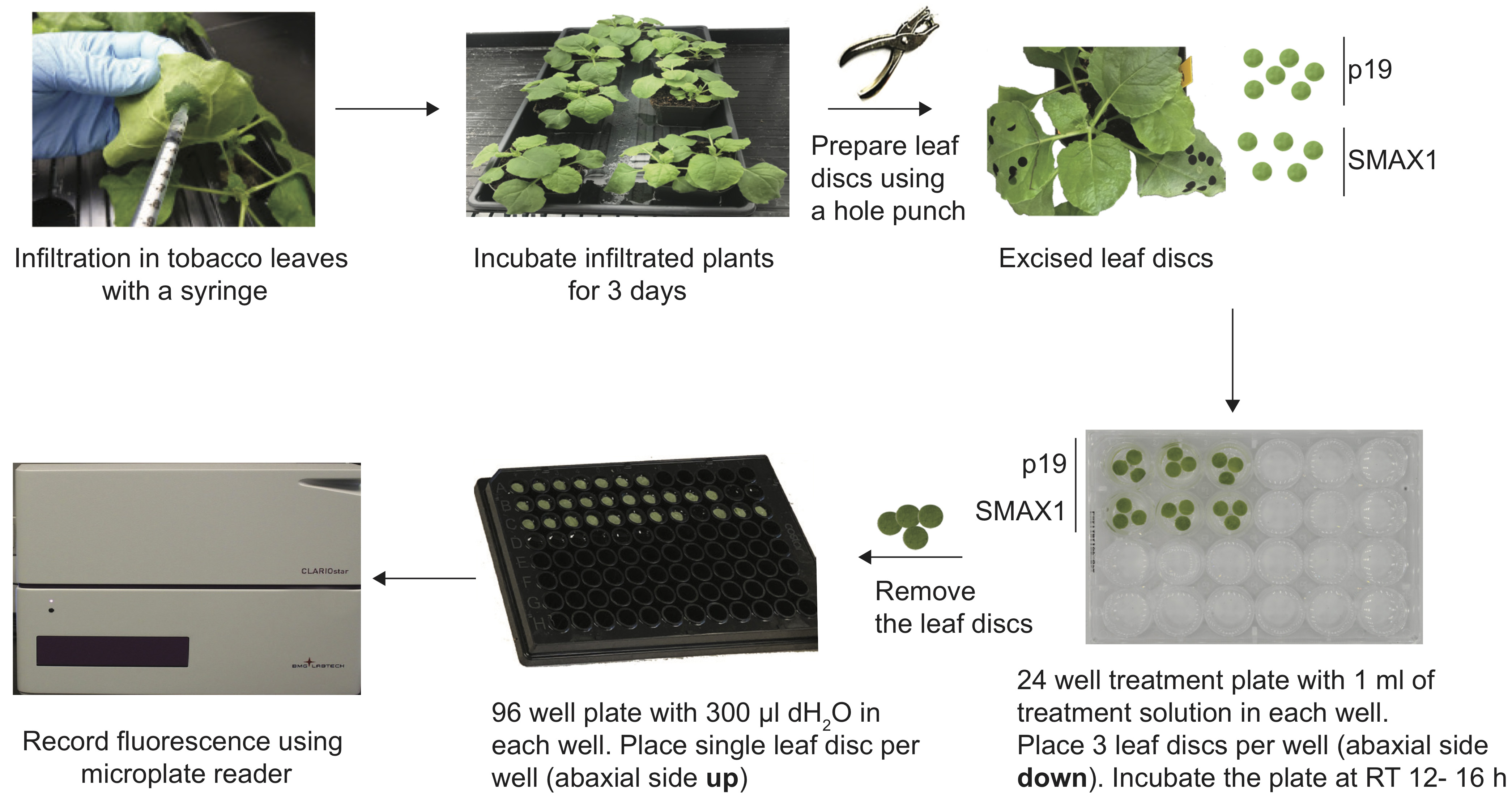
Figure 1. Workflow for ratiometric measurement of SMAX1 abundance in Nicotiana benthamiana. Schematic of the degradation assay using pRATIO3212-SMAX1 ratiometric reporter system in N. benthamiana.
- Quantification of fluorescence intensity
We suggest using the following optimized settings on the CLARIOstar plate reader to measure mScarlet-I and Venus fluorescence.
Note: It would be possible to use these settings with other dual-fluorescent pRATIO reporters (Khosla et al., 2020a).- Basic settings
- Measurement method: Fluorescence intensity
- Reading mode: Well scanning
- Microplate type: The CLARIOstar software contains a database with dimensions of microplates from most microplate manufacturers. In this case, we select “COSTAR BLACK 96” from the pull-down menu.
Note: For measuring fluorescence, we recommend using black plates as they absorb light and reduce background and crosstalk.
- Well scan settings
Note: Well scanning measures different points of a well in a matrix style. The output is an average measurement value of all scan points.- Number of flashes per scan point: 8
Note: For fluorescence measurements using pRATIO reporters., 8-10 flashes are usually adequate. Increasing the number of flashes also increases the reading time. - Scan mode: matrix scan
- Scan matrix dimension: 9 x 9
Note: You can use this option to define the matrix size for the scanning procedure and, therefore, the number of measurement (scan) points. - Scan width [mm]: 4
Note: This value is optimized to reduce the edge effect (see Note 3).
- Number of flashes per scan point: 8
- Optic settings
Note: We used spectral scanning to identify excitation and emission wavelengths for each fluorescent protein (FP) that produced the strongest signal above background autofluorescence of tobacco leaves (Table 1) (Khosla et al., 2020a).
Table 1. Optimal filter settings for tobacco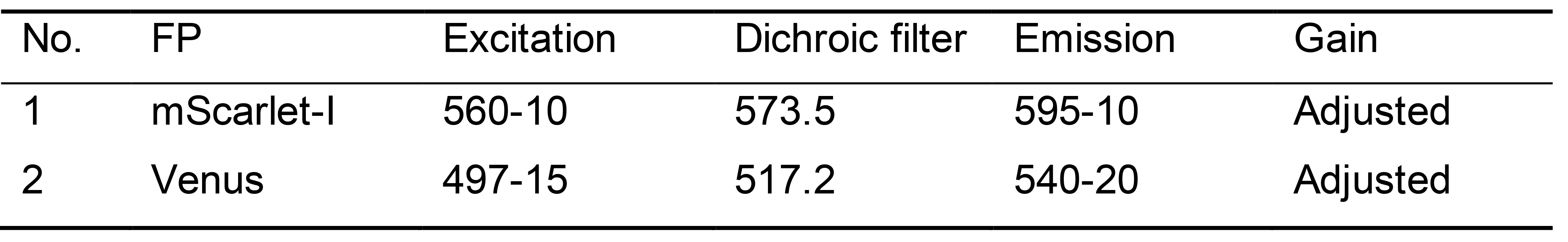
- Gain adjustment: For each fluorophore, we performed an automatic gain adjustment on the entire plate before the measurement. The instrument finds the well with the highest intensity and determines an appropriate gain.
Notes:- The purpose of a gain adjustment is to optimize the signal amplification so that the results have the maximum sensitivity and dynamic range.
- It is preferred to have the highest gain for each signal coming from the same well.
- Focal height [mm]: Perform automatic focal height adjustment on the well with the optimal gain (identified above). For a disc floating on 300 μl water, this is typically ~13 mm.
- Gain adjustment: For each fluorophore, we performed an automatic gain adjustment on the entire plate before the measurement. The instrument finds the well with the highest intensity and determines an appropriate gain.
- General settings
Optic: Top optic used
Note: Top reading relies on measuring reflected light above the wells. For our protocol, we expect top reading to be more sensitive than bottom measurements. Floating leaf discs on the water in 96 well microplates ensure they do not get soaked and sit at the right height for accurate measurement.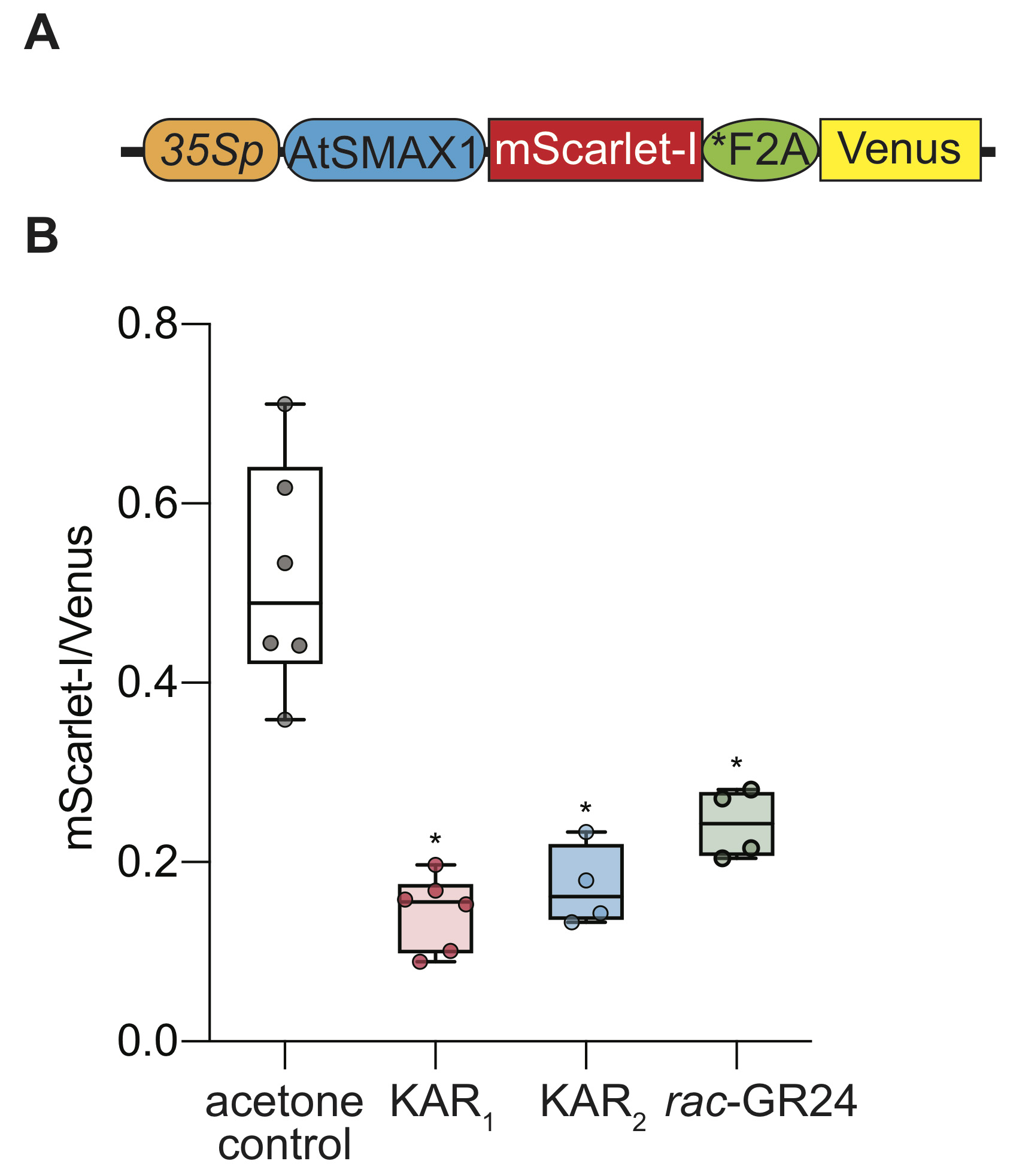
Figure 2. Ratiometric detection of SMAX1 degradation in tobacco. A. Diagram of the pRATIO3212-SMAX1 ratiometric reporter system. The 35S promoter drives expression of a multicistronic transcript encoding an SMAX1-mScarlet-I fusion that is separately translated from the Venus fluorescent protein due to the *F2A peptide. B. Relative fluorescence of SMAX1 in the pRATIO3212 system after transient expression in tobacco and overnight treatment with 10 μM KAR1, KAR2, rac-GR24, or 0.02% (v/v) acetone control. n = 5-6 leaf discs. *P < 0.01, Mann-Whitney U test comparisons to control treatment. The data shown is representative of one leaf out of three independent leaves with similar results.
- Basic settings
Data analysis
- Normalize the data with the following four steps.
- Average the fluorescence signals obtained with mScarlet-I detection settings for negative control leaf discs transformed with Agrobacterium carrying p19 (or untransformed). Similarly, average the Venus fluorescence values from the wells containing p19 (or untransformed) leaf discs. These two values represent the background signal contributed by leaf autofluorescence for each reporter.
- Subtract the mScarlet-I background value from the mScarlet-I signals obtained for each sample well on the microplate.
- Subtract the Venus background value from the Venus signals obtained for each sample well on the microplate.
- Calculate the fluorescence intensity using the following formula: ([mScarlet-I signals]-[mScalrlet-I background])/([Venus signals]-[Venus background]). After 12 h treatment with 10 μM KAR1, KAR2, or rac-GR24, we observed a > 2-fold decline in the mScarlet-I/Venus ratio in response to all three treatments, indicating degradation of SMAX1 (Figure 2, Supplemental file 1).
- Prism (v8.2.0, GraphPad Software Inc.) is used to perform data visualization and statistical analysis. The Centerline of the box-whisker plot indicates the median, and the colored box represents the interquartile range. The whiskers go down to the smallest value and up to the largest, and each individual value depicted as a point superimposed on the graph.
Notes
- p19 GV3101 strain
If you want to co-express multiple proteins, we recommend transforming the pRATIO construct into a strain of GV3101 that is already carrying p19. This minimizes the amount of Agrobacterium used for infiltration. It can also lead to less variation than you may get from mixing several strains of bacteria. - We do not recommend using leaves that show mScarlet-I/Venus ratio less than 0.3 for degradation analysis. To calculate mScarlet-I/Venus ratio, perform data normalization (see DATA ANALYSIS section) using a negative control leaf with the lowest signal for each mScarlet-I and Venus for background subtraction.
- The gain and focal height settings determined in this step must be the same settings used when reading discs after overnight chemical treatment.
- Compatibility of other pRATIO vectors
Although we use the pRATIO3212 ratiometric system as an example in this study, the protocol works equally well with pRATIO vectors containing bioluminescent reporters (Khosla et al., 2020a) in both tobacco and Arabidopsis protoplast transient assays. When using bioluminescent reporters, we suggest using white or gray plates as they reflect light, which will maximize the light output signal. - Comparison of different reading modes
In this protocol, we measured fluorescence with well scan mode. Besides well scanning, fluorescence intensity can be measured by CLARIOstar using endpoint, orbital, and spiral averaging modes. A center endpoint reading excites and measures only in the center of the well, giving a single number. This can lead to inaccuracies as the disc may move between time points. When using spiral and orbital averaging modes, the plate reader takes multiple measurements for each well on a defined orbit, giving one averaged number over the entire area of the well. Well scanning measures different points of a well in a matrix style. Although both spiral averaging and well scanning yielded comparable sensitivity, well scanning is more versatile as it provides raw data value from single scan points. It is also possible to define thresholds to exclude scan points valid for all wells. Furthermore, the leaf disc harvest can sometimes induce edge damage, resulting in higher background signals. Using the scanning mode, you can define the diameter, such as measurement points that are inside the circle with the defined diameter will be measured, thereby excluding potential inaccurate readings. One limitation, however, is that well scanning is slower than orbital or spiral averaging modes. - Performing a time course assay
Our protocol is also suitable for performing a time course study. To examine the time course of degradation, set up a black 96-well microplate with 300 µl chemical treatments and transfer one leaf disc per well with the abaxial side up. Use at least 12 leaf discs (4 discs per leaf x 3 leaves). Measure relative fluorescence at the indicated time points in the plate mode (slow kinetics) with spiral averaging option. Define number of cycles as the number of times the entire plate will be measured and cycle time, which is the amount of time it takes to measure the plate during one cycle. For example, if you want to read a plate every 15 min per hour for 6 h, then cycle time will be 300 s and the number of cycles 24. However, there are likely to be some limitations when performing a longer time course experiment (> 6 h) using this method, foremost of which is the leaf discs sinking to the bottom of the well, and therefore decreasing the efficiency of measurement in top reading microplate readers. Alternatively, perform the assay as described in this study. At the indicated time points, discs are transferred to 96-well plate for fluorescence measurement and then returned to 24-well treatment plates until the next time point. Careful handling is important to minimize damage to the discs.
Recipes
- MgCl2 (1 M stock)
- Dissolve 101.65 g of MgCl2·6H2O in 400 ml of deionized H2O
- Adjust the volume to 0.5 L with deionized water
- Filter-sterilized by vacuum filtration
- MES (0.5 M stock)
- Dissolve 19.52 g MES in 200 ml deionized water
- Adjust to pH 5.7 with 1 N KOH
- Filter-sterilized by vacuum filtration
- Acetosyringone (1 M stock)
- Dissolve 0.196 g acetosyringone in 1 ml of DMSO
- Divide into single use aliquots are store at -20 °C
- Infiltration buffer
Note: Infiltration medium is made fresh from the stock solutions on the day of infiltration and kept at room temperature until required.
10 mM MgCl2
10 mM MES
150 μM acetosyringone - KAR1 (50 mM stock)
Dissolve 3.75 mg of KAR1 in 500 µl acetone and store at -20 °C. - KAR2 (50 mM stock)
Dissolve 3.4 mg of KAR2 in 500 µl acetone and store at -20 °C. - rac-GR24 (50 mM stock)
Dissolve 7.45 mg of rac-GR24 in 500 µl acetone and store at -20 °C.
Note: Acetone stocks (5-7) may evaporate during storage, so we use amber color glass vials with screw thread cap and wrap the vials with Parafilm for long term storage. Allow the tubes to warm up to room temperature before opening to avoid introduction of atmospheric moisture, which can hydrolase GR24. - 10 µM working chemical stocks (freshly prepared)
2 µl 50 mM stock
Dilute with 9,998 µl dH2O - 0.02% (v/v) acetone control (freshly prepared)
2 µl of acetone
Dilute with 9,998 µl dH2O
Acknowledgments
This protocol is a detailed expansion of that described in Khosla et al., 2020b. We gratefully acknowledge funding support from NSF IOS-1737153 and NSF IOS-1740560 to DCN. We thank Dr. Gavin Flematti and Dr. Adrian Scaffidi (University of Western Australia) for chemical synthesis and purification of KARs and rac-GR24. We thank Dr. Mark Waters (University of Western Australia) for valuable feedback on an early draft of this protocol.
Competing interests
We declare no competing interests.
References
- Conn, C. E. and Nelson, D. C. (2015). Evidence that KARRIKIN-INSENSITIVE2 (KAI2) Receptors may Perceive an Unknown Signal that is not Karrikin or Strigolactone. Front Plant Sci 6(1219).
- Flematti, G., Ghisalberti, E., Dixon, K. and Trengove, R. (2004). A Compound from Smoke That Promotes Seed Germination. Science 305(5686): 977.
- Flematti, G. R., Scaffidi, A., Waters, M. T. and Smith, S. M. (2016). Stereospecificity in strigolactone biosynthesis and perception. Planta 243(6): 1361-1373.
- Goddard-Borger, E. D., Ghisalberti, E. L. and Stick, R. V. (2007). Synthesis of the germination stimulant 3-methyl-2h-furo[2,3-c]pyran-2-one and analogous compounds from carbohydrates. Eur J Organic Chemistry 38(49): 3925-3934.
- Guo, Y., Zheng, Z., La Clair, J. J., Chory, J. and Noel, J. P. (2013). Smoke-derived karrikin perception by the α/β-hydrolase KAI2 from Arabidopsis. Proc Natl Acad Sci U S A 110(20): 8284-8289.
- Hamiaux, C., Drummond, R., Janssen, B., Ledger, S., Cooney, J., Newcomb, R. and Snowden, K. (2012). DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Current Biology 22(21): 2032-2036.
- Habibi, P., Soccol, C. R., O’Keefe, B. R., Krumpe, L. R. H., Wilson, J., de Macedo, L. L. P., Faheem, M., Dos Santos, V. O., Prado, G. S., Botelho, M. A., Lacombe, S. and Grossi-de-Sa, M. F. (2018). Gene-silencing suppressors for high-level production of the HIV-1 entry inhibitor griffithsin in Nicotiana benthamiana. Process Biochemistry 70: 45-54.
- Jiang, L., Liu, X., Xiong, G., Liu, H., Chen, F., Wang, L., Meng, X., Liu, G., Yu, H., Yuan, Y., Yi, W., Zhao, L., Ma, H., He, Y., Wu, Z., Melcher, K., Qian, Q., Xu, H. E., Wang, Y. and Li, J. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504(7480): 401-405.
- Khosla, A., Rodriguez-Furlan, C., Kapoor, S., Van Norman, J. M. and Nelson, D. C. (2020a). A series of dual-reporter vectors for ratiometric analysis of protein abundance in plants. bioRxiv: 2020.2002.2010.939363.
- Khosla, A., Morffy, N., Li, Q., Faure, L., Chang, S. H., Yao, J., Zheng, J., Cai, M. L., Stanga, J. P., Flematti, G. R., Waters, M. and Nelson, D. C. (2020b). Structure-Function Analysis of SMAX1 Reveals Domains that Mediate its Karrikin-Induced Proteolysis and Interaction with the Receptor KAI2. The Plant Cell. Doi.org/10.1105/tpc.19.00752.
- Kim, M. J., Baek, K. and Park, C.-M. (2009). Optimization of conditions for transient Agrobacterium-mediated gene expression assays in Arabidopsis. Plant Cell Reports 28: 1159-1167.
- Koncz, C . and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Molecular General Genetics 204(3): 383-396.
- Luke, G.A. and Ryan, M.D. (2018). Using the 2A Protein Coexpression System: Multicistronic 2A Vectors Expressing Gene(s) of Interest and Reporter Proteins. Methods Mol Biol 1755: 31-48.
- Mersereau, M., Pazour, G. J. and Das, A. (1990). Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene 90(1): 149-151.
- Nelson, D. C., Flematti, G. R., Ghisalberti, E. L., Dixon, K. W. and Smith, S. M. (2012). Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annu Rev Plant Biol 63(1): 107-130.
- Nelson, D. C., Flematti, G. R., Riseborough, J. A., Ghisalberti, E. L., Dixon, K. W. and Smith, S. M. (2010). Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc Natl Acad Sci U S A 107(15): 7095-7100.
- Nelson, D. C., Riseborough, J.-A., Flematti, G. R., Stevens, J., Ghisalberti, E. L., Dixon, K. W. and Smith, S. M. (2009). Karrikins discovered in smoke trigger arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol 149(2): 863-873.
- Nelson, D. C., Scaffidi, A., Dun, E. A., Waters, M. T., Flematti, G. R., Dixon, K. W., Beveridge, C. A., Ghisalberti, E. L. and Smith, S. M. (2011). F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci U S A 108(21): 8897-8902.
- Scaffidi, A., Waters, M. T., Sun, Y. K., Skelton, B. W., Dixon, K. W., Ghisalberti, E. L., Flematti, G. R. and Smith, S. M. (2014). Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol 165(3): 1221-1232.
- Soundappan, I., Bennett, T., Morffy, N., Liang, Y., Stanga, J. P., Abbas, A., Leyser, O. and Nelson, D. C. (2015). SMAX1-LIKE/D53 family members enable distinct max2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27(11): 3143-3159.
- Stanga, J. P., Smith, S. M., Briggs, W. R. and Nelson, D. C. (2013). SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol 163(1): 318-330.
- Sun, X. D. and Ni, M. (2011). HYPOSENSITIVE to LIGHT, an alpha/beta fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de-etiolation. Mol Plant 4: 116-126.
- Wang, L., Wang, B., Jiang, L., Liu, X., Li, X., Lu, Z., Meng, X., Wang, Y., Smith, S. M. and Li, J. (2015). Strigolactone signaling in Arabidopsis regulates shoot development by targeting d53-like smxl repressor proteins for ubiquitination and degradation. Plant Cell 27(11): 3128-3142.
- Waters, M. T., Gutjahr, C., Bennett, T. and Nelson, D. C. (2017). Strigolactone signaling and evolution. Annu Rev Plant Biol 68(1): 291-322.
- Waters, M., Nelson, D., Scaffidi, A., Flematti, G., Sun, Y., Dixon, K. and Smith, S. (2012). Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285-1295.
- Waters, M. T., Scaffidi, A., Moulin, S. L. Y., Sun, Y. K., Flematti, G. R. and Smith, S. M. (2015). A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27(7): 1925-1944.
- Zhou, F., Lin, Q., Zhu, L., Ren, Y., Zhou, K., Shabek, N., Wu, F., Mao, H., Dong, W., Gan, L., Ma, W., Gao, H., Chen, J., Yang, C., Wang, D., Tan, J., Zhang, X., Guo, X., Wang, J., Jiang, L., Liu, X., Chen, W., Chu, J., Yan, C., Ueno, K., Ito, S., Asami, T., Cheng, Z., Wang, J., Lei, C., Zhai, H., Wu, C., Wang, H., Zheng, N. and Wan, J. (2013). D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 504(7480): 406-410.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Khosla, A. and Nelson, D. C. (2020). Ratiometric Measurement of Protein Abundance after Transient Expression of a Transgene in Nicotiana benthamiana. Bio-protocol 10(17): e3747. DOI: 10.21769/BioProtoc.3747.
Category
Plant Science > Plant molecular biology > Protein
Molecular Biology > Protein > Targeted degradation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









