- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Neutrophil Extracellular Trap Killing Assay of Candida albicans
Published: Vol 10, Iss 16, Aug 20, 2020 DOI: 10.21769/BioProtoc.3716 Views: 6275
Reviewed by: Kristin L. ShinglerEmmanuel Orta-ZavalzaSuresh Panthee

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Infection Process Observation of Magnaporthe oryzae on Barley Leaves
Xiao-Lin Chen
May 5, 2018 9765 Views

Qualitative and Quantitative Methods to Measure Antibacterial Activity Resulting from Bacterial Competition
Boris Taillefer [...] Eric Cascales
Jul 5, 2023 3545 Views

Utilizing EdU to Track Leukocyte Recruitment to the Brain
Zoie K. Lipfert [...] David P. Sullivan
Dec 5, 2025 1586 Views
Abstract
Fungal pathogen Candida albicans is one of the top leading causes of overall healthcare-associated bloodstream infections worldwide. Neutrophil is the major effector cell to clear C. albicans infection. Our study showed that mouse neutrophils utilize two independent mechanisms to kill C. albicans: one is CR3 downstream NADPH oxidase-dependent mechanism that kills opsonized C. albicans; the other one is dectin-2-mediated NADPH oxidase-independent neutrophil extracellular trap (NET) that kills unopsonized C. albicans. Neutrophil killing of opsonized C. albicans requires phagocytosing the organism and production of reactive oxygen species production (ROS). Most existing protocols that assay for neutrophil killing of C. albicans requires a washing step after allowing neutrophils to phagocytose the organism. By definition, NET kills organisms extracellularly. Therefore, it is important to skip the washing step and add an optimal ratio of neutrophils and C. albicans to the wells. To demonstrate the effect of NET, it is necessary to compare killing ability of neutrophils treated with micrococcal nuclease (MNase), an enzyme that digests NET, to that treated with heat-inactivated MNase. MNase is also applied to release NET-bound fungal elements for counting. This protocol can be applied to assay NET killing of other biofilm-forming organisms.
Keywords: Neutrophil extracellular trapBackground
Candida albicans is an opportunistic fungal pathogen that resides as a commensal on mucosal surface and the skin in most humans. Environmental changes in temperature, nutrition, or the presence of serum induces its transformation from yeast form to hyphae. Candida infection is one of the top leading causes of overall healthcare-associated bloodstream infections in medical centers as well as regional hospitals. Mortality among patients with invasive candidiasis is as high as 40% even after receiving antifungal therapy (Brown et al., 2012; Chen et al., 2014; Kullberg and Arendrup, 2015). Patients with neutropenia and neutrophil dysfunction are at high risk for invasive candidiasis, suggesting the importance of neutrophil anti-Candida functions in host defense (Antachopoulos et al., 2007; Horn et al., 2009; Lionakis and Netea, 2013). Our work showed that mouse neutrophils utilize two independent mechanisms, one NADPH oxidase-dependent and the other NADPH oxidase-independent, to kill C. albicans. NADPH oxidase-dependent killing of opsonized C. albicans requires phagocytosis mediated by CR3, and NADPH oxidase-independent killing of unopsonized C albicans is through NET formation mediated by dectin-2 (Wu et al., 2017 and 2019). MNase is an enzyme that digests NET. To demonstrate the effect of NET, it is necessary to compare the killing ability of neutrophils that is treated with MNase to that treated with heat-inactivated MNase. Difference between the two treatments denotes killing by NET and not by other factors (Wu et al., 2019). Neutrophils are phagocytic. Taking up microorganisms through receptors triggers robust ROS production that kills the engulfed organism. To assay for NADPH oxidase-dependent killing of opsonized C. albicans, one of the important steps is to wash off un-engulfed microorganisms after allowing time for phagocytosis to take place (Vonk et al., 2012; Wu et al., 2017). In contrast, since NET kills microorganisms extracellularly, it is critical not to wash off un-engulfed organisms to assay for NET-mediated killing of C. albicans. Moreover, instead of lysing neutrophils by ddH2O at high pH (pH 11.0) to release ingested microorganisms, NET-forming neutrophils and fungal hyphal elements are detached from the wells by mini cell scraper and DNA digesting enzyme MNase (Wu et al., 2017 and 2019). The method described here were published in 2019 (Wu et al., 2019) to observe killing of C. albicans by NET. This method can be adapted to quantify NET-mediated killing of other biofilm-forming organisms.
Materials and Reagents
- Pipette tips
- Mini cell scraper (Biotium, catalog number: 22003 )
- Flat-bottomed 96-well plates (Corning, catalog number: 3599 )
- 15 ml conical tube (Corning, catalog number: 430791 )
- 90 mm x 15 mm Petri dish (Alpha Plus, catalog number: 16001 )
- 1.5 ml microcentrifuge tube (Corning, catalog number: MCT-150-C )
- 3 ml syringe with 23-gauge needle (BD, catalog number: 302111 )
- Parafilm (Bemis, catalog number: PM996 )
- C. albicans SC5314 strain (ATCC MYA-2876)
- Female inbred mice in C57BL/6JNarl background, 6-12 weeks of age (National Laboratory Animal Center, RMRC11005)
- Percoll (GE Healthcare, catalog number: 17-0891-01 )
- 10x Dulbecco's Phosphate Buffered Saline (DPBS) (Biological Industries, catalog number: 02-023-1A )
- 10x Hank's Balanced Salt Solution (HBSS) (Biological Industries, catalog number: 02-015-5A )
- 0.4% Trypan blue (Sigma-Aldrich, catalog number: 93595 )
- 7.5% sodium bicarbonate (Biological Industries, catalog number: 03-040-1B )
- Yeast-peptone-dextrose (YPD) broth (Bioshop, catalog number: YPD002.500 )
- Agar A (BIO BASIC, catalog number: FB0010 )
- 2 x 106 U/ml Micrococcal nuclease (MNase) (NEB, catalog number: M0247S )
- Sterile double distilled H2O (ddH2O), adjusted to pH 11 by NaOH
- 1x Dulbecco's Phosphate Buffered Saline (DPBS) (see Recipes)
- 1x Hank’s Balanced Salt Solution (HBSS) (see Recipes)
- YPD agar plate (see Recipes)
- Red blood cell (RBC) lysis buffer (see Recipes)
- 100% Percoll (see Recipes)
- 55%, 62%, and 81% Percoll (see Recipes)
- Heat-inactivated micrococcal nuclease (MNase, see Recipes)
Equipment
- FinnpipetteTM F1 Variable Volume Pipettes (Thermo Scientific, catalog numbers: 4641050N , 4641080N , and 4641100N )
- Counting chamber Neubauer-improved (HAUSSER, catalog number: CB73811-01490 )
- Autoclave Machine (see Note 2)
- 37 °C, 5% CO2 incubator (see Note 2)
- 30 °C incubator for growing C. albicans yeasts (see Note 2)
- Table top general-purpose centrifuge with swinging bucket rotor (KUBOTA, Model 4000, Rotor Name: ST-720M and PT-89M)
- CellGard ES Energy Saver Class II Type A2 Biological Safety Cabinet (NuAire, model: NU-475-400 )
- Vortex-Genie 2 (Scientific Industries)
- Light Microscope (see Note 2)
- Surgical operating dissecting scissors (sharp/sharp) (see Note 2)
- Semken forceps (see Note 2)
- Euthanasia chamber that is used to administer CO2 for euthanasia (see Note 2)
Software
- Prism GraphPad Software (CA, USA)
Procedure
- Mouse bone marrow neutrophil enrichment
- Mice at 6-12 weeks of age are placed in euthanasia chamber and euthanized by carbon dioxide for 4 min according to the AVMA Guidelines on Euthanasia (Cima, 2013). Obtain femur and tibia by using surgical operating dissecting scissors and Semken forceps.
Note: All animal procedures and experimental protocols need to be approved by AAALAC-accredited facility of the host institute. - Soak the bones in 1 ml ice cold 1x HBSS.
- Cut both ends of the bone, insert to one end the needle of a 3 ml syringe that is filled with ice cold 1x HBSS, push the plunger to flush out bone marrow cells into a 15 ml conical tube.
- Flush the bone one more time as in Step A3.
- Centrifuge the tube at 300 x g in room temperature for 10 min and discard the supernatant.
- Resuspend cell pellet in 1 ml of ice cold RBC lysis buffer and leave it in room temperature for 1 min.
- Add 1 ml of 1x DPBS to the tube.
- Centrifuge the tube at 500 x g in room temperature for 5 min, discard the supernatant and suspend the pellet in 1 ml of 1x DPBS.
- Prepare three layers of discontinuous Percoll solutions (55%, 62%, and 81% Percoll in the order of top to bottom in a 15 ml conical tube. Each layer is consisted of 3 ml as shown in Figure 1).
- Overlay bone marrow cell suspension on top of the discontinuous Percoll gradient as shown in Figure 1.
- Centrifuge at 1,400 x g in room temperature for 30 min in a swing-out rotor, with the brake off.
- “Gently” remove the top 6 ml of the solution by micropipette and collect 1.0-1.5 ml of the solution containing the band of neutrophils at the interface of 62 and 81% as shown in Figure 1 to a new 15 ml conical tube.
- Add 8 ml of 1x DPBS to the tube containing neutrophils and centrifuge at 1,000 x g in room temperature for 5 min and discard the supernatant.
- Suspend the pellet in 2 ml of DPBS and centrifuge at 500 x g in room temperature for 3 min and discard the supernatant.
- Suspend the pellet in 1 ml of HBSS.
- Dilute 10 µl of well-mixed cell suspension in 390 µl of 0.4% trypan blue solution and count the number of viable cells by loading 10 µl of the mixture to a Neubauer counting chamber. While viable cells are bright and shiny, dead cells stain blue. There is approximately 0.8 x 107-1 x 107 bone marrow cells per mouse.
- Leave cells in room temperature until use.
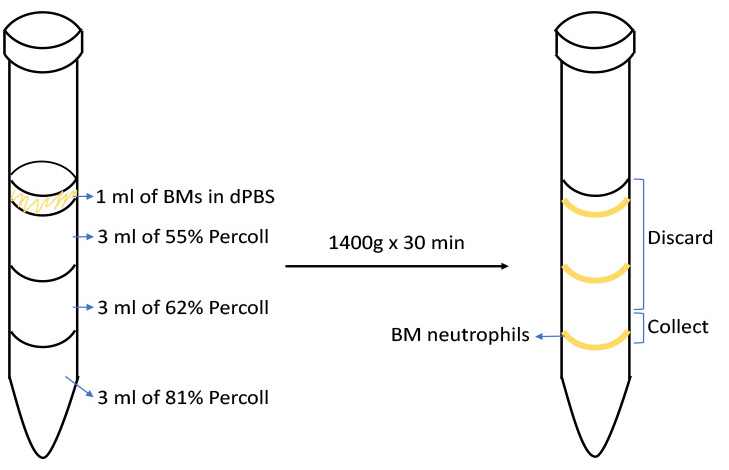
Figure 1. Enrichment of bone marrow neutrophils by discontinuous Percoll gradient centrifugation. Overlay bone marrow cell suspension on top of the 55/62/81% Percoll gradient (Step A10). After centrifugation in swing-out rotor (Step A11), there will be three visible bands in the gradient solution. Neutrophils are located at the interface of 62% and 81% Percoll (the lowest band). Carefully remove the top 6 ml of the solution and collect the top 1.0-1.5 ml of the remaining solution that contains cells at the lowest band (between the 2.5-4.0 mark on the scale) (Step A12). - Mice at 6-12 weeks of age are placed in euthanasia chamber and euthanized by carbon dioxide for 4 min according to the AVMA Guidelines on Euthanasia (Cima, 2013). Obtain femur and tibia by using surgical operating dissecting scissors and Semken forceps.
- Preparation of fresh C. albicans
- Thaw a frozen aliquot of C. albicans and plate it onto a YPD agar plate.
- Incubate the dish at 30 °C overnight.
- Scrape C. albicans colonies with a sterile1,000 µl tip and streak the yeasts on YPD agar plate.
- Incubate the plate at 30 °C overnight.
- Scrape C. albicans colonies with a sterile 200 µl tip and suspend in 1 ml HBSS.
- Take 10 µl of well-mixed C. albicans solution, dilute it in 390 µl of 4% trypan blue solution and count the number of viable yeast cells by loading 10 µl of the mixture to a Neubauer counting chamber. While viable yeast cells are bright and shiny, dead cells stain blue (approximately 0.5 x 107-2 x 107 C. albicans yeasts per ml).
- Leave C. albicans on ice until use.
- Neutrophils and C. albicans preparations
- Dilute bone marrow neutrophils obtained from Step A16 to 2 x 105 per 1 ml of HBSS solution.
- Dilute freshly harvested C. albicans yeasts obtained from Step B7 to 4 x 105 per 1 ml of HBSS.
- Prepare 4 x 103 U/ml MNase by diluting 0.5 µl of enzyme stock in 250 µl of HBSS.
- NET killing assay of C. albicans
- Seed 2 x 104 neutrophils (100 µl) in the wells of flat-bottomed 96-well plate and let adhere by incubation at 37 °C (5% CO2 incubator) for 30 min.
- Leave wells for 100 µl of HBSS without adding neutrophils as control as in Figure 2.
- Add 4 x 104 unopsonized C. albicans yeasts (100 µl) to all the wells (including control well) as shown in Figure 2.
- Add 0.5 µl of MNase (4 x 103 U/ml) at a final concentration of 10 U/ml or heat-inactivated MNase at otherwise equivalent concentration to appropriate wells as shown in Figure 2. Centrifuge the plate at 800 x g in room temperature for 3 min to spin down the yeasts.
- Incubate the plate at 37 °C (5% CO2 incubator) for 3 h.
- Add 0.5 µl of MNase to all the wells.
- Leave the plate at 37 °C (5% CO2 incubator) for 15 min.
- Collect the content (about 200 µl) of each individual well into a microcentrifuge tube.
- Add 200 µl of ice cold H2O (pH = 11) to each well.
- Use mini cell scraper to scrape the bottom of each well followed by vigorous pipetting to dislodge C. albicans hyphae.
- Collect the content of each well and add to the same microcentrifuge tubes.
- Repeat steps 9 to 11 once (total of 600 µl solution in the microcentrifuge tube now).
- Add 400 µl of HBSS to each microcentrifuge tube.
- Vortex microcentrifuge tubes vigorously for 20 s.
- Make 1:10 serial dilutions of the supernatant and plate 100 µl of the diluted solution on YPD plate in duplicate.
- Seal the plates with paraffin and leave in 30 °C incubator for 2 days.
- Enumerate colony forming units (CFUs) as shown in Figure 3.
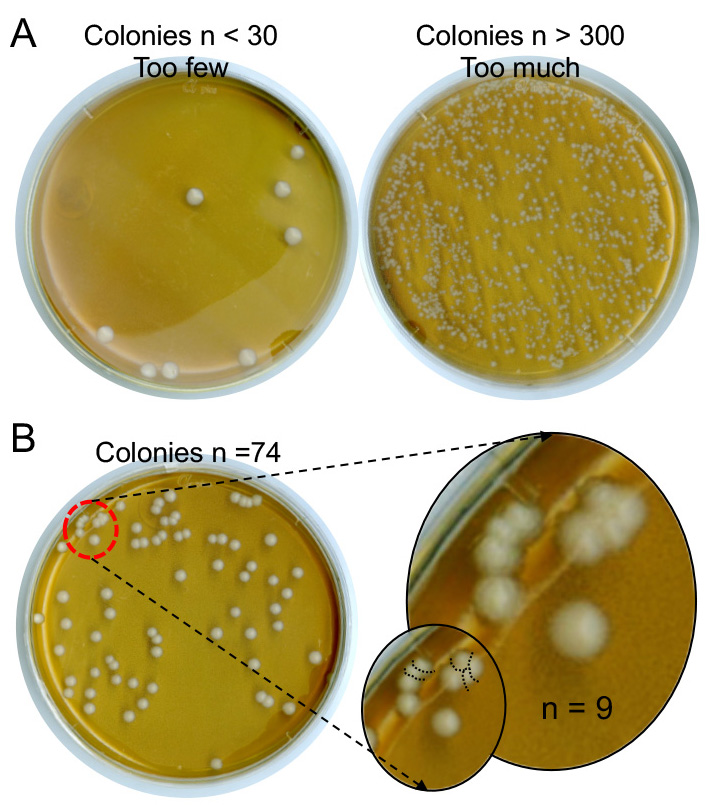
Figure 3. YPD plates with different C. albicans colony forming units (CFUs). Supernatants containing C. albicans are serially diluted (Step D15) and plated on YPD plates. The plates are incubated at 30 °C for 2 days (Step D16). C. albicans colonies are smooth and creamy white in color. There will be 3 different numbers of colonies from three 1:10 serially diluted supernatants. Select the plates with colonies fall between 30 and 300 (B) for counting and disregard the ones outside of this range (A). Carefully count each single colony even when they are very close to each other (B, red dotted circle).
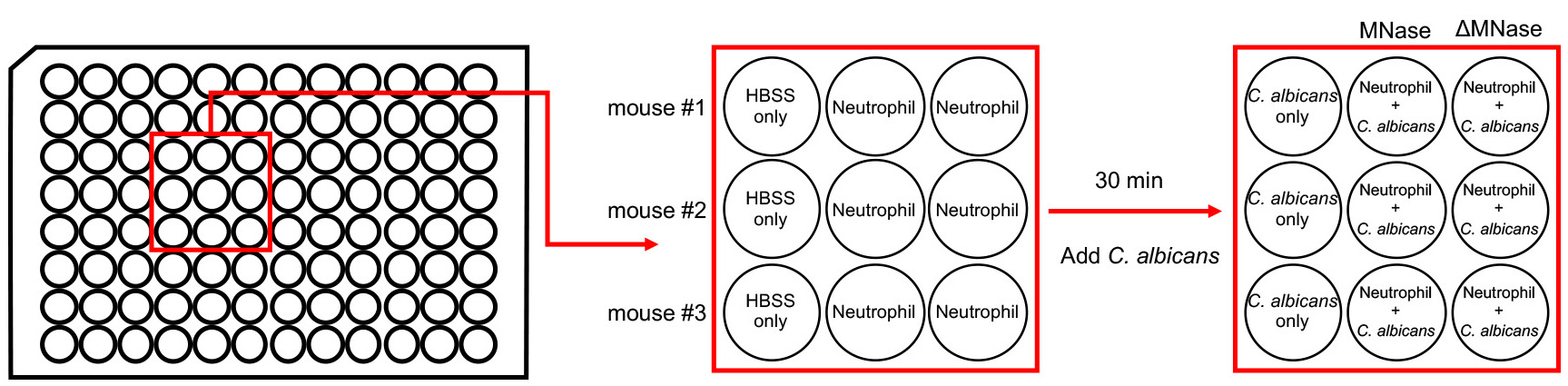
Figure 2. Layout for NET killing assay of C. albicans. Bone marrow neutrophils obtained from three different mice are suspended in 1x HBSS separately. Neutrophils from each mouse are seeded in two separate wells (Step D1). Leave one well for 1x HBSS only (Step D2). After incubation of the plate at 37 °C for 30 min, C. albicans suspensions are added to all three wells (Step D3). At the same time, MNase and heat-inactivated MNase (∆MNase) are added to separate wells separately (Step D4).
Data analysis
- CFU of control well is N0; CFU of wells containing MNase is NM; CFU of wells containing heat-inactivated MNase is N∆M as shown in Figure 4.
The average of CFU counts on the YPD dish in duplicate will be used as N0, NM and N∆M. - % of killing in the MNase group = (N0 - NM)/N0; % of killing in the heat-inactivated MNase group = (N0 - N∆M)/N0 as shown in Figure 4.
- To estimate the % of yeast cells killed by NET, data are analyzed by Mann-Whitney test by comparing the % of killing in MNase group to that in heat-inactivated MNase group.
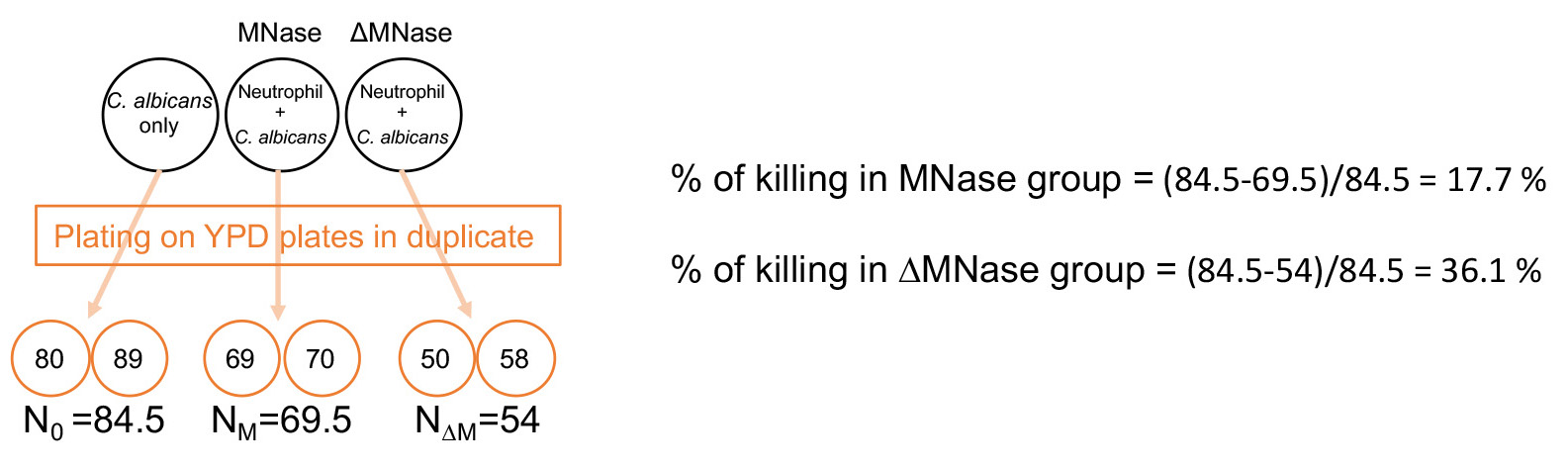
Figure 4. Calculation of % NET killing of C. albicans. N0, NM and N∆M are determined by the average of CFU counts of duplicate control wells (C. albicans only), wells containing neutrophils, C. albicans, and MNase and wells containing neutrophils, C. albicans, and heat-inactivated MNase, respectively. % of killing is calculated as shown
Notes
- MNase digests NET. To include both MNase and inactivated MNase in Step D4 is to confirm that the killing is mediated by NET but not by other factors. To add MNase in Step D6 is to release NET-bound C. albicans from NET for subsequent plate count.
- The reagents and materials from the manufacturers listed are those that have been used for this assay in the authors’ laboratory. We have not tried reagents and materials from different manufacturers. The equipments that do not have their manufactures listed are common laboratory equipments.
- It is important to completely lyse red blood cells because RBC contamination may interfere NET formation. One should also note that repeating or prolonging RBC lysis in Step A6 may result in neutrophil death. Therefore, if 1 ml of ice cold RBC lysis buffer does not lyse all the RBCs, increase the volume but do not prolong the time for lysis.
- Germination-defective strain of C. albicans (e.g., HLC 54 strain) does not induce NET formation (Wu et al., 2019).
- Colony counts on YPD plates between 30 and 300 ensures accurate counting. Plating of several 1:10 serial dilutions of the supernatants collected in microcentrifuge tubes will ensure that plating one of the dilutions will result in counts within the range.
Recipes
- 1x Dulbecco's Phosphate Buffered Saline (DPBS)
5 ml of 10x DPBS is diluted in 45 ml of ddH2O - 1x Hanks' Balanced Salt Solution (HBSS)
5 ml of 10x HBSS is diluted in 44.64 ml of ddH2O supplemented with 360 µl of sodium bicarbonate - YPD agar plate
- Add 50 g of YPD broth and 20 g of agar A into 1 L of ddH2O
- Stir the solution until YPD is dissolved
- Cool to 50 °C after autoclaving, and pour 23 ml of the medium into sterile 10-cm Petri dishes
- Store YPD dishes in refrigerator until use
- RBC lysis buffer
- 0.61 g of Tris-HCl and 4.15 g of NH4Cl are dissolved in 500 ml of ddH2O
- Adjust the pH value to 7.4 and autoclave
- 100% Percoll
Mix 45 ml of Percoll stock with 5 ml of 10x DPBS - 55%, 62%, and 81% Percoll
55%, 62%, and 81% Percoll are prepared by diluting 100% Percoll (prepared in Recipe 5) with 1x DPBS - Heat-inactivated MNase
Heat MNase (4 x 103 U/ml) at 65 °C for 4 h and store in refrigerator until used
Acknowledgments
AcknowledgmentThis work was supported by Academia Sinica thematic project AS-105-TP-B08 to BWH and the Ministry of Science and Technology research grants 104-2320-B-002-052-MY2 and 107-2321-B-002-053-MY3 to BWH and SYW, respectively. This protocol was adapted from a publication by Vonk et al. (2012) and modified according to protocols that quantified the viability of biofilm-forming microorganisms in other studies ( Morici et al., 2016; Mohammed et al., 2017).
Competing interests
NO financial competing interests.
Ethics
Mouse study was carried out in strict accordance with the recommendations in the Guidebook for the Care and Use of Laboratory Animals, The Third Edition published by The Chinese-Taipei Society of Laboratory Animal Sciences in 2007. All animal procedures and experimental protocols were approved by AAALAC-accredited facility, the Committee on the Ethics of Animal Experiments of the National Taiwan University College of Medicine (Permit Number: 20140304, 20140533 and 20180013).
References
- Antachopoulos, C., Walsh, T. J. and Roilides, E. (2007). Fungal infections in primary immunodeficiencies. Eur J Pediatr 166(11): 1099-1117.
- Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G. and White, T. C. (2012). Hidden killers: human fungal infections. Sci Transl Med 4(165): 165rv113.
- Chen, P. Y., Chuang, Y. C., Wang, J. T., Sheng, W. H., Yu, C. J., Chu, C. C., Hsueh, P. R., Chang, S. C. and Chen, Y. C. (2014). Comparison of epidemiology and treatment outcome of patients with candidemia at a teaching hospital in Northern Taiwan, in 2002 and 2010. J Microbiol Immunol Infect 47(2): 95-103.
- Cima, G. (2013). AVMA Guidelines for the Euthanasia of Animal: 2013 Edition. Javma-J Am Vet Med A 242: 715-716.
- Horn, D. L., Neofytos, D., Anaissie, E. J., Fishman, J. A., Steinbach, W. J., Olyaei, A. J., Marr, K. A., Pfaller, M. A., Chang, C. H. and Webster, K. M. (2009). Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis 48(12): 1695-1703.
- Kullberg, B. J. and Arendrup, M. C. (2015). Invasive Candidiasis. N Engl J Med 373(15): 1445-1456.
- Lionakis, M. S. and Netea, M. G. (2013). Candida and host determinants of susceptibility to invasive candidiasis. PLoS Pathog 9(1): e1003079.
- Mohammed, M. M. A., Pettersen, V. K., Nerland, A. H., Wiker, H. G. and Bakken, V. (2017). Quantitative proteomic analysis of extracellular matrix extracted from mono- and dual-species biofilms of Fusobacterium nucleatum and Porphyromonas gingivalis. Anaerobe 44: 133-142.
- Morici, P., Fais, R., Rizzato, C., Tavanti, A. and Lupetti, A. (2016). Inhibition of Candida albicans biofilm formation by the synthetic lactoferricin derived Peptide hLF1-11. PLoS One 11(11): e0167470.
- Vonk, A. G., Netea, M. G. and Kullberg, B. J. (2012). Phagocytosis and intracellular killing of Candida albicans by murine polymorphonuclear neutrophils. Methods Mol Biol 845: 277-287.
- Wu, S. Y., Huang, J. H., Chen, W. Y., Chan, Y. C., Lin, C. H., Chen, Y. C., Liu, F. T. and Wu-Hsieh, B. A. (2017). Cell intrinsic Galectin-3 attenuates neutrophil ROS-dependent killing of Candida by modulating CR3 downstream Syk activation. Front Immunol 8: 48.
- Wu, S. Y., Weng, C. L., Jheng, M. J., Kan, H. W., Hsieh, S. T., Liu, F. T. and Wu-Hsieh, B. A. (2019). Candida albicans triggers NADPH oxidase-independent neutrophil extracellular traps through dectin-2. PLoS Pathog 15(11): e1008096.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wu, S. and Wu-Hsieh, B. A. (2020). Neutrophil Extracellular Trap Killing Assay of Candida albicans . Bio-protocol 10(16): e3716. DOI: 10.21769/BioProtoc.3716.
Category
Immunology > Immune cell function > Neutrophil
Microbiology > Antimicrobial assay > Killing assay
Cell Biology > Cell-based analysis > Fungal infection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link