- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Flow Cytometry of CD14, VDR, Cyp27 and Cyp24 and TLR4 in U937 Cells
(*contributed equally to this work) Published: Vol 10, Iss 15, Aug 5, 2020 DOI: 10.21769/BioProtoc.3695 Views: 4393
Reviewed by: Xiaoyi ZhengYing ShiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protocol for the Isolation and Analysis of Extracellular Vesicles From Peripheral Blood: Red Cell, Endothelial, and Platelet-Derived Extracellular Vesicles
Bhawani Yasassri Alvitigala [...] Lallindra Viranjan Gooneratne
Nov 5, 2025 1442 Views

Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1337 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1466 Views
Abstract
Chronic Kidney Disease (CKD) patients present a micro inflammation state due to failure renal function. The calcitriol has been described as an anti-inflammatory factor that might modulates the inflammatory response in CKD patients. However, these patients have deficiency of Calcitriol due to failure renal function. But, synthesis of this vitamin has been reported in extra renal production, as in monocytes. In this context, it has been reported that the supplementation with 25 vitamin D (calcidiol or inactive form of vitamin D) induces monocytes to downregulate inflammation, due to the intracellular 1α-hidroxilase that converts calcidiol to calcitriol in these cells. Besides some reports used RT-qPCR, Western Blot or immunofluorescence techniques to investigate the expression of inflammatory and vitamin D machinery biomarkers in several disease, in the present study we used flow cytometry technique to evaluate the effect of 25 vitamin D on CD14, Toll-like receptor 4 (TLR4), vitamin D receptor (VDR), 1-α hydroxylase (CYP27), 24 hydroxylase (CYP24) in monocytes lineage (U937). The U937 culture was incubated with healthy or CKD serum and treatment with/without 25-vitamin D (50 ng/ml for 24 h) to evaluate CD14, TRL4, VDR, CYP27 and CYP24 expression. This protocol showed the advantage to investigate the effect of treatment with 25 vitamin D on the intracellular and cell membrane biomarkers expression quickly and simultaneously. In addition, this technique is not laborious, but easy to perform and to interpret compared to RT-qPCR, western blot or immunofluorescence.
Keywords: 25 vitamin DBackground
Chronic Kidney Disease (CKD) is characterized by the failure filtration rate resulting in serum accumulation of toxic substances, called uremic toxins (Vanholder et al., 2003). It has been shown that uremic toxins induce an inflammatory response in patients with CKD (Rossi et al., 2014; Borges et al., 20 16; Kaminski et al., 2017). Several studies have been reported a relationship between uremic toxins as indoxyl sulfate, p-cresyl sulfate, and indole-3-acetic acid with inflammation (Rossi et al., 2014; Lau et al., 2018; Onal et al., 2019). These toxins signalize NF-ĸB, oxidative stress, upregulated cytokines, intercellular adhesion molecule-1 (ICAM-1) (Bolati et al., 2013; Shimizu et al., 2013; Stockler-Pinto et al., 2018), and induce endothelial injury, contributing to endothelial dysfunction (Carmona et al., 2017). In addition, with the loss of renal function, there is no synthesis of the 1α-hidroxilase enzyme that converts calcidiol (25(OH)D/inactive form of vitamin D) to calcitriol (1,25(OH)2D–active form of vitamin D). The uremic toxins and deficiency of calcitriol have been described to contribute to inflammation in this population (Franca Gois et al., 2018). The calcitriol has been described as anti-inflammatory (Tsoukas et al., 1984; Mathieu and Adorini, 2002). However, in the last 20 years, it has been reported that there is an extra renal production of this vitamin, mainly in monocytes (Stumpf et al., 1979; Schwartz et al., 1998; Cross et al., 2001; Holick, 2007, Gombart et al., 2007). The monocytes have the enzyme 1α-hydroxylase (CYP27) that convert calcidiol (25(OH)D–inactive form of vitamin D) to calcitriol (1,25(OH)2D) and 24 hidroxilase (CYP24) that regulate 1,25(OH)2D over production. The extrarenal synthesis of calcitriol might be linked to autocrine and paracrine biologic regulatory mechanisms, mainly in cells of the immunologic system (Aranow, 2011; Holick, 2012). Besides, the monocytes have TLR-4 and TLR-2 receptors that recognize antigens and activation of these receptors increased expression of VDR (vitamin D receptor) and CYP27, both regulate the conversion of inactive to active form of vitamin D (Kreutz et al., 1993).
As uremic toxins and hypovitaminosis D are important in inflammatory state in CKD environment, the aim of our study was to evaluate in vitro the effect of 25-vitamin D on TLR4, VDR, CYP27 and CYP24 in human monocyte lineage (U937). For this purpose, we evaluated TLR4 and the vitamin D intracellular mechanisms by flow cytometry.
In fact, in our previous study, we have already used this protocol and observed that 25(OH)D improved CYP27 and decreased IL-6, IFN-γ, TLR7 and TLR9 intracellular expression in lymphocytes from dialysis patients (Carvalho et al., 2017).
The VDR, CYP27 and CYP24 can be detected by other techniques such as RT-qPCR, Western-blot and immunohistochemistry as reported by Zou et al. (2019). In this study, authors investigated the effect of Shenyuan granules on the Klotho/FGFR23/Egr1 signaling pathway and calcium-phosphorus metabolism in diabetic nephropathy. Similarly, Chang et al. (Chang and Kim, 2019; Chang, 2019) investigated the effect of vitamin D on muscle cell (C2C12) proliferation and differentiation via VDR, CYP24 and CYP27 by RT-qPCR.
Although RT-qPCR, Western Blot and Immunofluorescence are techniques with high sensitivity and specificity to assess the expression of the TLR4, VDR, CYP27 and CYP24 gene, these are laborious techniques, requiring several stages for their performance (which can contribute to the failure of the techniques), require a large number of cells, high cost compared to the protocol described in the present study using flow cytometry, which is less laborious and easy to perform. Besides CKD patients, this protocol can also be applied in other researches and clinical areas such as oncology, auto immune disease, immune response, since vitamin D may modulate several cell types as lymphocytes that expression VDR, CYP27 and CYP24.
Materials and Reagents
- 25 cm2 cell culture flask (Sigma-Aldrich, catalog number: SIAL0168 ), storage: room temperature
- Coverslip: 18 x 18 mm Thickness 0.13-0.17 mm (Amscope, catalog number: CS-S18-100 , storage: room temperature
- U937 human monocytic lineage cell (ATCC, catalog number: TIB-202 ), storage: Liquid Nitrogen
- APC-VDR secondary antibody, clone mouse (Invitrogem, catalog number: Z25151 ), storage: 8 °C
- CD14-FITC anti-human monoclonal antibody, clone 61D3 (eBioscience, catalog number: 11014942 ), storage: 8 °C
- RPMI 1640 culture medium, pH 7.4 (Sigma-Aldrich, catalog number: R4130 ), storage: 8 °C
- Pen Strep Solution 100 ml (Penicillin Streptomycin) (Gibco by Life Technologies, catalog number: 15140-122 ), storage: -20 °C
- PMA (Phorbol 12-myristate 13-acetate) (Sigma-Aldrich, catalog number: P1585 ), storage: -20 °C
- PBS (Phosphate buffered saline pH 7.4) (Sigma-Aldrich, catalog number: P3744-1 PAK ), storage: room temperature
- Trypsin EDTA solution (Sigma-Aldrich, catalog number: T4049 ), storage: -20 °C
- FBS (Fetal Bovine Serum) (Sigma-Aldrich, catalog number: 12103C ), storage: -20 °C
- Trypan Blue solution (Sigma-Aldrich, catalog number: T8154 ), storage: room temperature
- 25(OH)D (25-Hydroxyvitamin D3 solution 50 µg/ml in ethanol) (Sigma-Aldrich, catalog number: 739642 ), storage: -20 °C
- TLR4-PE anti-human monoclonal antibody, clone HTA125 (eBioscience, catalog number: 12991742 ), storage: 8 °C
- VDR anti-human monoclonal antibody, clone H1512 (Santa Cruz, catalog number: SC13133 ), storage: 8 °C
- CYP27B1 anti-human monoclonal antibody, clone K2911 (Santa Cruz, catalog number: SC49642 ), storage: 8 °C
- Alexa Fluor 647-CYP27B1 secondary antibody, clone goat (Invitrogem, catalog number: Z25608 ), storage: 8 °C
- CYP24 anti-human monoclonal antibody, clone D0811 (Santa Cruz, catalog number: SC66851 ), storage: 8 °C
- Alexa Fluor 488-CYP24 secondary antibody, clone rabbit (Invitrogem, catalog number: Z5302 ), storage: 8 °C
- Fixation/permeabilization buffer–Cytofix/Cytoperm (1x) (BD Pharmingen, catalog number: 554722 ), storage: 8 °C
- Wash buffer containing saponin–Perm/Wash buffer (BD Pharmingen, catalog number: 554723 ), storage: 8 °C
- DMSO (dimetilsulfoxide) (Synth, catalog number: 01D1011.01.BJ ), storage: room temperature
- Sodium azide (Merckmillipore, catalog number: 247-852-1 ), storage: room temperature
- Anticoagulants EDTA (Disodium ethylenediaminetetraacetate dihydrate) (Sigma-Aldrich, catalog number: ED2SS-100g ), storage: room temperature
- Azide sodium (Sigma-Aldrich, catalog number: S2002-100g ), storage: room temperature
- PMA (PhorboI12-Myristate 13-Acetate) (see Recipes)
- 500 ng/ml 25(OH)D (see Recipes)
- 1% Azide PBS solution (see Recipes)
Equipment
- -80 °C freezer
- Hemocytometer/Neubauer chamber (Microyntech, catalog number: MT-BCC-BL , storage: room temperature)
- Flow Citometer: FacsCanto I four parameters (BD Biosciences, San Diego, USA)
- CO2 incubator 4110 (Thermo Fisher Scientific, USA)
- Centrifuge 5804 R (Eppendorf, Germany)
Software
- BD FACSDivaTM (BD Biosciences, San Diego, USA; www.bdbiosciences.com)
- SPSS version 22 statistics software (SPSS, Chicago, USA)
Procedure
- Preparation of healthy and uremic human serum pool
- Healthy Serum Pool: Collect blood samples (20 ml) from 20 healthy volunteers in tubes with anticoagulants and centrifuge at 500 x g for 10 min at 4 °C.
Note: The supernatant should be separated in aliquots, put in a box and storage in vertical freezer at -80 °C. - Uremic Serum Pool: Collect blood samples with anticoagulant tubes from 30 patients in hemodialysis treatment immediately before the beginning of the second session of the week. Centrifuge at 500 x g for 10 min at 4 °C. Store the supernatant in a -80 °C freezer.
Exclusion criteria: patients that had any type of infections, have used any immunosuppressive drugs, vitamin D supplementation or had diabetes mellitus or neoplasia.
- Healthy Serum Pool: Collect blood samples (20 ml) from 20 healthy volunteers in tubes with anticoagulants and centrifuge at 500 x g for 10 min at 4 °C.
- U937 (human monocytic lineage) culture assay
- Culture 1 x 105 U937 cells (human monocytic lineage) in 25 cm cell culture flask (Sigma) with 10 ml of RPMI 1640 culture medium, pH 7.4, supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco, BRL, Life Technologies) and 10% FBS (fetal bovine serum). And then incubate in an incubator with 5% CO2 at 37 °C. Change the culture medium (RPMI 1640) each 24 h until the culture achieves confluence.
- Remove old RPMI from cell culture flask and add a 10 ml of fresh culture medium RPMI with 5 µM/ml of PMA (add 10 µl of 5 mM/ml PMA-stock solution to 10 ml fresh RPMI 1640 culture medium). Maintain the culture in a 5% CO2 incubator at 37 °C for 24 h until U937 cells differentiate into monocytes cells.
- After differentiation, wash the cells with 10 ml of PBS (Sigma) to remove the PMA. Then incubate in 5 ml of trypsin 0.5% for 3 min at room temperature, in order to detach the cells from the surface of the cell culture flask to obtain a homogeneous cell suspension.
- Next wash with 5 ml of RPMI 1640 culture medium for trypsin neutralization. Transfer cells from the cell culture flask to a 15 ml tube and centrifuge for 5 min at 500 x g, 4 °C.
- Remove the supernatant and add 10 ml of fresh RPMI 1640 to test cell viability using Trypan Blue.
- Trypan blue viability assay
- Clean the hemocytometer.
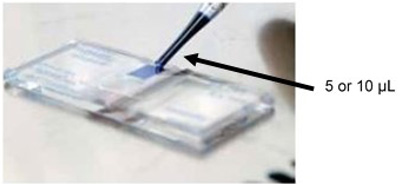
Figure 1. Fill 5 or 10 µl cell suspendsion into both sides of the chamber in Hemocytometer - Moisten the coverslip with water or exhaled breath. Slide the coverslip over the chamber back and forth using slight pressure until Newton’s refraction rings appear (Newton’s refraction rings are seen as rainbow-like rings under the coverslip).
- Under sterile conditions take out 100-200 µl of cell suspension, transfer it in a micro tube and staining with an equal volume of Trypan Blue solution (dilution factor = 2) and mix by gentle pipetting. The maximum time of trypan blue staining is until 30 seconds (Figure 1).
- Fill both sides of the chamber with cell suspension (approximately 5-10 µl) (Figure 1) and view under an inverted phase contrast microscope using 20x magnification.
- Count the number of viable (seen as bright cells) and non-viable cells (stained blue). Ideally >100 cells should be counted in order to increase the accuracy of the cell count (see note below) (Figure 2).
Note: The number of squares counted to obtain your count of > 100.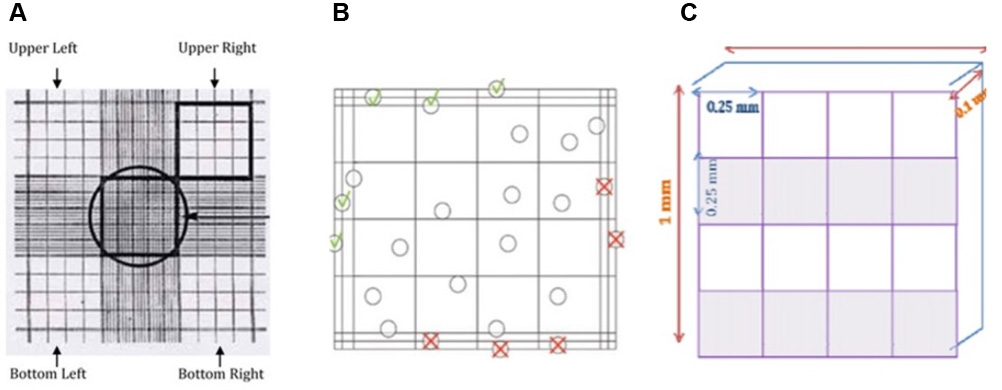
Figure 2. Hemocytometer design. A. Image from hemocytometer by inverted phasecontrast. B. Count viable Cells (bright/clear cells) and dead cells (blue) on four squares (upper left + right + bottom left + right ( ), but not the upper and bottom right border (
), but not the upper and bottom right border ( ) and not the upper and bottom left border (
) and not the upper and bottom left border ( ). C. Hemocytometer area
). C. Hemocytometer area - Calculate the concentration of viable and non-viable cells and the percentage of viable cells using the example below and as illustrated above:
Volume hemacytometer = 1 × 1 × 0.1 = 0.1 mm3
Calculate the concentration of viable and non-viable cells per ml.
For example: there are 120 counted viable cells or non-viable cells on the hemacytometer, then
the concentration of viable cells per ml = 120 cells /0.1 mm3 × 2 × 1,000 mm3/1 ml =2.4 × 106 cells/ml
which 2 is the dilution factor (V cell suspension/V trypan Blue);
1 ml corresponds 1,000 mm3.Percentage Viability=(N0 of viable cells)/(Total N0 of cells)×100
- Clean the hemocytometer.
- Treatment with or without 25(OH)D and with or without uremic pool serum assay
After calculating the viability (that must be around 95%), use the tube of 15 ml with cell suspension in the RPMI (that is used to calculate viability) and transfer 1 x 106 cells for each condition to cell culture flask described allow: (In order to adjust the 1 x 106 cell to each condition of treatment described below, it is needed cell count by hemocytometer).- Treatment with healthy pool serum: Transfer 1 x 106 cells to 25 cm2 cell culture flask with 2.5 ml of RPMI 1640 and with 2.5 ml of healthy pool serum. Incubate for 24 h with 5% CO2 at 37 °C.
- Treatment with 25(OH)D and healthy pool serum: Transfer 1 x 106 cells to a 25 cm2 cell culture flask with 2.5 ml of RPMI 1640 and 2.5 ml of healthy pool serum and add 50 ng/ml of 25(OH)D. Incubate for 24 h, 5% CO2 at 37 °C.
- Treatment with uremic pool serum: Transfer 1 x 106 cells to a 25 cm2 cell culture flask with 2.5 ml of RPMI 1640 and 2.5 ml of uremic pool serum. Incubate for 24 h, 5% CO2 at 37 °C.
- Treatment with 25(OH)D and uremic pool serum: Transfer 1 x 106 cells to a 25 cm2 cell culture flask with 2.5 ml of RPMI 1640 and with 2.5 ml of uremic pool serum and add 50 ng/ml of 25(OH)D. Incubate for 24 h, 5% CO2 at 37 °C.
- Detection of TLR4, VDR, CYP27 and CYP24 human monoclonal antibodies
- After incubation for each condition, transfer cells of each condition of treatment with or without 25(OH)D described above, centrifuge 10 min, 500 x g at 4 °C. Discard supernatant and add to pellet 1 ml of PBS.
- To each conditions of treatment with or without 25(OH)D and healthy pool serum or uremic pool serum, add 3 x 105/100 µl of PBS in 3 (three) tubes to stain with antibodies, respectively. Add each monoclonal antibody according to Table 1.
Table 1. Monoclonal antibody protocol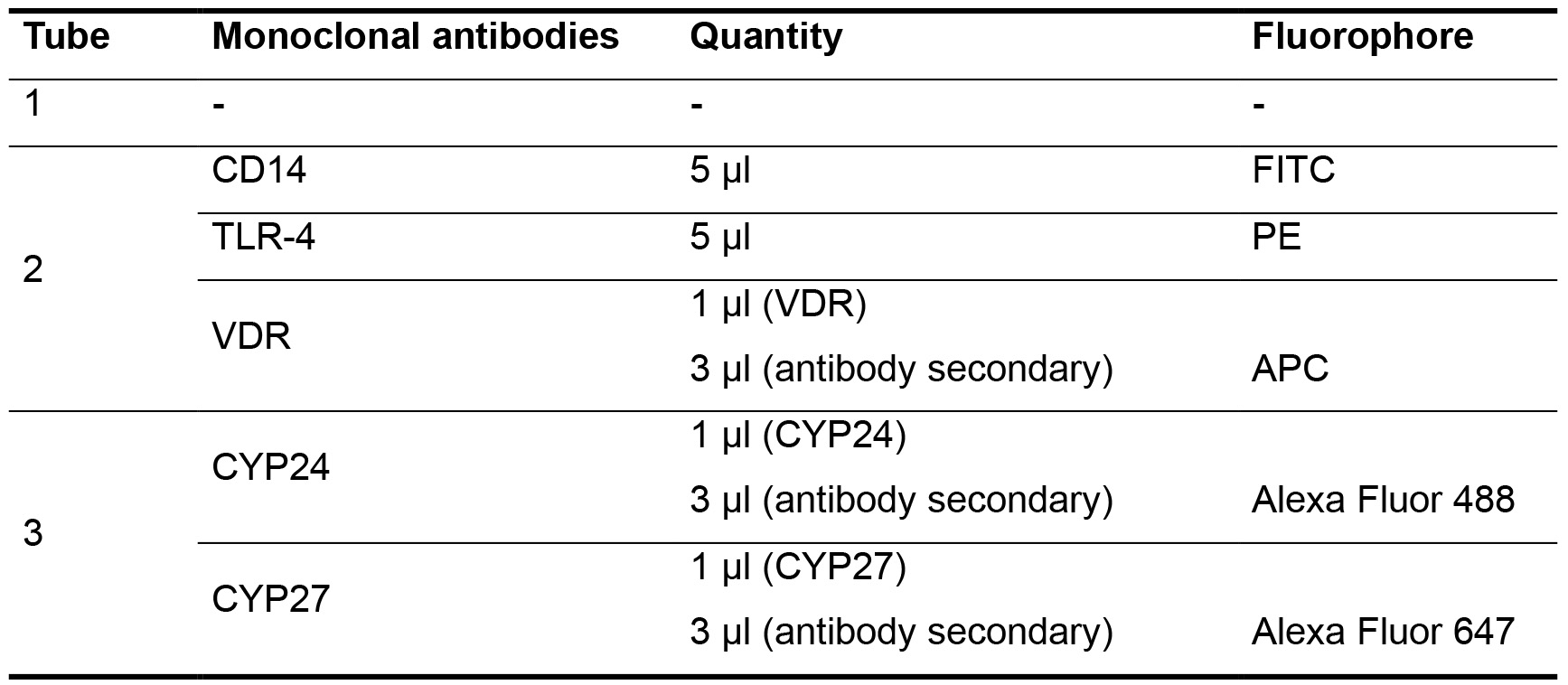
- Tube 1: 3 x 105 cells + 100 µl of PBS. Ready to analyze in flow cytometry.
- Tube 2: 3 x 105 cells + 100 µl of PBS + 5 µl CD14 + 5 µl TLR4, incubate for 15 min in the dark at room temperature. After, centrifuge 5 min, 500 x g at 4 °C. Remove supernatant and add 1 ml with Fixation/permeabilization buffer for 20 min. After, add 1 µl of VDR antibodies at concentration of 1 μg/ml (Santa Cruz, San Diego, USA) and incubate for 30 min at fridge (2-8 °C). After, washing 3 times with Wash buffer containing saponin–Perm/Wash buffer and incubate with 3 μg/ml of respective secondary antibodies (APC-conjugated anti-human VDR) for 15 min at fridge (2-8 °C) Centrifuge and discard the supernatant. Add 300 µl of 1% azide PBS solution. Ready to analyze in flow cytometry.
- Tube 3: 3 x 105 cells + 100 µl of PBS, add 1 ml with Fixation/permeabilization buffer for 20 min. After, add 1 µl of CYP27B1 + 1 µl CYP24 antibodies at concentration of 1 μg/ml (Santa Cruz, San Diego, USA) and incubate for 30 min at fridge (2-8 °C). After, washing 3 times with Wash buffer containing saponin–Perm/Wash buffer and incubate with 3 μg/ml of respective secondary antibodies (Alexa-Fluor 647-conjugated anti-human CYP27B1 and Alexa-Fluor 488-conjugated anti-human CYP24)) for 15 min at fridge (2-8 °C). Centrifuge and discard the supernatant. Add 300 µl of 1% azide PBS solution. Ready to analyze in flow cytometry.
- Flow cytometry analysis (BD FacsCanto I)
- Use a forward scatter plot versus side scatter plot to make a gate for the CD14+ (monocytes cells) and to exclude debris (Figures 3A-3D). Use DOT-PLOT graphs to analyze the CD14+ (Figure 3E), CD14+TLR4+ (Figure 3F) and CD14+VDR+ (Figure 3G).
- Use DOT-PLOT graphs to analyze the CD14+ (Figure 4A) and use histograms to analyze the CYP27+ (Figure 4D) and CYP24+ (Figure 4E). Describe results as mean fluorescence intensity (MFI), such that the higher the MFI the greater the expression of these receptors. For each sample, acquire 30,000 events/cells.
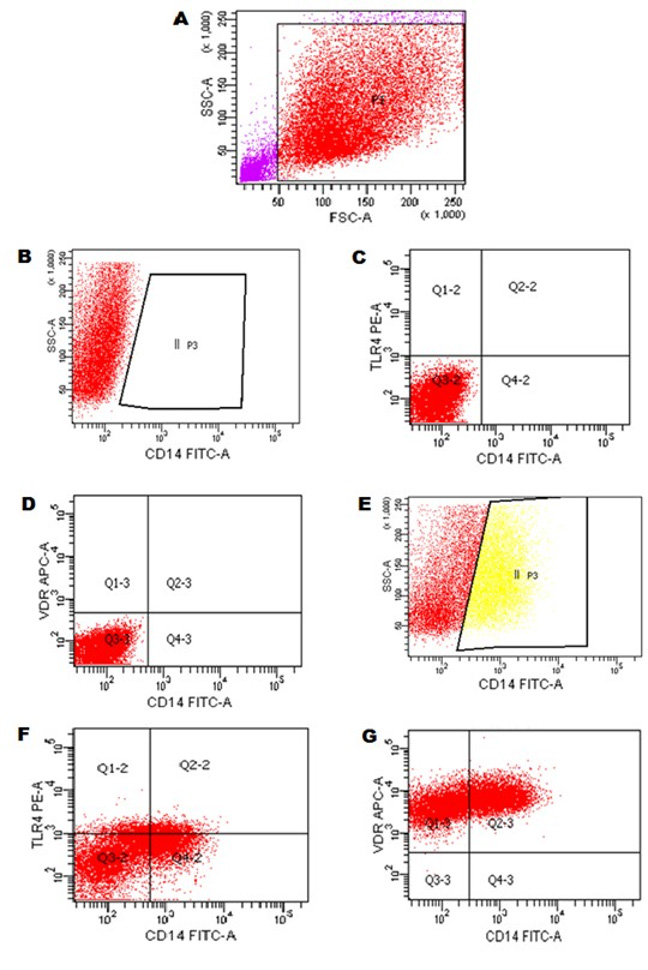
Figure 3. Flow cytometry gating strategy of CD14+, TRL4 and VDR expression in monocytes cells. A. Monocytes cells were selected at gate P1 according to the characteristics of size (Foward Scatter–FSC) and internal complexity (Side Scatter–SSC). B-D. Adjustment of the autofluorescence of monocytes cells in the detection channels for the respective antibodies marked with the respective fluorochromes FITC, PE and APC. E. II P3 = Expression of CD14 FITC+ in monocytes cells. F. Q2-2 = Expression of TLR4 PE+ and CD14 FITC+ in monocytes cells. G. Q2-3 = Expression of VDR APC+ and CD14 FITC+ in monocytes cells.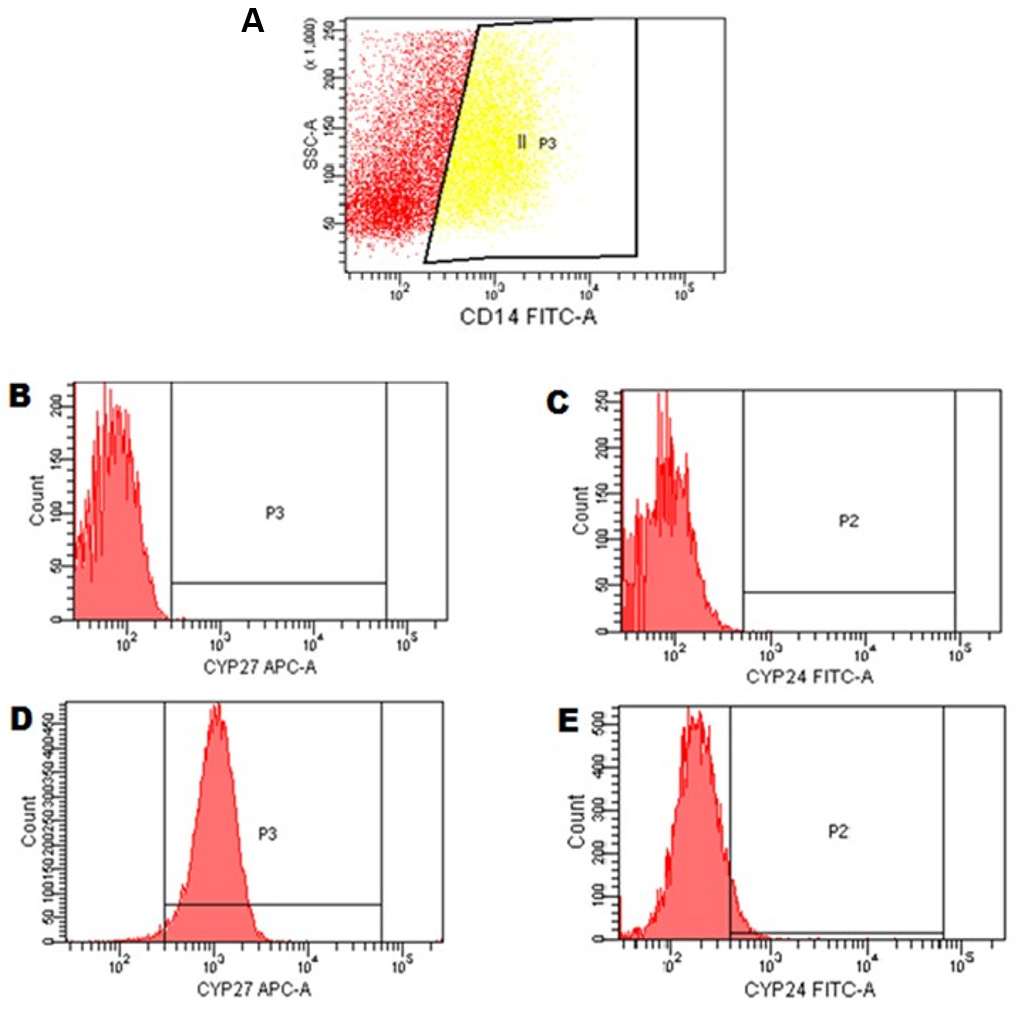
Figure 4. Flow cytometry strategy for CYP27 and CYP24 in monocytes cells. A. Gate II P3 = monocytes cells CD14+ were selected based on previous experiments. Histogram type graphics were created (number of cells in relation to the detected fluorescence intensity). B and C. Cell autofluorescence adjustment for monoclonal antibody detection channels conjugated to the respective fluorochromes: Alexa fluor 647 is detected in APC flow cytometry channel and Alexa Fluor 488 is detected in FITC flow cytometry channel. D and E. monocytes positive for expression of CYP27 and CYP24 as shown in the quadrants P3 and P2, respectively.
Data analysis
Twenty (20) replicates were used in each experiment. The results according to each group (healthy, uremic, with 25-vitamin D supplementation and without 25-vitamin D supplementation) were expressed as median (minimum and maximum), and the differences tested by Kruskal Wallis. Relationships between independent variables were performed in the entire group. Kolmogorov-Smirnov test was used to test normality of data. We used Pearson or Spearman correlation coefficient, as appropriate. The value of P < 0.05 was considered for statistical significance. The analyses were performed using SPSS (Statistical Package Social Sciences) software version 22.0 for Windows. These analyses have already appeared in the text from original article (Brito et al., 2020).
Recipes
- PMA (PhorboI12-Myristate 13-Acetate); 5 mg; FW 616.8
1 M dilution = 616 mg/ml
5 x 10-3 M dilution (5 mM) = 5 mg in 1.62 ml in DMSO- To prepare a 5 mM dilution of PMA stock, dilute 5 mg in 1.62 ml of DMSO
- Light protect during preparation as PMA is light sensitive
- Store at -20 °C in 10 µl aliquots (approx. 160 Eppendorfs) till use (shelf life: 6 months)
- For the protocol, a further 10-3 dilution of this stock aliquot is made in RPMI to achieve a final dilution of 5 µM, which is then used in the experiments)
- 500 ng/ml 25(OH)D
Dilute the purchased solution in ethanol (Sigma-Aldrich, 50 µg/ml) at a ratio of 1:100. For example: dilute 50 µl of stock solution in 5 ml of ethanol. Store at -20 °C in 120 µl aliquots (approx. 40 Eppendorfs) untill use
Note: For this protocol, a further 10-1 dilution of this stock aliquot is made directly in Cell culture flask with RPMI to achieve a final dilution of 50 ng/ml, which is then used in the experiments for 25(OH)D treatment. - 1% Azide PBS solution (storage in refrigerator 2-8 °C)
Dissolve 1 g of azide sodium in 10 ml of PBS
Acknowledgments
The authors thank all the professionals who provided essential technical support during the conduction of this work. This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo, FAPESP (Grant Number 2014-14192-1).
Data collection, protocol assay, flow cytometry data acquisition, analysis, interpretation, and approval of the final version of the article: Rebello J.F.; Brito, R.; and Grabulosa, C.C. Interpretation and significant intellectual insights and review and approval of the final version of the article: Elias, R.M. and Moysés, R.M. Study design, data analysis, interpretation, intellectual insights, manuscript writing, review, and approval of the final version of the article, approval of the final version of the article: Dalboni, M.A.
Competing interests
The authors declare no competing interests.
Ethics
This study was approved by the Institutional Review Board of the Federal University of São Paulo (#0727/10).
Healthy and uremic serum samples were obtained after written informed consent of participants or legal representative, according to the Helsinki Declaration and local regulations.
References
- Aranow, C. (2011). Vitamin D and the immune system. J Investig Med 59(6): 881-886.
- Bhalla, A. K., Amento, E. P., Clemens, T. L., Holick, M. F. and Krane, S. M. (1983). Specific high-affinity receptors for 1,25-dihydroxyvitamin D3 in human peripheral blood mononuclear cells: presence in monocytes and induction in T lymphocytes following activation. J Clin Endocrinol Metab 57(6): 1308-1310.
- Bolati, D., Shimizu, H., Yisireyili, M., Nishijima, F. and Niwa, T. (2013). Indoxyl sulfate, a uremic toxin, downregulates renal expression of Nrf2 through activation of NF-kappaB. BMC Nephrol 14: 56.
- Borges, N. A., Barros, A. F., Nakao, L. S., Dolenga, C. J., Fouque, D. and Mafra, D. (2016). Protein-bound uremic toxins from gut microbiota and inflammatory markers in chronic kidney disease. J Ren Nutr 26(6): 396-400.
- Brito, R. B. O., Rebello, J. F., Grabulosa, C. C., Pinto, W., Morales, A., Jr., Elias, R. M., Moyses, R. M. A. and Dalboni, M. A. (2020). 25-vitamin D reduces inflammation in uremic environment. Sci Rep 10(1): 128.
- Carmona, A., Guerrero, F., Buendia, P., Obrero, T., Aljama, P. and Carracedo, J. (2017). Microvesicles derived from indoxyl sulfate treated endothelial cells induce endothelial progenitor cells dysfunction. Front Physiol 8: 666.
- Carvalho, J. T. G., Schneider, M., Cuppari, L., Grabulosa, C. C., D, T. A., BM, Q. R., M, C. B., Cendoroglo, M., Maria Moyses, R. and Dalboni, M. A. (2017). Cholecalciferol decreases inflammation and improves vitamin D regulatory enzymes in lymphocytes in the uremic environment: A randomized controlled pilot trial. PLoS One 12(6): e0179540.
- Chang, E. and Kim, Y. (2019). Vitamin D ameliorates fat accumulation with AMPK/SIRT1 activity in C2C12 skeletal muscle cells. Nutrients 11(11): 2806.
- Chang, E. (2019). 1,25-Dihydroxyvitamin D decreases tertiary butyl-hydrogen peroxide-induced oxidative stress and increases AMPK/SIRT1 activation in C2C12 muscle cells. Molecules 24(21): 3903.
- Cross, H. S., Bareis, P., Hofer, H., Bischof, M. G., Bajna, E., Kriwanek, S., Bonner, E. and Peterlik, M. (2001). 25-Hydroxyvitamin D3-1α-hydroxylase and vitamin D receptor gene expression in human colonic mucosa is elevated during early cancerogenesis. Steroids 66(3-5): 287-292.
- Franca Gois, P. H., Wolley, M., Ranganathan, D. and Seguro, A. C. (2018). Vitamin D deficiency in chronic kidney disease: recent evidence and controversies. Int J Environ Res Public Health 15(8).
- Gombart, A. F., O'Kelly, J., Saito, T. and Koeffler, H. P. (2007). Regulation of the CAMP gene by 1,25(OH)2D3 in various tissues. J Steroid Biochem Mol Biol 103(3-5): 552-557.
- Holick, M. F. (2012). Vitamin D: extraskeletal health. Rheum Dis Clin North Am 38(1):141-60.
- Holick, M. F. (2007). Vitamin D deficiency. N Engl J Med 357(3): 266-281.
- Kaminski, T. W., Pawlak, K., Karbowska, M., Mysliwiec, M. and Pawlak, D. (2017). Indoxyl sulfate - the uremic toxin linking hemostatic system disturbances with the prevalence of cardiovascular disease in patients with chronic kidney disease. BMC Nephrol 18(1): 35.
- Kreutz, M., Andreesen, R., Krause, S. W., Szabo, A., Ritz, E. and Reichel, H. (1993). 1,25-dihydroxyvitamin D3 production and vitamin D3 receptor expression are developmentally regulated during differentiation of human monocytes into macrophages. Blood 82(4): 1300-1307.
- Lau, W. L., Savoj, J., Nakata, M. B. and Vaziri, N. D. (2018). Altered microbiome in chronic kidney disease: systemic effects of gut-derived uremic toxins. Clin Sci (Lond) 132(5): 509-522.
- Mathieu, C. and Adorini, L. (2002). The coming of age of 1,25-dihydroxyvitamin D3 analogs as immunomodulatory agents. Trends Mol Med 8(4): 174-179.
- Onal, E. M., Afsar, B., Covic, A., Vaziri, N. D. and Kanbay, M. (2019). Gut microbiota and inflammation in chronic kidney disease and their roles in the development of cardiovascular disease. Hypertens Res 42(2): 123-140.
- Rossi, M., Campbell, K. L., Johnson, D. W., Stanton, T., Vesey, D. A., Coombes, J. S., Weston, K. S., Hawley, C. M., McWhinney, B. C., Ungerer, J. P. and Isbel, N. (2014). Protein-bound uremic toxins, inflammation and oxidative stress: a cross-sectional study in stage 3-4 chronic kidney disease. Arch Med Res 45(4): 309-317.
- Schwartz, G. G., Whitlatch, L. W., Chen, T. C., Lokeshwar, B. L. and Holick, M. F. (1998). Human prostate cells synthesize 1,25-dihydroxyvitamin D3 from 25-hydroxyvitamin D3. Cancer Epidemiol Biomarkers Prev 7(5): 391-395.
- Shimizu, H., Yisireyili, M., Higashiyama, Y., Nishijima, F. and Niwa, T. (2013). Indoxyl sulfate upregulates renal expression of ICAM-1 via production of ROS and activation of NF-kappaB and p53 in proximal tubular cells. Life Sci 92(2): 143-148.
- Stockler-Pinto, M. B., Soulage, C. O., Borges, N. A., Cardozo, L., Dolenga, C. J., Nakao, L. S., Pecoits-Filho, R., Fouque, D. and Mafra, D. (2018). From bench to the hemodialysis clinic: protein-bound uremic toxins modulate NF-kappaB/Nrf2 expression. Int Urol Nephrol 50(2): 347-354.
- Stumpf, W. E., Sar, M., Reid, F. A., Tanaka, Y. and DeLuca, H. F. (1979). Target cells for 1,25-dihydroxyvitamin D3 in intestinal tract, stomach, kidney, skin, pituitary, and parathyroid. Science 206(4423): 1188-1190.
- Tsoukas, C. D., Provvedini, D. M. and Manolagas, S. C. (1984). 1,25-dihydroxyvitamin D3: a novel immunoregulatory hormone. Science 224(4656): 1438-1440.
- Vanholder, R. C. and Glorieux, G. L. (2003). An overview of uremic toxicity. Hemodialysis International 7(2): 156-161.
- Zou, X. R., Zhan, L. R., Chen, L., Long, Q. H., Yuan, J., Wang, L. and Wang, X. Q. (2019). Influence of the Klotho/FGF23/Egr1 signaling pathway on calcium-phosphorus metabolism in diabetic nephropathy and the intervention of Shenyuan granules. J Biol Regul Homeost Agents 33(6): 1695-1702.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Rebello, J. F., de Oliveira Brito, R. B., Grabulosa, C. C., Moyses, R. M. A., Elias, R. M. and Dalboni, M. A. (2020). Flow Cytometry of CD14, VDR, Cyp27 and Cyp24 and TLR4 in U937 Cells. Bio-protocol 10(15): e3695. DOI: 10.21769/BioProtoc.3695.
Category
Immunology > Immune mechanisms > In vitro model
Cell Biology > Cell-based analysis > Flow cytometry
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









