- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Microtubule Seeded-assembly in the Presence of Poorly Nucleating Nucleotide Analogues
Published: Vol 10, Iss 16, Aug 20, 2020 DOI: 10.21769/BioProtoc.3714 Views: 3701
Reviewed by: Manoj B. MenonEsteban Paredes-OssesAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Proximity Labelling to Quantify Kv7.4 and Dynein Protein Interaction in Freshly Isolated Rat Vascular Smooth Muscle Cells
Jennifer van der Horst and Thomas A. Jepps
Mar 20, 2024 1564 Views

Determination of Ligand-Target Interaction in vitro by Cellular Thermal Shift Assay and Isothermal Dose-response Fingerprint Assay
Danyu Du [...] Jing Xiong
Aug 5, 2024 4917 Views

Ub-POD: A Ubiquitin-Specific Proximity-Dependent Labeling Technique to Identify E3 Ubiquitin Ligase Substrates in Human Cells
Urbi Mukhopadhyay [...] Sagar Bhogaraju
Jun 20, 2025 2435 Views
Abstract
Microtubule dynamic instability is driven by the hydrolysis of the GTP bound to the β-subunit of the α-β tubulin heterodimer. Nucleotide analogues are commonly used to mimic the different steps of the tubulin GTPase cycle, but most of them are poor microtubule nucleators. Usually, microtubule assembly is seeded by guanylyl-(α, β)-methylene-diphosphonate (GMPCPP) or glycerol that can be limiting factors in monitoring the effect of other nucleotide analogs on their polymerization. Here, we describe a protocol that allows the assembly of microtubules in the presence of nucleotide analogues without the need of heterogeneous seeds and at a low final glycerol concentration. Microtubules are first assembled in the presence of the analogue of interest and glycerol to promote assembly. These microtubules are then sonicated to produce seeds that will be used to assemble microtubules in the absence of glycerol. This strategy produces homogeneous nucleotide-bound microtubules that can be further analyzed by biochemical or structural methods such as cryo-electron microscopy.
Background
Nucleotide analogues are commonly used to investigate the conformational changes that the α-β tubulin heterodimer undergoes during microtubule assembly and hydrolysis of the GTP bound to its exchangeable (E) site. However, apart from GMPCPP, most analogues are poor microtubule nucleators. To overcome this difficulty, GMPCPP-stabilized seeds are classically used to elongate microtubules in the presence of other analogues (Maurer et al., 2011 and 2014; Zhang et al., 2015), resulting in the assembly of mixed nucleotide-bound lattices. Alternatively, glycerol at relatively high concentration (e.g., 25%) can be added to the reaction mix to stimulate tubulin self-assembly. However, high glycerol concentrations might not be desirable, for instance to analyze microtubule structure by cryo-electron microscopy, since this compound matches the protein density and reduces the image contrast.
To avoid the use of GMPCPP-seeds or high glycerol concentrations in the final reaction mix, we have developed a protocol that uses seeds assembled in the presence of the same nucleotides that will be used to assemble the microtubules. As a first step, glycerol is used to polymerize microtubules in the presence of the nucleotide analogue. These microtubules are then fragmented by sonication to produce seeds, which are further diluted in the tubulin mix in the absence of glycerol. Microtubules elongated onto these seeds can be analyzed by cryo-electron microscopy without the risk to mistake microtubules assembled in the presence of GMPCPP with those assembled in the presence of the nucleotide analogue of interest.
Our protocol describes the step-by-step preparation of the seeds and the assembly of microtubules, which is monitored by turbidimetry at 350 nm. It includes a cooling step at the end of the assembly process to assess the presence of depolymerizing microtubules. These conditions were used to assemble GDP-BeF3--microtubules, whose lattices were further characterized by cryo-electron microscopy (Estévez-Gallego et al., 2020). We have successfully applied this protocol to microtubules assembled in the presence of other analogues such as GTPγS, GDP-AlF3, GMPPCP and GMPCP, all of them being poor microtubule nucleators.
Materials and Reagents
- Pipette tips
- Open-top thinwall polypropylene centrifuge tubes, 175 μl, 5 x 20 mm (Beckman Coulter, catalog number: 342630 )
- Beryllium sulfate (BeSO4·4H2O) (Sigma-Aldrich, catalog number: 14270 )
- Ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- Guanosine 5’-triphosphate sodium salt hydrate (GTP) (Sigma-Aldrich, catalog number: G8877 )
- Magnesium chloride solution (MgCl2) (Sigma-Aldrich, catalog number: 63069 )
- Piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (Sigma-Aldrich, catalog number: P6757 )
- Potassium hydroxide (KOH) (Sigma-Aldrich, catalog number: 30603 )
- Sodium Fluoride (NaF) (Sigma-Aldrich, catalog number: S7920 )
- BRB80 buffer solution 5x (see Recipes)
- GDP solution (see Recipes)
- Tubulin stocks (see Recipes)
- NaF solution (see Recipes)
- BeSO4 solution (see Recipes)
- G-BRB80-BeF3- (see Recipes)
- BRB80-BeF3- solution (see Recipes)
Equipment
- Pipetman Classic P100, P20, P10 (Gilson, catalog numbers: F123615 , F123600 , F144802 )
- Airfuge Air-Driven (Beckmann Coulter, catalog number: 340401 )
- Fixed-angle rotor A-110 (Beckman Coulter, catalog number: 347596 )
- Refrigerator MEDIKA 100 ECT-F (fiocchetti)
- Compressed air cooler BEKOBLITZZ LC12 (Beko Technologies, catalog number: 4020103 )
- Compressed air filter Clearpoint, 1 μm (Beko Technologies, catalog number: S040SWT )
- Membrane dryer Drypoint M plus (Beko Technologies, catalog number: 4007626 )
- Condensate drain ECD model ED12
- B2000 Ultrasonic Cleaner (Branson)
- Dri-Block heater DB-2D (Techne)
- Spectrophotometer Uvikon XS (Secomam, Bio-Tek Instruments)
- Quartz cuvette Ultra-Micro cells (Hellma, catalog number: 105-201-15-40 )
- Julabo F30-C (Julabo) with Thermal HY fluid, -60 °C to +65 °C, (Julabo, catalog number: 8940104 )
- Julabo MP-5 (Julabo) with Thermal HY fluid, -60 °C to +65 °C, (Julabo, catalog number: 8940104 )
- Three-way valves, PFTE (Bürkle, catalog number: 8610-0006 )
Software
- LabPower v3.50 (Secomam, Bio-Tek Instruments)
- Excel (Microsoft)
- Affinity Designer (Serif Ltd)
Procedure
- Spectrophotomer preparation
- Switch on the spectrophotometer and make sure that it uses the UV lamp at 350 nm.
Note: The default value to switch from the UV (D2) to the halogen lamp is 340 nm on the Uvikon XS. This value must be changed to, e.g., 360 nm, since the UV lamp must be used with the Ultra-Micro-cell quartz cuvettes at 350 nm. - Select the autorate mode at 350 nm.
- Switch on the warm water bath, set the temperature to 35 °C and let equilibrate for 15 min.
Note: The temperatures of the warm and cold circulating baths must be adjusted using a small thermocouple inside the quartz cuvette filled with water. The goal is to reach 35 °C and 4 °C for polymerization and depolymerization conditions, respectively. Variations in ambient temperature ( the cold water bath, set the temperature to 2 °C and let equilibrate for 30 min.e.g., between summer and winter) may require periodic control and adjustment of these settings. Do not exceed 35 °C during polymerization, since tubulin denaturates easily above this temperature (Weis et al., 2010). - Switch on the cold water bath, set the temperature to 2 °C and let equilibrate for 30 min.
- Circulate the 35 °C fluid inside the spectrophotometer thermostated cell holder (Figure 1).
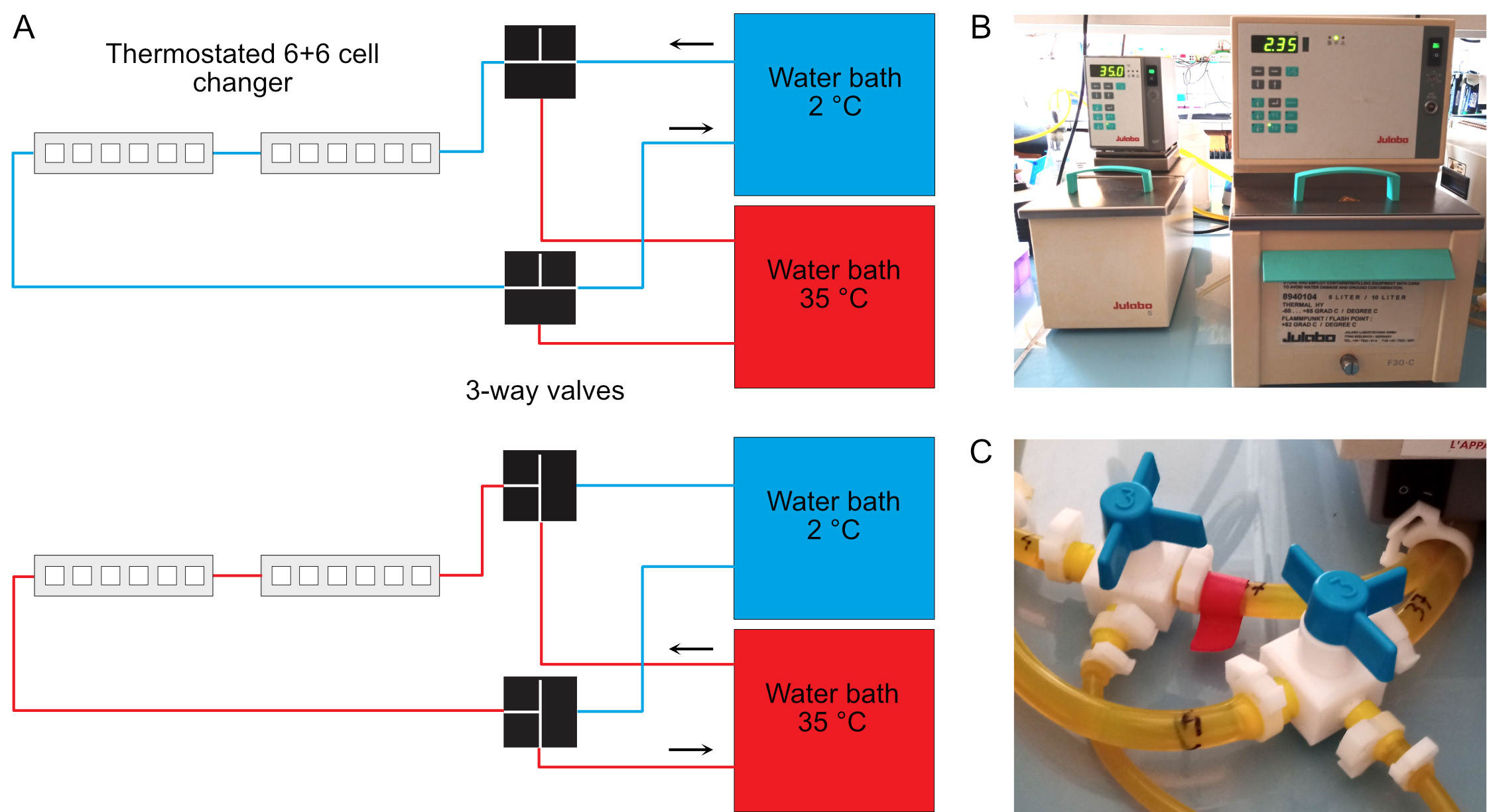
Figure 1. Temperature control of the spectrophotometer. A. Two water baths regulated at 35 °C and 2 °C are connected to the thermostated 6+6 cell changer of the spectrophotometer. Three-ways valves allow rapid switch of the temperature from 35 °C (bottom) to 4 °C (top). The temperature of the cold water bath must be set below 4 °C due to warming up of the fluid in the tubing system. The actual warm and cold temperatures must be checked regularly inside the cell holder using a small thermocouple. B. Warm (left) and cold (right) bather baths. C. Three-way valves. These must be switched simultaneously.
- Switch on the spectrophotometer and make sure that it uses the UV lamp at 350 nm.
- Microtubule seeds preparation (for 30 µl)
- Thaw rapidly between fingers a 10 μl aliquot of tubulin (initial concentration 22 mg/ml) and keep at 4 °C until use.
Note: Tubulin was purified from porcine brains by two cycles of assembly in a high-molarity buffer (Castoldi and Popov, 2003), followed by a cycling step to obtain tubulin liganded with GDP at the E-site (Ashford et al., 1998). Cycled tubulin is commercially available, e.g., from PurSolution (https://puresoluble.com/cycled-tubulin/).Tubulin at a final concentration of 22 mg/ml was aliquoted in 100 µl and 10 µl aliquots, snap frozen in liquid nitrogen, and stored at -80 °C until use. Once tubulin has been defrost, try to minimize all steps until polymerization since it will rapidly denaturates if kept in its unassembled form, even at 4 °C. - Add 5.5 µl of the tubulin solution to 24.5 μl of G-BRB80-BeF3- in a polypropylene centrifuge tube.
- Centrifuge the tube in the Airfuge at 141,000 x g (A-110 rotor, 22 psig), 5 min, 4 °C (Figure 2).
Note: The airfuge is not regulated in temperature. It must be placed in a cold room, or alternatively in a freezer such as in Figure 2. In both cases, the incoming air flow must also be cooled, otherwise the rotor will warm up (see manufacturer’s instructions if installed in a cold room). - Incubate the supernatant in the dry block heater, 1 h, 35 °C.
- Sonicate the microtubules in the ultrasonic cleaner, 1 min.
- Keep microtubule seeds at 35 °C until use in the following step.
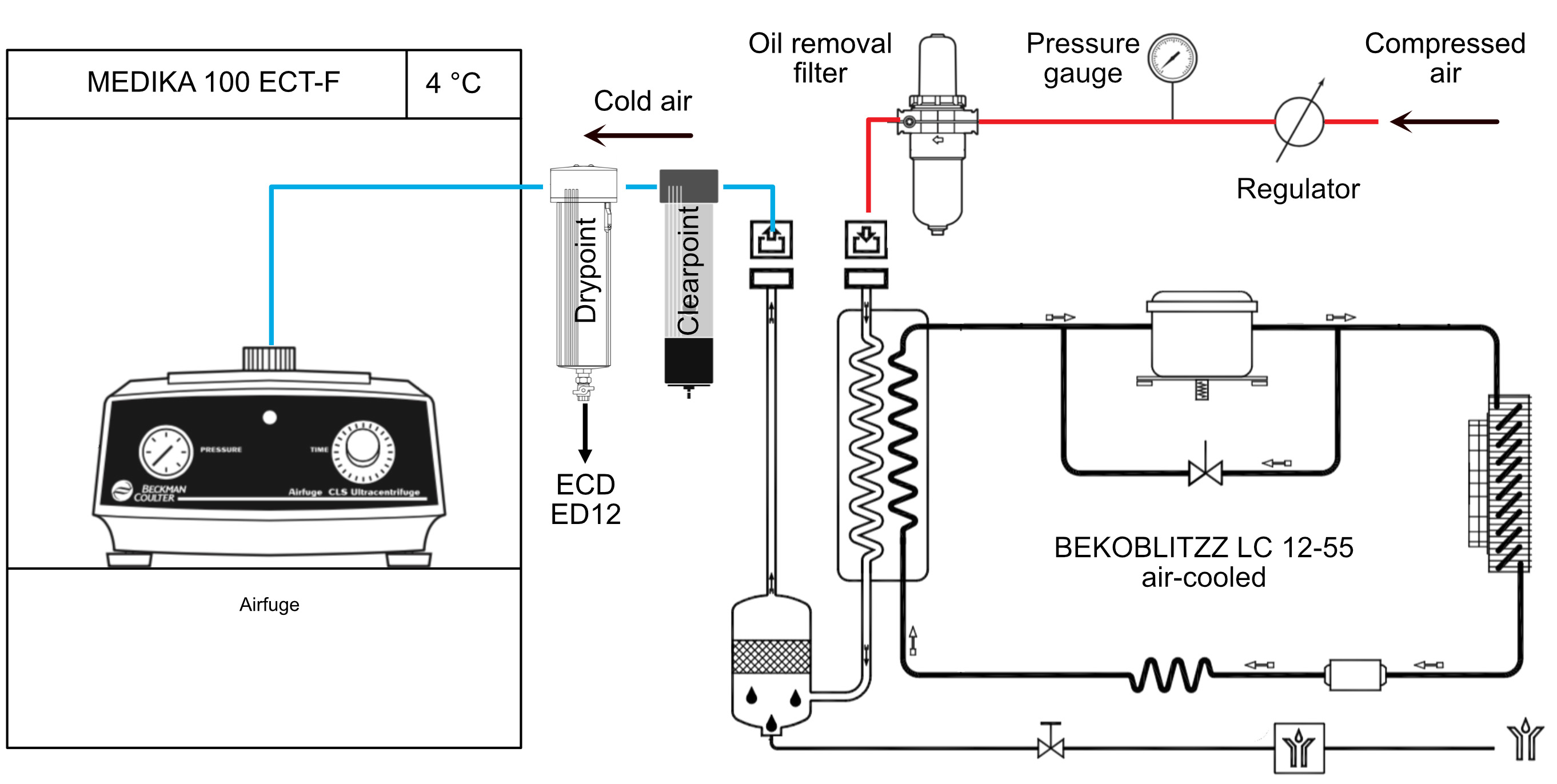
Figure 2. Temperature control of the Airfuge. The Airfuge is maintained at 4 °C inside a refrigerator. The incoming compressed air must also be cooled before reaching the centrifuge to avoid warming-up of the rotor during centrifugation.
- Thaw rapidly between fingers a 10 μl aliquot of tubulin (initial concentration 22 mg/ml) and keep at 4 °C until use.
- Microtubule seeded assembly followed by spectrophotometry at 350 nm (for 100 µl)
- Thaw rapidly a 20 µl aliquot of tubulin (initial concentration 22 mg/ml).
- Add 18 µl of this tubulin solution to 82 μl of BRB80-BeF3- in a polypropylene centrifuge tube.
- Centrifuge the tube in the Airfuge at 141,000 x g (A-110 rotor, 22 psig), 5 min, 4 °C.
- Incubate the supernatant in the dry block heater, 5 min, 35 °C.
- Pipet 90 µl of the pre-warmed tubulin solution and add 10 µl of pre-warmed seeds.
- Transfer the mix to the quartz cuvette and place it inside the spectrophotometer.
- Press autozero and start recording at 2 points/min.
- After about 1 h of polymerization, switch the fluid to the 2 °C thermostated bath to induce microtubule depolymerization (Figure 1).
- Stop recording when the OD baseline trace remains stable.
- Save data in Excel format (.xls) and plot the variation in optical density at 350 nm (∆OD 350 nm, a.u.) versus time (min) in Excel (Figure 3).
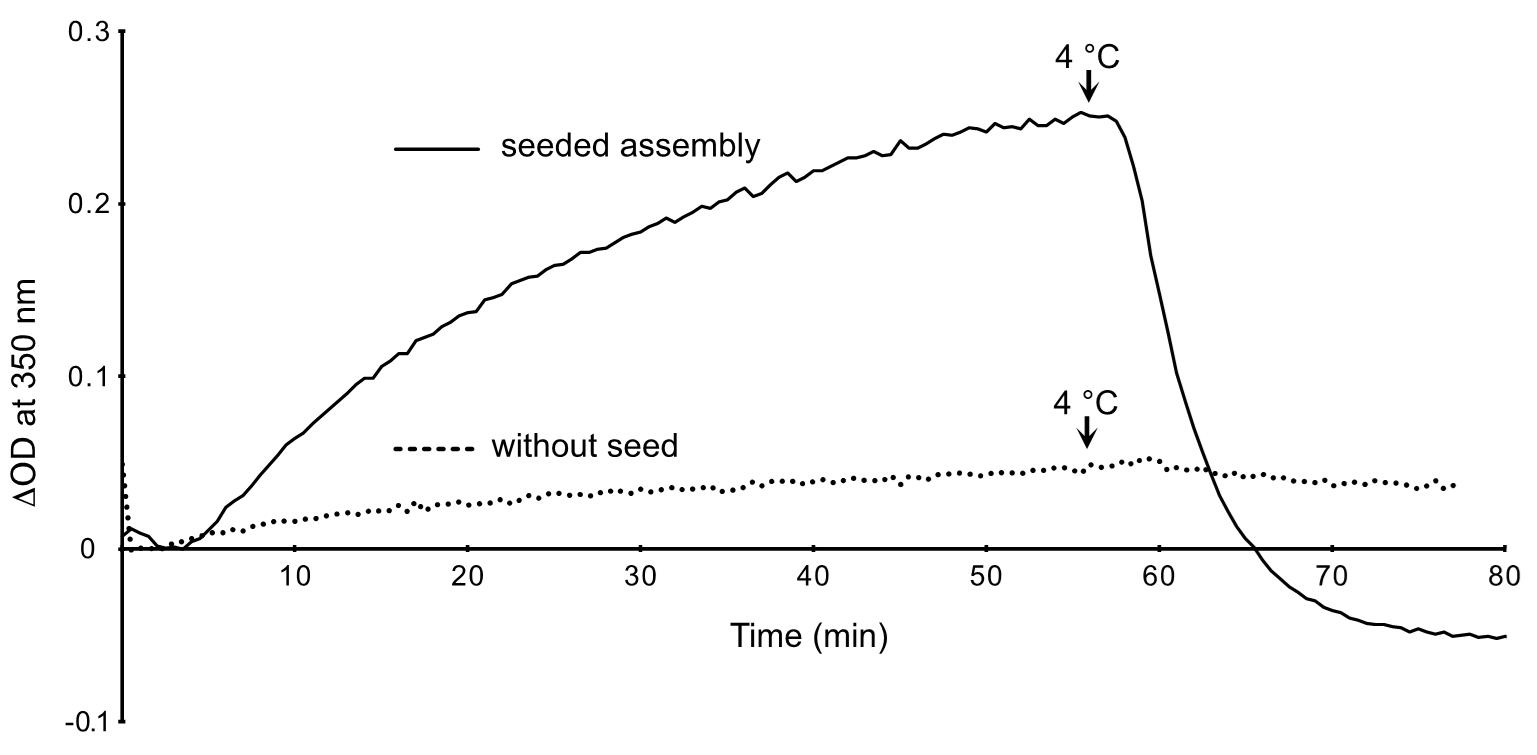
Figure 3. Assembly of GDP-BeF3--microtubules in the presence and absence of seeds. The OD decreases below zero after cold depolymerization of the microtubules assembled in the presence of seeds (plain line), since these latter contributed to the initial turbidity of the specimen. In the absence of seeds (dotted line), the OD remains almost stable after switching to 4 °C. This indicates that the small increase in OD was essentially due to the formation of tubulin aggregates.
Data analysis
The LabPower software was used to control the Uvikon XS spectrophotometer and acquire data in autorate mode, which were further saved in Excel (.xls) format. Data were imported into Excel for visualizing the variations in optical density versus time plots. Figures were prepared in Affinity Designer.
Recipes
- BRB80 buffer solution 5x
0.4 M PIPES, adjusted to pH 6.8 with KOH
5 mM MgCl2
5 mM EGTA
Store at 4 °C for up to one month - GDP solution
20 mM in BRB80 1x
Split into 20 µl aliquots
Store at -20 °C for up to one year - Tubulin stocks
22 mg·ml-1 purified porcine brain tubulin
Store at -80 °C for up to one year - NaF solution
200 mM in BRB80 1x
Store at 4 °C for up to one month - BeSO4 solution
20 mM in BRB80 1x
Store at 4 °C for up to one month - G-BRB80-BeF3-
1x BRB80
20 mM NaF
5 mM BeSO4
1 mM GDP
25% Glycerol
Keep at 4 °C until use. - BRB80-BeF3- solution
1x BRB80
20 mM NaF
5 mM BeSO4
1 mM GDP
Keep at 4 °C until use.
Note: Prepare fresh BRB80-BeF3- solution since BeF3- salts are not stable.
Acknowledgments
This work was funded by the Agence Nationale de la Recherche (ANR-16-C11-0017-01). This protocol was derived from Estévez-Gallego et al., 2020.
Competing interests
The authors declare no competing interests.
References
- Ashford, A. J., Anderson, S. S. L. and Hyman, A. A. (1998). Cell Biology: A Laboratory Handbook. 2nd edition. Vol 2. In: Celis, J. E. (Ed.). Academic Press, 205-212.
- Castoldi, M. and Popov, A. V. (2003). Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif 32(1): 83-88.
- Estévez-Gallego, J., Josa-Prado, F., Ku, S., Buey, R. M., Balaguer, F. A., Prota, A. E., Lucena-Agell, D., Kamma-Lorger, C., Yagi, T., Iwamoto, H., Duchesne, L., Barasoain, I., Steinmetz, M. O., Chrétien, D., Kamimura, S., Díaz, J. F. and Oliva, M. A. (2020). Structural model for differential cap maturation at growing microtubule ends. Elife 9: e50155.
- Maurer, S. P., Bieling, P., Cope, J., Hoenger, A. and Surrey, T. (2011). GTPγS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs). Proc Natl Acad Sci U S A 108(10): 3988-3993.
- Maurer, S. P., Fourniol, F. J., Hoenger, A. and Surrey, T. (2014). Seeded microtubule growth for cryoelectron microscopy of end-binding proteins. Methods Mol Biol 1136: 247-260.
- Weis, F., Moullintraffort, L., Heichette, C., Chrétien, D. and Garnier, C. (2010). The 90-kDa heat shock protein Hsp90 protects tubulin against thermal denaturation. J Biol Chem 285(13): 9525-9534.
- Zhang, R., Alushin, G. M., Brown, A. and Nogales, E. (2015). Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell 162(4): 849-859.
Article Information
Copyright
Ku et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ku, S., Heichette, C., Duchesne, L. and Chrétien, D. (2020). Microtubule Seeded-assembly in the Presence of Poorly Nucleating Nucleotide Analogues. Bio-protocol 10(16): e3714. DOI: 10.21769/BioProtoc.3714.
- Estévez-Gallego, J., Josa-Prado, F., Ku, S., Buey, R. M., Balaguer, F. A., Prota, A. E., Lucena-Agell, D., Kamma-Lorger, C., Yagi, T., Iwamoto, H., Duchesne, L., Barasoain, I., Steinmetz, M. O., Chrétien, D., Kamimura, S., Díaz, J. F. and Oliva, M. A. (2020). Structural model for differential cap maturation at growing microtubule ends. Elife 9: e50155.
Category
Molecular Biology > Protein > Protein-protein interaction
Biochemistry > Protein > Self-assembly
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









