- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
RETRACTED: Paper Lateral Flow Biosensor for Nodavirus Reverse Transcribed RNA Detection
Published: Vol 10, Iss 15, Aug 5, 2020 DOI: 10.21769/BioProtoc.3711 Views: 4375
Reviewed by: Alexandros AlexandratosTien Anh NgoRajesh Thippeshappa

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Novel PCR-Based Methodology for Viral Detection Utilizing Mechanical Homogenization
Zachary P. Morehouse [...] Rodney J. Nash
Mar 5, 2022 2761 Views

Direct RNA Sequencing of Foot-and-mouth Disease Virus Genome Using a Flongle on MinION
Lizhe Xu [...] Bonto Faburay
Jun 20, 2024 2144 Views

Single Molecule Fluorescence In Situ Hybridization Using RNAscope to Study Hematopoietic and Vascular Interactions in the Zebrafish Embryo and Larva
Léa Torcq and Anne A. Schmidt
Apr 5, 2025 3229 Views
Abstract
Paper nanobiosensors have been established as an excellent platform for analysis of veterinary and human pathogens causing various diseases. Especially, lateral flow assays or biosensors ideal for sensitive, rapid, robust and accurate analysis in laboratory setups and on-site analysis. Viral RNA detection is of great importance for public health as well as animal health protection. In that aspect, the present protocol focuses on the development of functionalized gold nanoparticle-based lateral flow biosensor for fish nervous necrosis virus (Nodavirus) nucleic acids detection. Total viral RNA, isolated from fish samples was subjected to reverse transcription PCR amplification and the amplification products were mixed with specific oligonucleotide probe. A red test line was formed when nodavirus product was present. The proposed assay has great implications on basic research since it eliminates the need for time-consuming, cumbersome electrophoresis protocols and could be adjusted for use on the site of fish culture by fish farmers. Disease monitoring by such bioanalytical platforms without time consuming and costly procedures would have great impact on the aquaculture and environmental safety.
Keywords: Lateral flow biosensorBackground
Point-of-care and/or on-site bioanalysis has been the ultimate goal of research efforts focusing on the well being of humans and animals. Sensing platforms based on paper substrates are very attractive as analytical platforms because they are functionalized easily, with great reproducibility and low fabrication costs. Paper-based analytical devices have been applied on the analysis of small molecules, proteins and all kinds of nucleic acids (Parolo and Merkoçi, 2013; Bahadir and Sezgintürk, 2016; Jiang et al., 2019). Lateral flow biosensors (LFBs) are prefabricated strips of materials with dry reagents and they are activated by fluid samples. They are designed for disposable single use where an on/off signal is sufficient (Posthuma-Trumpie et al., 2009). Lateral flow nanobiosensors incorporate nanoparticles and are optimal for rapid analysis in research laboratories, along with their potential for use on point-of-care. Gold nanoparticles, with size smaller than 100 nm, are mainly used as LFB labels due to their unique optical properties, i.e., vivid red color, easy surface functionalization with a variety of biomolecules, intense optical signal, high surface-to-volume ratio and exceptional chemical stability (Cheng et al., 2019; Posthuma-Trumpie et al., 2009).
In that frame, our research efforts are focused on the development of gold nanoparticle-based lateral flow biosensors for virus assessment. The present protocol was developed for fish nervous necrosis virus or nodavirus nucleic acids detection (Toubanaki et al., 2015), in order to increase the detection accuracy, simplify and speed up the total time of PCR-based analysis. In brief, total viral RNA, isolated from fish samples was subjected to reverse transcription PCR (RT-PCR) amplification. The PCR products were mixed with specific oligonucleotide probe and applied next to oligonucleotide conjugated gold nanoparticles (Au NPs). A red test line was formed when nodavirus product was present. The visual detection of the RT-PCR product was completed within 20 min. The present detection platform has been utilized in other pathogens detection (e.g., hepatitis C virus [Glynou et al., 2003]; bacterial infections [Kalogianni et al., 2007]; Leishmania parasite DNA [Toubanaki et al., 2016]) and could be adjusted for other pathogens detection with proper primers and probe design.
Materials and Reagents
- Materials
- 0.22 μm filter
- Pipettes tips: 0.5-10 μl, 10-200 μl (Greiner Bio-One, catalog numbers: 771291 , 739290 )
- Filtered tip racks 0.5-10 μl, 5-100 μl, 5-200 μl and tip racks 0.5-20 μl, 2-200 μl, 50-1,000 μl (Brand, catalog numbers: 732624 , 732630 , 732632 , 732124 , 732128 , 732132 )
- Siliconized tubes 1.7 ml (Sigma-Aldrich, catalog number: T3406 )
- Stainless steel beads 5 mm (Qiagen, catalog number: 69989 )
- 1.5 ml polypropylene tubes, attached flat-cap (Greiner Bio-One, catalog number: 616201 )
- 15 ml polypropylene tubes, screw cap (Greiner Bio-One, catalog number: 188271 )
- 50 ml polypropylene tubes, screw cap (Greiner Bio-One, catalog number: 227261 )
- Glass fiber pad (Millipore, catalog number: GFCP000800 ), store at RT
- Reagents
- Agarose (HT Biotechnology, catalog number: SB11a ), store at RT
- β-Mercaptoethanol (β-ME) (Sigma-Aldrich, catalog number: 444203 ), store at RT
- Cellulose fiber pad (Millipore, catalog number: CFSPOO1700 ), store at RT
- Dextran cross-linked G-25 (Sigma-Aldrich, catalog number: 92639 ), store at RT
- dNTPs: dATP, dTTP, dCTP, dGTP (HT Biotechnology, catalog number: SB23 ), store at -20 °C
- Ethanol absolute (Sigma-Aldrich, catalog number: 32205-M ), store at RT
- Ethidium bromide (Sigma-Aldrich, catalog number: E1510 ), store at RT
- Glycerol (Applichem, catalog number: A2926 ,1000), store at RT
- Gold nanoparticles 40 nm (Sigma-Aldrich, catalog number: 741981 ), store at 4 °C
- GoTaq Flexi DNA polymerase (Promega, catalog number: M8301 ), store at -20 °C
- Hydrochloric acid (HCl) solution, 1 M (Sigma-Aldrich, catalog number: 150696 ), store at RT
- Methanol (Sigma-Aldrich, catalog number: 322415 ), store at RT
- Nitrocellulose membrane (Millipore, catalog number: HF180MC100 ), store at RT
- N-methylmaleimide (Sigma-Aldrich, catalog number: 389412 ), store at RT
- Oligonucleotides (Table 1): dA20, SH-dT30, UpNdv_B upstream primer, DpNdv downstream primer, probe_Ndv (VBC-Biotech, custom order), store at -20 °C after reconstitution
Table 1. Oligonucleotides used in the present protocol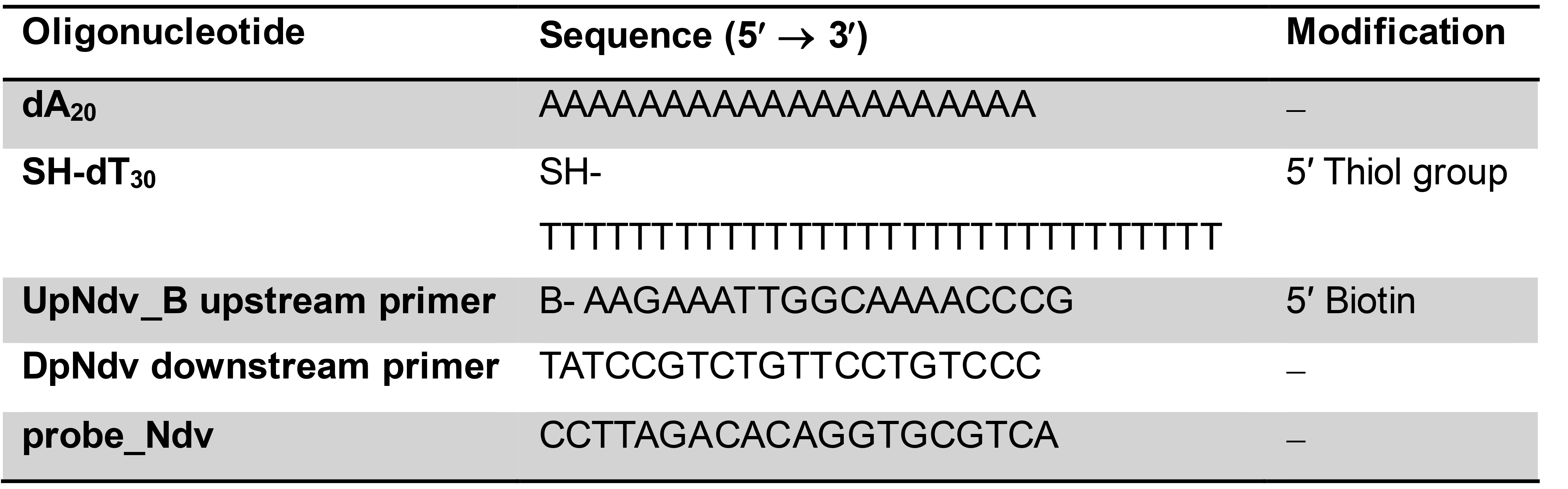
- PCR-grade water (Jena Bioscience, catalog number: PCR-258-500 ), store at 4 °C
- Pyridine (Sigma-Aldrich, catalog number: 270970 ), store at RT
- RNaseOUTTM Recombinant Ribonuclease Inhibitor (Invitrogen, catalog number: 10777019 ), store at -20 °C
- RNeasy Mini kit (Qiagen, catalog number: 74104 ), store at RT
- Sodium chloride (NaCl) (Applichem, catalog number: 381659 ), store at RT
- Sodium Dodecyl Sulfate (SDS) (Sigma-Aldrich, catalog number: L4509 ), store at RT
- Sodium hydroxide (NaOH) solution, 1 M (Sigma-Aldrich, catalog number: 79724 ), store at RT
- Streptavidin from Streptomyces avidinii (Sigma-Aldrich, catalog number: S4762 ), store at -20 °C
- Sucrose (Sigma-Aldrich, catalog number: S0389 ), store at RT
- SuperScript II (Invitrogen, catalog number: 18064014 ), store at -20 °C
- Terminal deoxynucleotidyl transferase (New England Biolabs, catalog number: M0315S ), store at -20 °C
- Tween-20 (Sigma-Aldrich, catalog number: P9416 ), store at RT
- φX174 DNA HaeIII digest DNA (New England Biolabs, catalog number: N3026 S ), store at -20 °C
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: 746336 ), store at RT
- Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: 795410 ), store at RT
- Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: 795488 ), store at RT
- Phosphate-buffered saline, 10× (PBS, pH 7.4) (see Recipes)
- LFB developing solution (see Recipes)
Equipment
- -20 °C freezer
- -80 °C freezer
- Desktop scanner (HP, HP Scanjet G4050)
- Dosage syringe 100 µl for Linomat (Camag, catalog number: 695.0014 )
- Microcentrifuge (Heraeus, Sepatech)
- Paper cutter Guillotine (Plaisio Office Supplies, catalog number: 800882 )
- Sonicator bath Bransonic (Sigma-Aldrich, catalog number: Z305359EU )
- PCR Workstation (Euroclone-Bioair, Aura)
- pH Meter (Thermo Fisher Scientific, catalog number: 13-644-928 )
- Pipettes:
Nichipet EXII 0.5-10 μl and 20-200 μl (Nichiryo, catalog numbers: NPX-10 , NPX-200 )
Transferpette S 0.5-10 μl, 10-100 μl, 20-200 μl and 100-1,000 μl (Brand, catalog numbers: 705870 , 705874 , 705878 , 705880 ) - Scissors and forceps, kept sterile with 70% ethanol
- Spectrophotometer (Thermo Fisher Scientific, model: NanoDropTM 1000, catalog number: ND-1000 )
- Thermal cycler (Applied Biosystems, GeneAmp PCR System 9700)
- Tissue homogenizer (TissueLyser LT) (Qiagen, catalog number: 85600 )
- Tissue homogenizer adapter (TissueLyser LT Adapter 12-Tube) (Qiagen, catalog number: 69980 )
- TLC applicator (Camag, Linomat, catalog number: 0 22.7808 )
- Vortex (Velp Scientifica, mdoel: Vortex ZX3)
- Water bath (LabTech, model: LSB-015S )
- Water distiller (Sartorius, model: Arium® Comfort I, catalog number: H2O-I-1-UV-T )
Software
- Adobe PhotoShop (Adobe Systems)
- CAMAG® HPTLC Software visionCATS Basic Version (Camag, catalog number: 028.0000)
- NanoDrop 1000 software (Thermo Fisher Scientific, NanoDrop ND-1000 )
- ImageJ software (National Institutes of Health (NIH), https://imagej.net/Welcome)
- Microsoft® Office Excel 2010 (Microsoft)
Procedure
- Tailing of dA20 oligonucleotide probes with dATP
- Dissolve the lyophilized oligonucleotide dA20 with the amount of PCR-grade water which is required to have 100 pmol/µl stock solution of the oligonucleotides, according to the manufacturer instructions.
- Set up the tailing reaction (20 µl) by mixing 2 µl of the terminal transferase reaction buffer, containing potassium acetate (50 mM), Tris-acetate (20 mM) and magnesium acetate (10 mM), in pH 7.9, 2 µl of CoCl2 (0.25 mM); 0.7 µl of dATP (3.5 mM), 0.5 µl of terminal deoxynucleotidyl transferase (TdT, 10 units) and 7 µl of dA20.
- Incubate for 1 h at 37 °C.
- Stop the reaction with incubation at 70 °C for 10 min.
- Store the poly(dA) tailed oligonucleotides at -20 °C.
- Preparation of the dry-reagent lateral flow biosensors
- Wash the TLC applicator syringe:
- Wash 10 times with distilled-deionized water (ddH2O).
- Wash 10 times with 50% ethanol (EtOH).
- Leave filled until use.
- Wash 20 times with distilled-deionized water (ddH2O).
- Prepare the poly(dA) oligonucleotide working solution by diluting the 35 pmol/µl poly(dA) tailed stock with the appropriate volume of a solution containing 50 ml/L methanol and 20 g/L sucrose in 1× PBS buffer, to a final concentration of 4 pmol/µl.
- Prepare the streptavidin (SA) working solution by diluting a 10 µg/µl SA stock solution with the appropriate volume of a solution containing 150 ml/L methanol and 20 g/L sucrose in 1× PBS buffer, to a final concentration of 4 µg/µl.
- Load 7 µl of the 4 pmol/µl poly(dA) tailed working solution to the syringe.
- Spray the poly(dA) tailed working solution to the immobilized membrane in the TLC applicator instrument with velocity of 60 nl/s, to result in the control zone (CZ) of 2.4 pmol poly(dA) per 4mm LFB.
- Spray the SA working solution to the immobilized membrane in the TLC applicator instrument with velocity of 250 nl/s, to result in the test zone (TZ) of 1.6 µg SA per 4 mm LFB.
- Dry the membrane at 80 °C for 1 h.
- Store the membrane at ambient temperature overnight.
- After each use wash the TLC applicator syringe as follows:
- Wash 5 times with NaOH 0.1 M.
- Leave the syringe filled for 15 min.
- Wash 20 times with ddH2O.
- Wash 5 times with HCl 0.1 M.
- Leave the syringe filled for 15 min.
- Wash 20 times with ddH2O.
- Assemble the LFB on a plastic adhesive backing (laminated card) as follows (Figure 1):
- First place the diagnostic membrane on the center of the laminated card.
- Then place the conjugate pad below the membrane, overlapping by 2 mm.
- Place the immersion pad below the conjugate pad, overlapping by 2 mm.
- Place the absorbent pad above the membrane, overlapping by 2 mm.
- Cut the LFBs to a 4mm with by the paper cutter.
- Store the LFBs at ambient temperature in the dark.
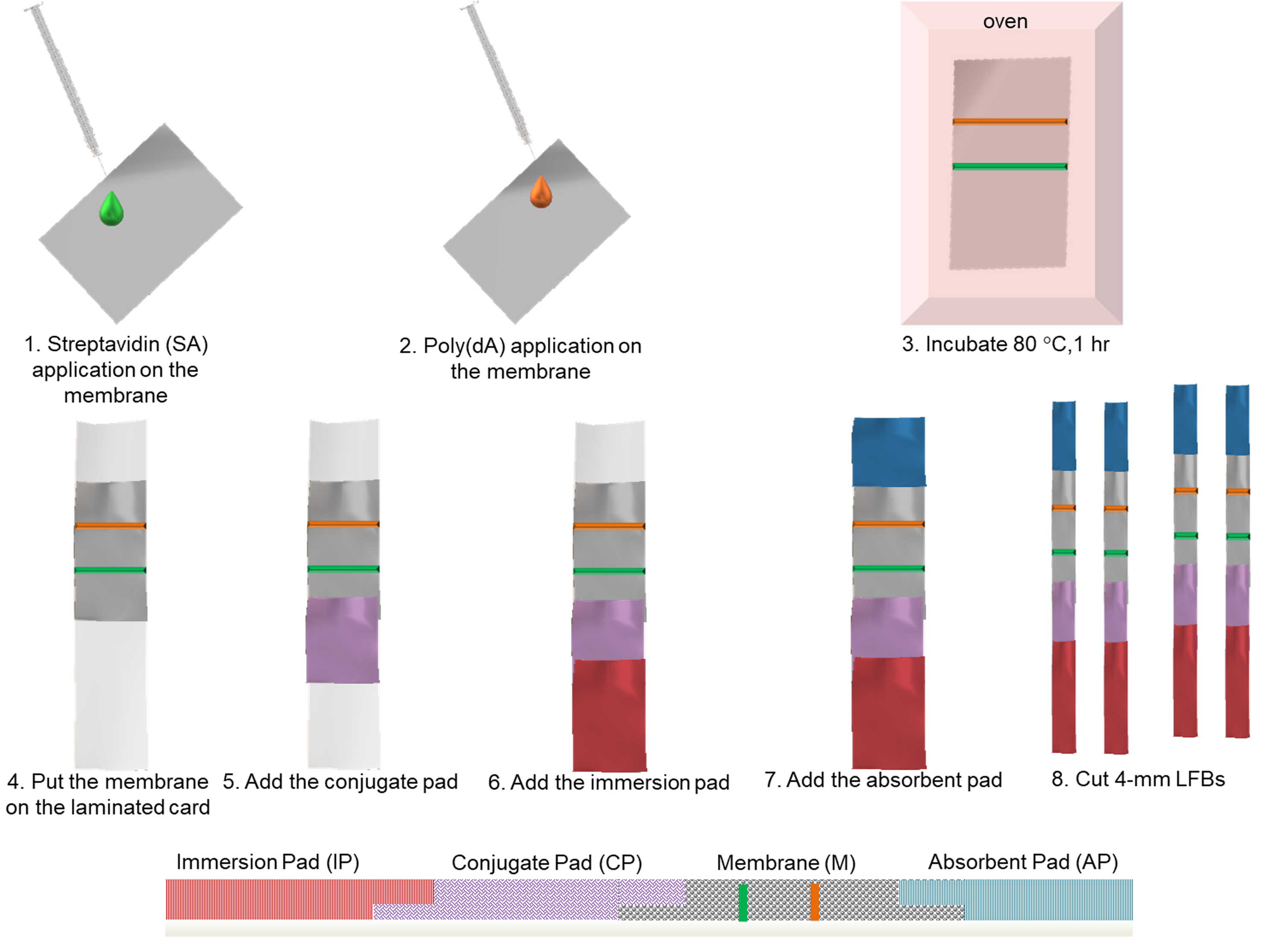
Figure 1. Schematic depiction of the lateral flow biosensor preparation procedure and side-view of the LFB assembling parts on a plastic backing
- Wash the TLC applicator syringe:
- Tailing of SH-dT30 with dTTP
- Dissolve the lyophilized oligonucleotides (SH-dT30) with PCR-grade water in order to have an 100 pmol/µl stock solution of the oligonucleotides.
- Set up the tailing reaction (20 µl) by mixing 2 µl of the terminal transferase reaction buffer (see A2), in pH 7.9; 2 µl of CoCl2 (0.25 mM); 0.7 µl of dTTP (3.5 mM); 0.5 µl of TdT (10 units) and 7 µl of SH-dT30.
- Incubate for 1 h at 37 °C.
- Stop the reaction with incubation at 70 °C for 10 min.
- Store the SH-poly(dT) tailed oligonucleotides at -20 °C.
- Gel Permeation Chromatography (GPC) for tailed SH-poly(dT) purification
- Preparation of dextran cross-linked G-25.
- Put 5 g of dextran particles in a glass bottle.
- Add ddH2O until all particles are covered, even after they are swallowed.
- Incubate overnight at 4 °C.
- Pour the pre-treated dextran G-25 in the GPC column.
- Centrifuge at 750 × g for 6 min.
- Put the tailed SH-poly(dT) oligonucleotides in the center of the column by a pipett tip.
- Centrifuge at 750 × g for 2 min.
- Collect the flowthrough (~50 µl) in a 1.5 ml tube.
- Store the purified tailed SH-poly(dT) oligonucleotides at -20 °C.
- Preparation of dextran cross-linked G-25.
- Preparation of oligonucleotide conjugated gold nanoparticles
- Put 1 ml of gold nanoparticles (Au NPs: 40 nm, 7.2 × 1010 particles) in siliconized tubes.
- Add 75 pmol of purified tailed SH-poly(dT) oligonucleotides (10 µl) and 0.8 ml of absolute pyridine.
- Incubate at 4 °C, for 24 h, in the dark.
- Subsequently, add 16.7 µl of 900 mM NaCl solution and incubate at 4 °C, for 2 h, in the dark.
- Repeat step E.4 for 6 times, up to a final concentration of 90 mM (“salt aging” protocol).
- Incubate at 4 °C, for 24 h, in the dark.
- Centrifuge at 1,300 × g for 30 min.
- Discard the supernatant carefully.
- Resuspend the Au NPs pellet in 100 µl of an aqueous solution containing 30% sucrose, 0.25% Tween-20, 0.25% sodium dodecyl sulfate (SDS), and 45 mM NaCl, by vortexing and brief sonication (5 min).
- Store the poly(dT) conjugated gold nanoparticles at 4 °C, in the dark.
- Fish brain dissection
- Put the fish onto a dissecting mat and carefully break the skull open with scissors and forceps.
- Dissect the whole brain and remove it carefully with the forceps.
- Transfer the samples in sterile polypropylene tubes and store at -80 °C, until use.
- Total RNA extraction
- Remove the excised fish tissue from storage and thaw it in ice.
- Weigh an empty sterile 1.5 ml tube and add the tissue. Determine its weight and cut 30 mg of tissue with sterile scissor. If the isolated brain is bigger than 30 mg, the remaining tissue can be stored at -80 °C for future use.
- Add 6 µl of β-mercaptoethanol (β-ME) to 600 µl of Buffer RLT before use.
Note: Dispense β-ME in a fume hood and wear appropriate protective clothing. - Place the tissue in a sterile 1.5 ml tube and add ~600 µl Buffer RLT with β-ME.
- Add one stainless steel bead (5 mm) to the tube and put it in the TissueLyser Adapter.
- Disrupt the tissue and homogenize the lysate on the TissueLyser LT for 5 min at 50 Hz.
Notes:- Do not exceed this time as it may result in nucleic acid shearing.
- The TissueLyser Adapter is used directly after storage at -20 °C and the tissue disruption is performed at RT. Therefore, due to the short disruption time the adapter temperature remains low throughout the procedure.
- Centrifuge the homogenized sample for 5 min at full speed to ensure that all the tissue debris is on the bottom of the tube.
- Carefully remove the supernatant by pipetting, and transfer it to a new 1.5 ml tube.
- Add 1 volume of 70% ethanol (stored at RT) to the cleared lysate, and mix by pipetting. Do not centrifuge.
Note: Precipitates may be visible after addition of ethanol. - Transfer 700 μl of the sample, including any precipitate that may have formed, to an RNeasy spin column placed in a 2 ml collection tube.
- Centrifuge for 30 s at 10,000 × g. Discard the flow-through.
- Repeat Steps G10 and G11 with the remaining sample.
- Add 700 μl Buffer RW1 to the RNeasy spin column to wash the spin column membrane.
- Centrifuge for 30 s at 10,000 × g. Discard the flow-through.
- Add 500 μl Buffer RPE to the RNeasy spin column to wash the spin column membrane.
- Centrifuge for 2 min at 10,000 × g. Discard the flow-through.
- Place the RNeasy spin column in a new 2 ml collection tube.
- Centrifuge at full speed for 1 min.
- Place the RNeasy spin column in a new 1.5 ml collection tube and add 30 μl RNase-free water directly to the spin column membrane.
- Centrifuge for 1 min at 10,000 × g to elute the RNA.
- Repeat Steps G19 and G20 using another 30 μl RNase-free water.
- Store the eluted RNA in -20 °C.
- Spectrophotometric quantification and quality determination of RNA with NanoDrop
- Open the NanoDrop 1000 software. Select nucleic acid analysis and then RNA in the respectives tabs.
- Put 1 µl of the eluted RNA sample on the sample loading position.
- Measure the absorbance.
- Reverse transcription of NNV RNA from fish samples
- Set up the reverse transcription reaction (20 µl) by mixing 5 µl of oligonucleotide dT20 (2.5 mM), 1 µl of dNTPs (dNTPs: dATP, dTTP, dCTP, dGTP; 0.5 mM each), 100 ng of purified total RNA and the appropriate volume of RNase-free H2O up to 13 µl.
- Heat the mixture to 65 °C for 5 min, and quickly chill on ice (0 °C, 1 min).
- Collect the contents of the tube by centrifugation and add 4 µl of first-strand buffer (1×), 1 µl of dithiothreitol (0.1 M), 1 µl RNase OUT RNase inhibitor (40 U) and 1 µl of Superscript II reverse transcriptase (200 U).
- Incubate at 50 °C for 60 min.
- Inactivate at 70 °C for 15 min.
- Store the produced cDNA at -20 °C.
- PCR of NNV cDNA
- Set up the PCR mastermix (18 µl) by mixing 4 µl of the DNA polymerase reaction buffer (1×), 1.6 µl of MgCl2 (2 mM), 0.4 µl of dNTPs pool (200 µM each), 1 µl of the upstream and downstream primers (0.5 mM), 0.1 µl of GoTaq Flexi DNA polymerase (0.5 U) and ddH2O.
- Add 2 µl of cDNA to the PCR mastermix.
- Put samples in the thermal cycler, with the following cycling conditions: 95 °C for 5 min, followed by 35 cycles at 95 °C for 30 s, 60 °C for 60 s, and 72 °C for 60 s.
- Incubate the samples reactions at 72 °C for 7 min and cool down to 4 °C.
- Store the produced amplification products at -20 °C.
- Visualize the PCR products by 2% agarose gel electrophoresis.
- Quantify the PCR products by ImageJ software based on the φX174 DNA HaeIII digest DNA molecular weight marker.
- Tailing of nodavirus-specific (probe_Ndv) oligonucleotide probes with dATP
- Dissolve the lyophilized probe_Ndv oligonucleotides with the proper amount of PCR-grade water for 100 pmol/µl stock solution preparation.
- Set up the tailing reaction (20 µl) by mixing 2 µl of the terminal transferase reaction buffer (see Step A2), in pH 7.9; 2 µl of CoCl2 (0.25 mM); 4 µl of dATP (2 mM); 0.5 µl of TdT (10 units) and 4 µl of probe_Ndv (400 pmol).
- Incubate for 1 h at 37 °C.
- Stop the reaction with incubation at 70 °C for 10 min.
- Mix the probe_Ndv with 1.5 µl N-methylmaleimide (40 nmol/µl).
- Store the probe_Ndv tailed oligonucleotides at -20 °C.
- Lateral flow biosensor detection assay of NNV amplification products
- Mix a 5 µl aliquot of PCR products solution with 1 µl of NaCl 0.9 M, 0.5 pmol of dATP-tailed probe_Ndv and ddH2O, to final volume of 10 µl.
- Heat the mixture at 95 °C for 3 min and place on ice for 30 s.
- Incubate the mixture to 37 °C for 10 min to allow hybridization to proceed.
- Apply the hybridization mixture to the conjugation pad next to the poly(dT)-functionalized gold nanoparticles.
- Dip the immersion pad of the biosensor into 250 µl of the LFB developing solution.
- Wait for signal formation. The visual detection is completed within 20 min.
- Scan the LFBs with a desktop scanner and quantify the band densities with ImageJ software.
Data analysis
- Visual signal detection of lateral flow biosensor
- After signal formation is competed (~20 min) take the LFB off the developing buffer and dry it by swipping carefully with plain paper towel.
- Scan the LFB in a desktop scanner with the higher analysis settings and save it as .tiff files.
- Open the image with any image processing software (e.g., PhotoShop) and crop the LFB pads in a way that only the membrane is visible. The results are shown in Figure 2, where representive images of a positive and a negative nodavirus sample, after assay completion are depicted.
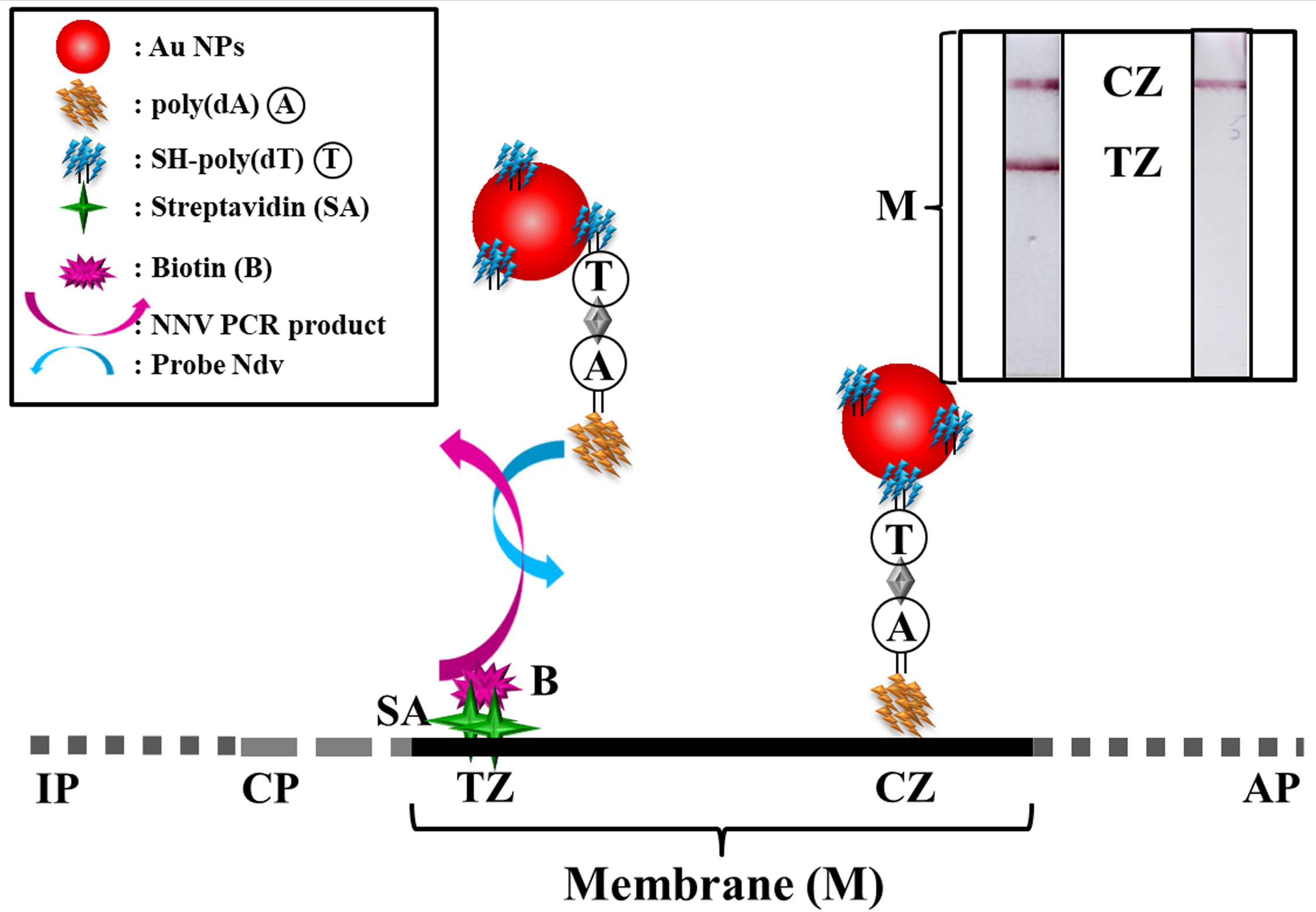
Figure 2. Representative lateral flow biosensor images for visual detection of a positive and a negative nodavirus sample and side view of the lateral flow biosensor presenting its working principle. IP: immersion pad; CP: conjugation pad; M: diagnostic membrane; AP: absorbent pad. (CZ: control zone; TZ: test zone). The assay components are not in scale.
- Semi-quantitative estimation of LFB signal by ImageJ
- Open your image with ImageJ. Then you can greyscale them with ImageJ.
- Go to Image → Type and click on 8-bit to convert the image to grayscale.
- Go to Process → Subtract Background and set rolling ball radius on 50.
- Go to Analyze → Set Measurements and click the boxes for Area, Mean Gray Value and Integrated Density.
- Go to Analyze → Set Scale and enter ‘pixels’ for Unit of length.
- Choose the Rectangular Selection tool and draw a rectangle around the mambrane.
- Press 1 and a window will pop up with the image and a blue label around the rectangular selection.
- Move the rectangle over the next LFB and press 2. Repeat for each LFB.
- When finished, press 3 and a new window with a profile plot of each membrane will pop-up.
- Choose the Straight Line selection tool. At the base of each peak, draw a line from one side of the peak to the other, enclosing the peak area.
- When each peak has been closed off choose the Magic Wand tool.
- With the wand, click inside each peak.
- The numerical results will pop up in a result window.
- Transfer the results to Excel workbook and draw the desired graph, e.g., results section on Toubanaki et al., 2015.
Notes
- Procedures A, C, D, G, I, J and K should be performed in DNA Workstation equipped with UV-Vis lamp to decontaminate reagents and equipment.
- All concentrations refer to the final concentration in 1× reaction.
Recipes
- Phosphate-buffered saline, 10× (PBS, pH 7.4)
- Dissolve 80 g NaCl, 2 g KCl, 11.5 g Na2HPO4 and 2 g KH2PO4 to 800 ml distilled water
- Adjust pH to 7.2 with 1 M HCl or 1 M NaOH depending on the acquired pH and bring volume to 1 L with distilled water
- Filter sterilize with 0.22 μm filter
- Store at 4 °C
- Dissolve 100 μl of stock solution (10×) in 1,000 ml of de-ionized water for the working solution of PBS (1×), when needed
- LFB developing solution
- Dissolve 4 ml glycerol and 1 g/L of SDS in 100 ml 1× PBS
- Store at 4 °C
Acknowledgments
The presented project was implemented within the framework of the Action «Supporting Postdoctoral Researchers» of the Operational Program “Education and Lifelong Learning” (Action's Beneficiary: General Secretariat for Research and Technology), and was co-financed by the European Social Fund (ESF) LS9 (448) and the Greek State. The protocols were adapted by Toubanaki et al., 2015.
Competing interests
The authors declare that they have no financial or non-financial competing interests. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Ethics
The collection of biological samples from fish farms was conducted by licensed personnel of the respective aquaculture facility. All qualified personnel were previously informed of the purpose of the study, the confidentiality of the data, and their voluntary participation. The present study was approved by the Hellenic Pasteur Institute Animal Bioethics Committee regulations according to Greek (PD 56/2013) and EU (Directive 63/2010) legislation for the protection, care and use of animals used for scientific purposes and all samples used were euthanized in the sites of fish farming according to the ethical principles and other requirements of the law.
References
- Bahadir, E. B. and Sezgintürk, M. K. (2016). Lateral flow assays: Principles, designs and labels. TrAC Trends Analyt Chem 82: 286-306.
- Cheng, N., Yang, Z., Wang, W., Wang, X., Xu, W. and Luo, Y. (2019). A variety of bio-nanogold in the fabrication of lateral flow biosensors for the detection of pathogenic bacteria. Curr Top Med Chem 19(27): 2476-2493.
- Glynou, K., Ioannou, P. C., Christopoulos, T. K. and Syriopoulou, V. (2003). Oligonucleotide-functionalized gold nanoparticles as probes in a dry-reagent strip biosensor for DNA analysis by hybridization. Anal Chem 75(16): 4155-4160.
- Jiang, N., Ahmed, R., Damayantharan, M., Ünal, B., Butt, H. and Yetisen, A. K. (2019). Lateral and vertical flow assays for point-of-care diagnostics. Adv Healthc Mater 8(14): e1900244.
- Kalogianni, D. P., Goura, S., Aletras, A. J., Christopoulos, T. K., Chanos, M. G., Christofidou, M., Skoutelis, A., Ioannou, P. C. and Panagiotopoulos, E. (2007). Dry reagent dipstick test combined with 23S rRNA PCR for molecular diagnosis of bacterial infection in arthroplasty. Anal Biochem 361(2): 169-175.
- Parolo, C. and Merkoçi, A. (2013). Paper-based nanobiosensors for diagnostics. Chem Soc Rev 42(2): 450-457.
- Posthuma-Trumpie, G. A., Korf, J. and van Amerongen, A. (2009). Lateral flow (immuno)assay: its strengths, weaknesses, opportunities and threats. A literature survey. Anal Bioanal Chem 393(2): 569-582.
- Toubanaki, D. K., Athanasiou, E. and Karagouni, E. (2016). Gold nanoparticle-based lateral flow biosensor for rapid visual detection of Leishmania-specific DNA amplification products. J Microbiol Methods 127: 51-58.
- Toubanaki, D. K., Margaroni, M. and Karagouni, E. (2015). Nanoparticle-based lateral flow biosensor for visual detection of fish nervous necrosis virus amplification products. Mol Cell Probes 29(3): 158-166.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Toubanaki, D. K. and Karagouni, E. (2020). RETRACTED: Paper Lateral Flow Biosensor for Nodavirus Reverse Transcribed RNA Detection. Bio-protocol 10(15): e3711. DOI: 10.21769/BioProtoc.3711.
Category
Microbiology > Pathogen detection > Biosensor
Molecular Biology > RNA > RNA detection
Molecular Biology > Nanoparticle > Sensor
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










