- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of the Cellular Ion Concentration in Saccharomyces cerevisiae Using ICP-AES
Published: Vol 10, Iss 16, Aug 20, 2020 DOI: 10.21769/BioProtoc.3727 Views: 3443
Reviewed by: Arvind PandayFabian den BraveFernando A Gonzales-Zubiate

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Determination of Elemental Concentrations in Lichens Using ICP-AES/MS
Liang-Cheng Zhao [...] Hua-Jie Liu
Mar 5, 2017 10934 Views

Induction and Quantification of Patulin Production in Penicillium Species
Yong Chen [...] Shiping Tian
Jun 5, 2017 9069 Views

Intracellular cAMP Measurements in Candida albicans Biofilms
Liuliu Jiang [...] Xin Wei
Dec 5, 2019 4678 Views
Abstract
The yeast Saccharomyces cerevisiae has been perceived over decades as a highly valuable model organism for the investigation of ion homeostasis. Indeed, many of the genes and biological systems that function in yeast ion homeostasis are conserved throughout unicellular eukaryotes to humans. In this context, measurement of the yeast cellular ionic content provides information regarding yeast response to gene deletion or exposure to chemicals for instance. We propose here a protocol that we tested for the analysis of 12 elements (Ba2+, Ca2+, Cd2+, Co2+, Cu2+, Fe2+, K+, Mg2+, Mn2+, Na+, Ni2+, Zn2+) in yeast using Inductively Coupled Plasma-Atomic Emission Spectrometry (ICP-AES). This technique enables determination of the cellular content of numerous ions from one biological sample.
Keywords: Ion contentBackground
Yeast has been extensively used to study the ionic response after gene deletion, modification of the extracellular environment, or heterologous protein production for example. In this context, having methods to monitor the ionic status of yeast cells is of high interest. ICP-AES uses high-energy plasma from an inert gas like argon to burn analytes very rapidly. The color that is emitted from the analyte is indicative of the elements present, and the intensity of the spectral signal is indicative of the concentration of these elements. Compared with other techniques (for example spectrophotometric methods, atomic absorption spectrometry and atomic fluorescence spectrometry), ICP-AES is a multi-ion analysis method involving simple and fast procedure with relatively low detection limits (Dahlquist and Knoll, 1978). The method described here applies to concentration determination of Ba2+, Ca2+, Cd2+, Co2+, Cu2+, Fe2+, K+, Mg2+, Mn2+, Na+, Ni2+, and Zn2+, with the possibility of measurement of numerous cellular ionic concentrations from one 100 ml yeast culture (Eide et al., 2005; Thines et al., 2018). While this protocol has been tested in our laboratory for these twelve ions, this protocol could most likely be transferred to other elements. Besides, although described here for whole-cell measurement of the yeast ionic content, this method could be transferred to other cell types or to isolated organelles by fractionation on sucrose gradient for instance.
Materials and Reagents
- Petri dishes (Sigma-Aldrich, catalog number: P5606-400EA )
- Toothpicks
- 5 ml pipette (Gilson, catalog number: F123607 )
- 25 mm diameter Whatman® glass microfiber filters, grade GF/F (GE Healthcare, catalog number: 1825-025 )
- Saccharomyces cerevisiae strains to be analyzed (performed here on BY4741 or BY4742 S. cerevisiae strains)
- Yeast extract KAT (Ohly, catalog number: OHLY® KAT)
- Glucose (Merck, catalog number: 1083469029 )
- EGTA (Acros Organic, catalog number: 409910250 )
- HCl 37% (VWR, catalog number: 20255.29 )
- Suprapur® nitric acid 65% (Merck, catalog number: 100441 )
- Certipur® single-element standards for inductively coupled plasma spectroscopy (Merck, catalog number: depends on the ion to be analyzed)
- Milli Q water
- NaOH (Sigma-Aldrich, catalog number: S5881 )
- YD plates (see Recipes)
- YD medium (see Recipes)
- 1 mM EGTA (1 L) (see Recipes)
Equipment
- 500 ml Erlenmeyer (VWR, catalog number: 734-1833 )
- 250 ml glass beakers (VWR, catalog number: 213-1124P )
- 100 ml graduated cylinders (VWR, catalog number: 612-3836 )
- HaldenwangerTM porcelain crucibles (Thermo Fisher Scientific, catalog number: 12306507 )
- Analytical balance (Mettler Toledo, catalog number: AB104-S )
- Filter holding manifold (Hoefer, catalog number: FH225V )
- Laboratory oven (Utest, catalog number: UTD )
- Desiccator (VWR, catalog number: 467-0071P )
- Carbolite® muffle furnace (Sigma-Aldrich, catalog number: Z654973 )
- iCAP 6500 ICP-OES CID spectrometer (Thermo Fisher Scientific, discontinued)
Procedure
- Sample preparation
- Streak the S. cerevisiae strains to be analyzed on YD plates (Recipe 1). Incubate for two days at 28 °C.
- Using a sterile toothpick, select individual colonies.
- Re-suspend cells in 100 ml YD medium (in 500 ml Erlenmeyer) (Recipe 2) and grow them at 28 °C under agitation (120 rpm) to an OD600 of 3 (OD600 = 1 corresponds to a density of 1.25 x 107 cells/ml).
Note: Wash the 500 ml Erlenmeyer with 10% HCl before yeast growth to avoid any impact of the detergents used for washing the glassware on the ionic content measurement. Rinse them with Milli Q water and allow them to dry. - Filter the 100 ml culture using glass microfiber filters on the filter holding manifold at 4 °C.
- Wash cells twice on the filter with 2 ml EGTA 1 mM (Recipe 3) at 4 °C.
- Wash cells twice with 2 ml Milli Q water at 4 °C.
- Immerge each filter in a heat-resistant porcelain crucible containing 10 ml Milli Q water for 10 min. Remove the filter from the crucible.
Note: Before introducing any sample in the porcelain crucibles, weigh them when empty for further determination of the mass of dry matter. - Place the porcelain crucibles in an oven at 95 °C overnight to dry the sample. Figure 1 summarizes the protocol Steps A4 to A8.
- Place the porcelain crucibles in a desiccator for 8 h for further drying.
- Weigh the porcelain crucibles for further determination of the mass of dry matter.
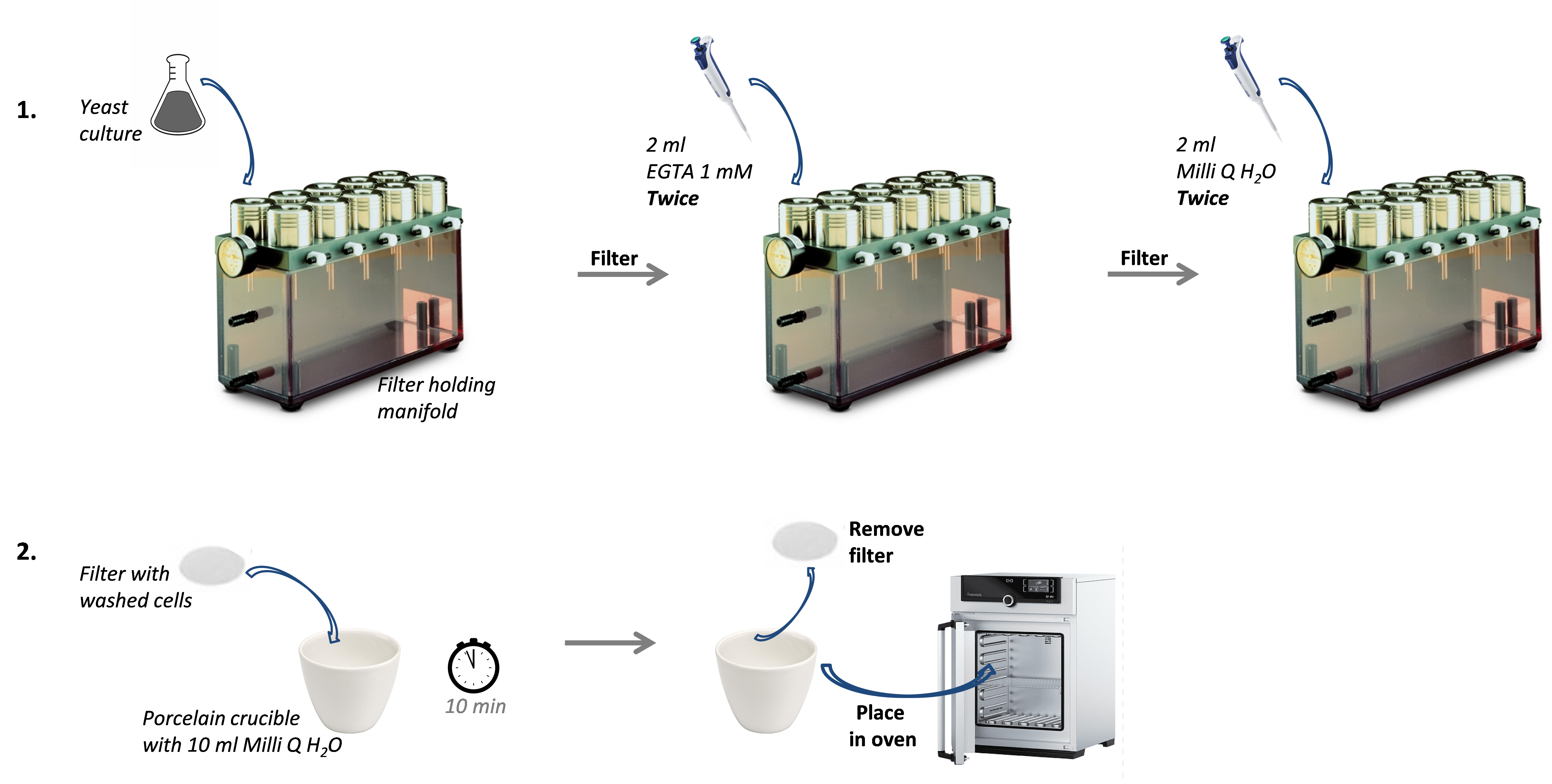
Figure 1. Experimental Steps A4 to A8. The 100 ml yeast culture is first filtered. Then, cells on the filter are washed with EGTA 1 mM (twice) and Milli Q water (twice). The filter with washed cells is subsequently immerged in a porcelain crucible containing 10 ml Milli Q water. Finally, the filter is removed and the porcelain crucible is placed in an oven for further drying. - Measurement of the ionic content
- Place the samples in the muffle furnace at 500 °C for 4 h.
- Resuspend the ashes in 6.5% nitric acid (0.5 ml nitric acid 65% + 9.5 ml Milli Q H2O).
- Dilute the ICP standards from Merck in a range of concentration that covers the expected ionic concentration in the samples to be analyzed. Acidify the standard solutions to 0.5% nitric acid (from 65% nitric acid). The standard concentrations used for measurement of yeast ionic content in our study are mentioned in Table 1.
- Measure the emitted light for each standard and sample. Typical instrument parameters are given in Table 2. Table 3 gathers the wavelengths and the viewing mode used for measurement of each cation.
- Deduce the ionic content in the samples using the calibration curve established from the standards (see Data analysis).
Table 1. Concentrations of the standards (St) used for the calibration line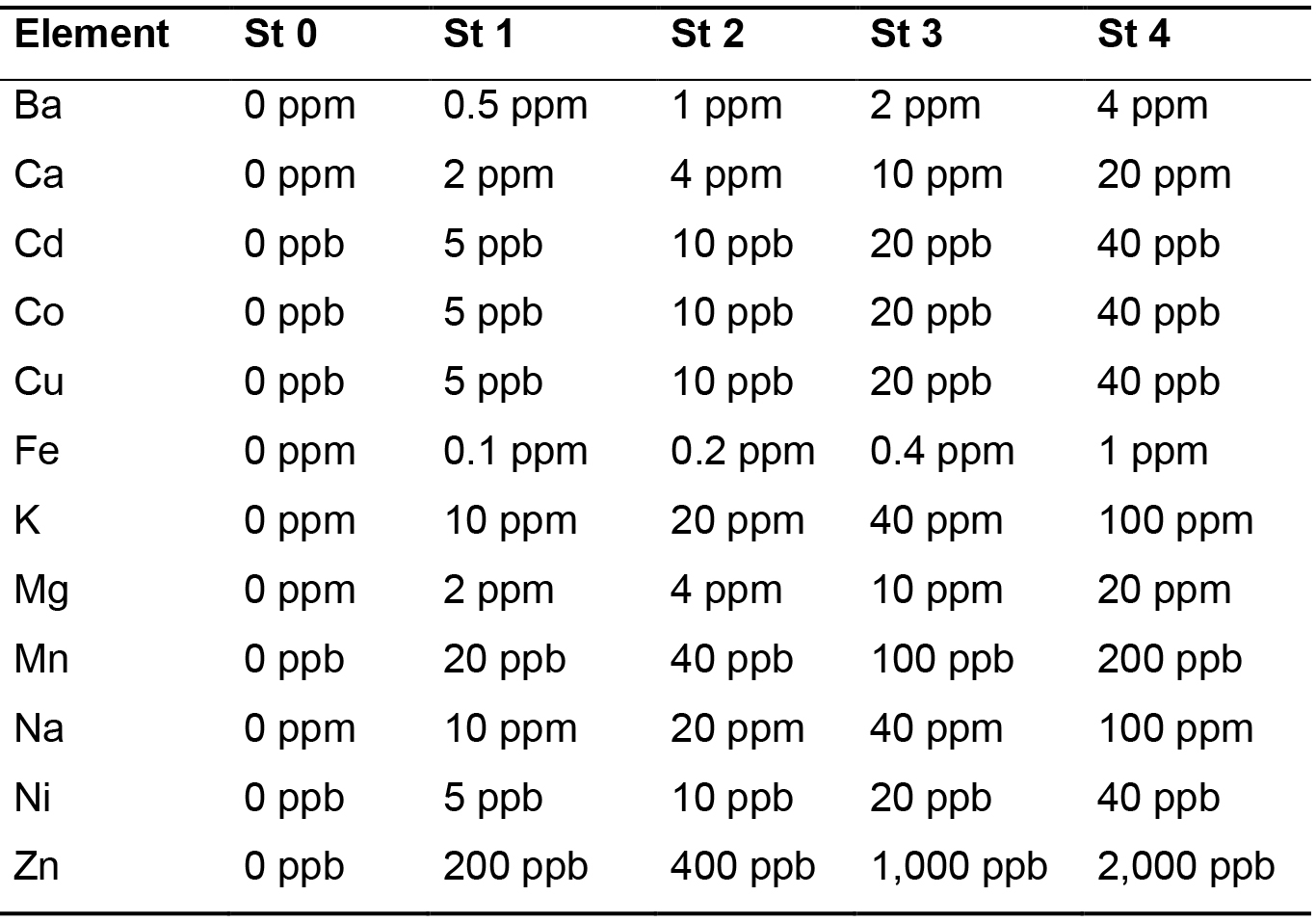
Table 2. Typical instrument parameters used on the Thermo Scientific ICAP 6500 ICP OES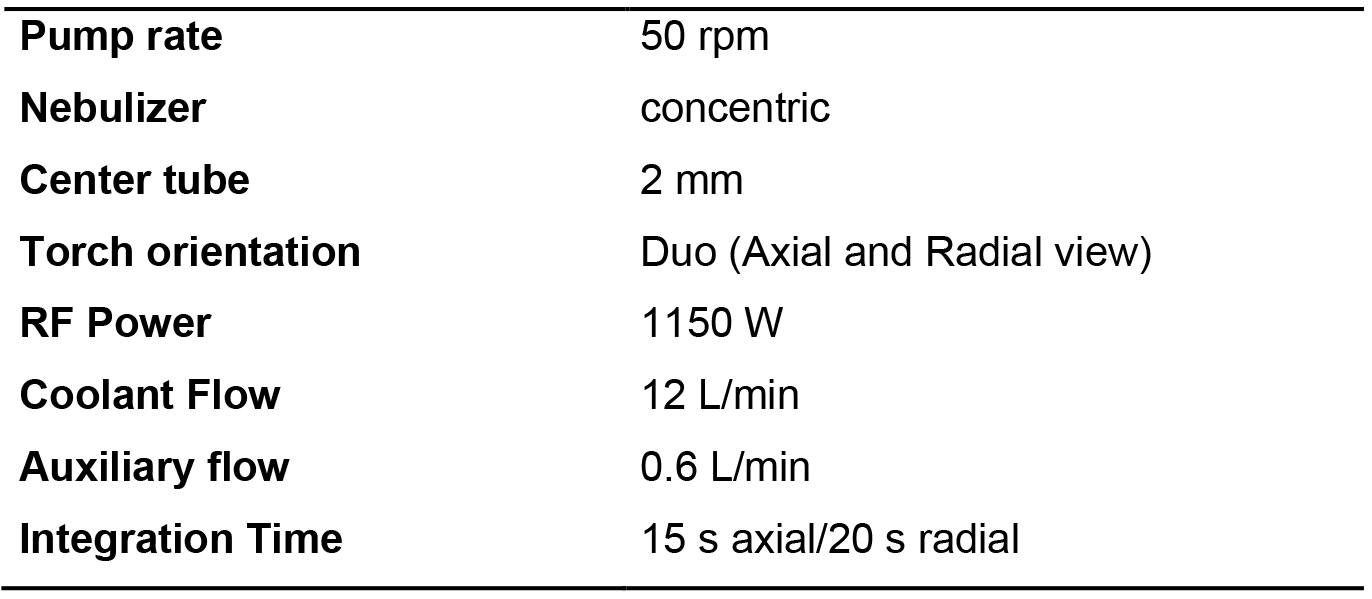
Table 3. Wavelengths and viewing mode used for the twelve elements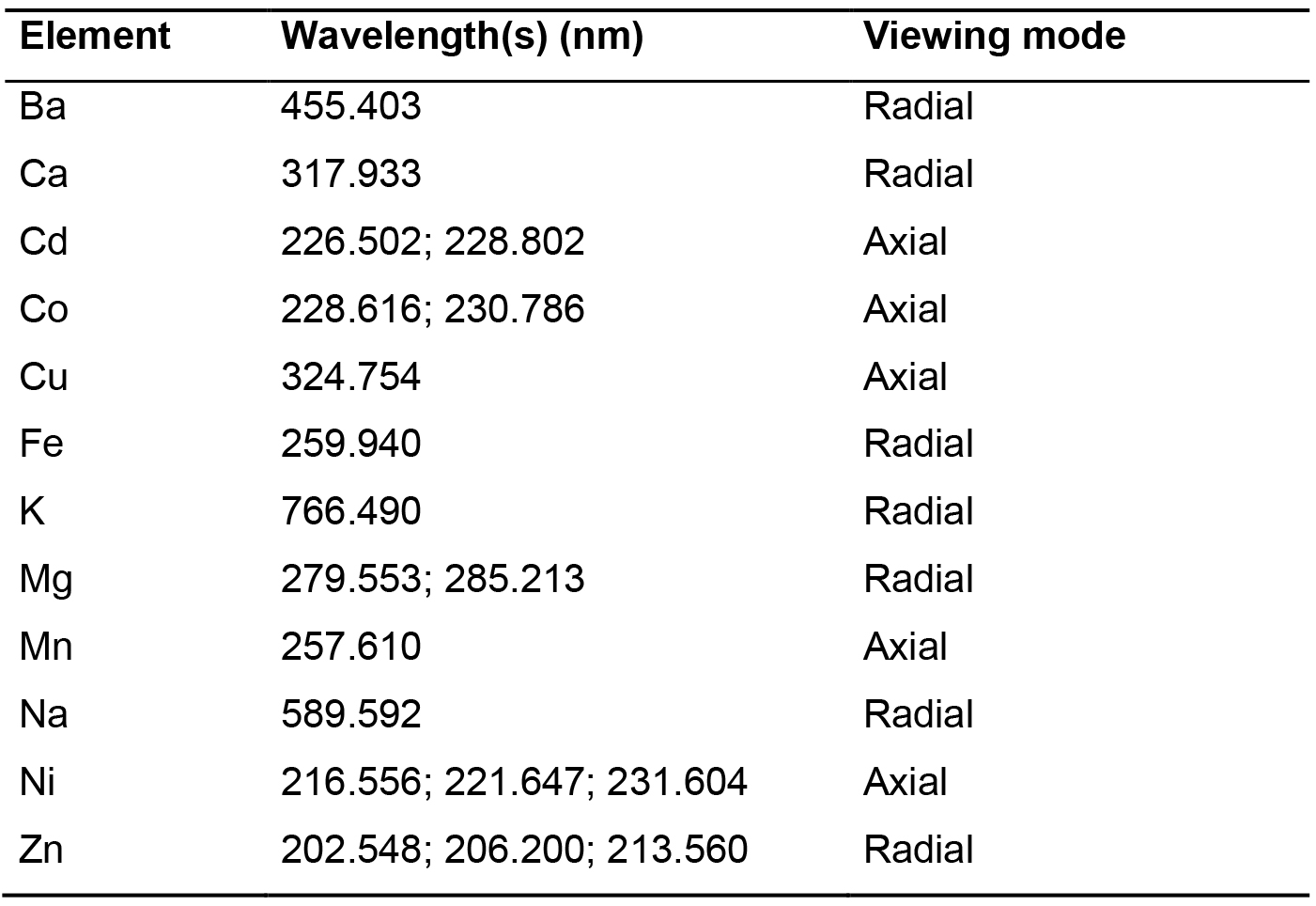
Data analysis
The calibration curve of the emitted light as a function of the concentration of the standards is first modelled using linear regression. The validation parameter for a calibration line is a R2 coefficient of minimum 0.999. An example of calibration line is provided in Figure 2.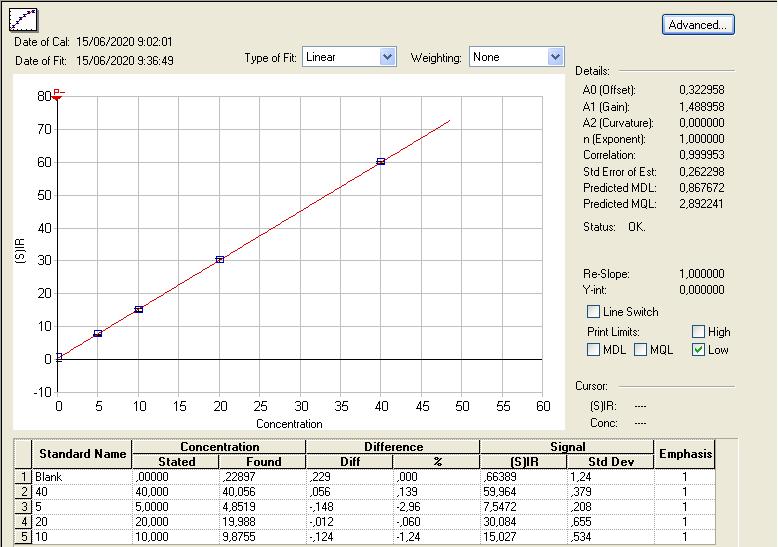
Figure 2. Calibration line of the emitted light according to the concentration of nickel (ppb) in the five standard solutions
The equation of the linear regression is determined as:![]()
with A the slope of the line and B its intercept. This equation is then used to deduce the concentration (in ppb or ppm) of the samples to be analyzed from the measurement of the intensity of the light emitted by the following equation:![]()
The ionic content obtained in ppm can then be converted into µmol of the ion per gram of yeast dry matter. To do so, first determine the mass of dry matter by subtracting the mass of the empty porcelain crucibles to that of the crucibles containing the cells after drying. Then convert the ionic concentration obtained in ppm to µmol of the ion per gram of dry matter according to the following formula: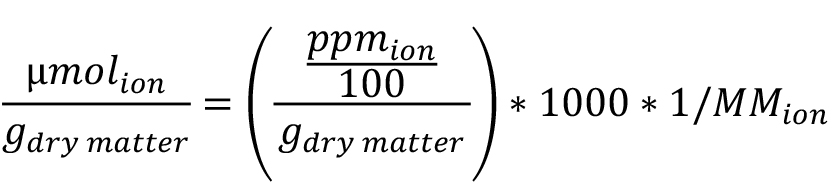
With MM the molar mass of the ion in g/mol.
The limit of quantification of each element is mentioned in Table 4.
Table 4. Limit of quantification for the twelve elements
Recipes
- YD plates
2 g (2% w/v) yeast extract KAT
2 g (2% w/v) glucose
2 g (2% w/v) agar
Adjust to 100 ml with Milli Q water and autoclave
Pour in Petri dishes - YD medium
2 g (2% w/v) yeast extract KAT
2 g (2% w/v) glucose
Adjust to 100 ml with Milli Q water and autoclave - 1 mM EGTA (1 L)
0.3804 g EGTA
Adjust pH to 8.0 with NaOH
Acknowledgments
The work was supported by grants from the Fonds National de la Recherche Scientifique (FNRS, grant PDR-T.0206.16). L.T. is a research fellow at the ‘Fonds pour le Formation à la Recherche dans l’Industrie et dans l’Agriculture’. We thank Anne-Sophie Colinet for prior protocol development.
Competing interests
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Thines, L., Deschamps, A., Sengottaiyan, P., Savel, O., Stribny, J. and Morsomme, P. (2018). The yeast protein Gdt1p transports Mn2+ ions and thereby regulates manganese homeostasis in the Golgi. J Biol Chem 293(21): 8048-8055.
- Dahlquist, R. L., and Knoll, J. W. (1978). Inductively Coupled Plasma-Atomic Emission Spectrometry: Analysis of Biological Materials and Soils for Major, Trace, and Ultra-trace Elements. Appl Spectrosc 32(1):1-30.
- Eide, D. J., Clark, S., Nair, T. M., Gehl, M., Guerinot, M. L., and Harper, J. F. (2005). Characterization of the yeast ionome: a genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae. Genome Biol 6(9):R77.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Thines, L., Iserentant, A. and Morsomme, P. (2020). Determination of the Cellular Ion Concentration in Saccharomyces cerevisiae Using ICP-AES. Bio-protocol 10(16): e3727. DOI: 10.21769/BioProtoc.3727.
- Thines, L., Deschamps, A., Sengottaiyan, P., Savel, O., Stribny, J. and Morsomme, P. (2018). The yeast protein Gdt1p transports Mn2+ ions and thereby regulates manganese homeostasis in the Golgi. J Biol Chem 293(21): 8048-8055.
Category
Microbiology > Microbial biochemistry > Other compound
Cell Biology > Cell-based analysis > Ion analysis
Biochemistry > Other compound > Ion
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










