- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Metabolomic and Lipidomic Analysis of Bone Marrow Derived Macrophages
Published: Vol 10, Iss 14, Jul 20, 2020 DOI: 10.21769/BioProtoc.3693 Views: 8987
Reviewed by: Parul MehrotraMarco LeporeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Differentiation of Bone Marrow Monocytes into Alveolar Macrophages-like Cells through Co-culture with Lung Epithelial Cells and Group 2 Innate Lymphoid Cells
Pauline Loos [...] Laurent Gillet
Sep 20, 2023 2461 Views
![Automated Sequential Derivatization for Gas Chromatography-[Orbitrap] Mass Spectrometry-based Metabolite Profiling of Human Blood-based Samples](https://en-cdn.bio-protocol.org/imageup/arcimg/20250107224024942.jpg?t=1770626474)
Automated Sequential Derivatization for Gas Chromatography-[Orbitrap] Mass Spectrometry-based Metabolite Profiling of Human Blood-based Samples
Akrem Jbebli [...] Elliott J. Price
Mar 5, 2025 2355 Views

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2234 Views
Abstract
Macrophages are highly plastic immune cells that are capable of adopting a wide array of functional phenotypes in response to environmental stimuli. The changes in macrophage function are often supported and regulated by changes in cellular metabolism. Capturing a comprehensive picture of metabolism is vital for understanding the role of metabolic rewiring in the immune response. Here we present a method for systematically quantifying the abundance of metabolites and lipids in primary murine bone marrow derived macrophages (BMDMs). This method simultaneously extracts polar metabolites and lipids from BMDMs using a rapid two-phase extraction procedure. The polar metabolite fraction and lipid fraction are subsequently analyzed by separate liquid chromatography-mass spectrometry (LC-MS) methods for optimized coverage and quantification. This allows for a comprehensive characterization of cellular metabolism that can be used to understand the impact of a variety of environmental stimuli on macrophage metabolism and function.
Keywords: LC-MSBackground
Macrophages, cells of the innate immune system, can adopt a multitude of functional phenotypes in response to cues within the local microenvironment. The activation of macrophages is coupled to, and highly reliant on, specific reprograming of cellular metabolism (Tannahill et al., 2013; Galván-Peña and O’Neill, 2014; Jha et al., 2015; Kelly and O'Neill, 2015; Cordes et al., 2016; Mills and O'Neill 2016; et al.; Mills et al., 2016; Liu et al., 2013; Lampropoulou et al., 2016; Van den Bossche et al., 2017; Williams et al., 2018; Martin et al., 2017). The metabolic reprogramming is wide-spread, involving changes in central metabolism, amino acid metabolism, and lipid remodeling (Galván-Peña and O’Neill, 2014; O’Neill and Pearce, 2016; Van den Bossche et al., 2017). These changes in different pathways are interdependent, allowing cells to produce energy, signaling molecules (e.g., eiconsanoids), and effector molecules (e.g., nitric oxide and reactive oxygen species) for immune functions. Systematic profiling of metabolites and lipids is an important tool to further understand the role these biomolecules play in macrophages.
Development of metabolomic and lipidomic methods has enabled reliable quantification of hundreds of metabolites and lipids simultaneously (Fiehn, 2002; Wenk, 2005). This protocol presents a method for isolation of murine bone marrow derived macrophages (BMDMs), extraction of their lipids and small molecule metabolites and subsequent quantification of their abundance using liquid chromatography–mass spectrometry (LC-MS). BMDMs are a frequently used model for understanding macrophage function and metabolism. Adaptation of the “BUME (butanol and methanol)” extraction method (Löfgren et al., 2012 and 2016), allows for simple and quick extraction of a wide range of both polar metabolites and lipids from a single sample of BMDMs. This reduces variability and gives good recovery. The cell extracts are analyzed by LC-MS, which provides the sensitivity and selectivity required for metabolomic and lipidomic analyses (Theodoridis et al., 2012; Gika et al., 2014). Utilization of parallel LC-MS methods for separate analysis of metabolites and lipids, as outlined in this protocol, provides the broad coverage of the metabolome and lipidome. The LC-MS data are then analyzed using previously developed and publically available analysis tools, MAVEN and LipiDex, giving confident identification and quantification of a wide range metabolite and lipid species (Melamud et al., 2010; Clasquin et al., 2012; Hutchins et al., 2018).
This method can be used to characterize how metabolism in macrophages is altered in response to different stimuli, and how it may be regulated by other microenviromental factors (e.g., nutrient availability). This LC-MS based metabolomics and lipidomics method can be further coupled with isotopic tracing approaches to determine the changes in metabolic flux during immune response, elucidate the mechanisms controlling metabolic rewiring, and facilitate the investigation of the mechanisms connecting altered metabolism to broader macrophage functions.
Materials and Reagents
- Isolation and culture of BMDM
- 27½ G needle (BD Biosciences, catalog number: 305109 )
- 10 ml plastic disposable syringe (Thermo Scientific, catalog number: S7515-10 )
- 70 μm cell strainer (Thermo Fisher Scientific, catalog number: 0 87712 )
- 0.45 μm PES filter unit (Thermo Fisher Scientific, catalog number: 162-0045 )
- 50 ml conical tubes (Thermo Fisher Scientific, catalog number: 339652 )
- Countess cell counting chambers (Invitrogen, catalog number: C10283 )
- Petri dishes (Fisherbrand, catalog number: FB0875712 )
- 6-well culture plates (Eppendorf, catalog number: 00 30720113 )
- 175 cm2 flasks (Eppendorf, catalog number: 00 30710029 )
- Sterile top filters (Nalgene, catalog number: 596-3320 )
- Cell scraper (Corning, catalog number: 3008 )
- 6+ weeks old mouse (C57BL/6J, Jackson Laboratory)*
- L929 cells (ATCC®, catalog number: CCL-1TM)
- 0.4% Trypan blue stain (Gibco, catalog number: 15250-061 )
- 70% Ethanol (Pharmco, catalog number: 1110002000 )
- DMEM High Glucose Medium (Sigma-Aldrich, catalog number: D1152 )
- Penicillin Streptomycin (Thermo Fisher Scientific, catalog number: 15-140-122 )
- Fetal Bovine Serum (FBS, Hyclone, catalog number: 89133-098 )
- Dialyzed Fetal Bovine Serum (dFBS, Hyclone, catalog number: 16777-212 )
- Recombinant mouse macrophage colony stimulating factor (mCSF, R&D Systems, catalog number: 416ML050 )
- RPMI 1640 Medium without Glutamine (Hyclone, catalog number: SH30096.01 )
- Glutamine (Thermo Fisher, catalog number: BP379-100 )
- HEPES (VWR, catalog number: 16777-032 )
- Lipopoylsaccharide from E. coli O111:B4 (LPS, Sigma-Aldrich, catalog number: L3024 )**
- Interferon-γ (IFNγ, R&D Systems, catalog number: 485-MI-100 )**
- Accutase (STEMCell Technologies, catalog number: 0 7922 )
- Trypsin-EDTA (0.05%) (Thermo Fisher Scientific, catalog number: 25300062 )
- Phosphate buffered saline (Thermo Fisher Scientific, catalog number: 14190144 )
- L929 conditioned media (see Recipes)
- BMDM maintenance media (see Recipes)
- BMDM differentiation media (see Recipes)
- L929 culture media (see Recipes)
- *Any mouse model with sufficiently healthy bone marrow can be used.
- **Optional based on desired stimulation protocol. LPS and IFN-γ stimulation will induce classical activation.
- Metabolite and Lipid Extraction and Sample Preparation
- Pipette tips 1,000 μl, 200 μl (VWR, catalog numbers: 89079-470 , 89079-478)
- 1.5 ml plastic tube (VWR, catalog number: 89000-028 )
- Glass bottles (used for LC-MS grade solvents only, see Notes)
- LC-MS polypropylene vials (Thermo Fisher Scientific, catalog number: 03-377-299 )
- LC-MS glass vials (Thermo Fisher Scientific, catalog number: 03-FIRVA )
- Snap caps (Thermo Fisher Scientific, catalog number: C4011-53 )
- Polypropylene microcentrifuge tubes (VWR, catalog number: 89000-028 )
- Glass vials (Wheaton, catalog number: 224740 )
- Nitrogen gas
- 1-Butanol LC-MS grade (Sigma-Aldrich, catalog number: 34867 )
- Methanol LC-MS grade (Fisher Chemical, catalog number: A456-4 )
- n-Heptane LC-MS grade (Sigma-Aldrich, catalog number: 1036541000 )
- Ethyl acetate LC-MS grade (Fisher Scientific, catalog number: E195-4 )
- Acetic acid LC-MS grade (Fisher Chemical, catalog number: A11350 )
- Water LC-MS grade (Thermo Fisher Scientific, catalog number: W64 )
- Tributylamine (Sigma-Aldrich, catalog number: 90780 )
- Ammonium Acetate LC-MS grade (Fisher Scientific, catalog number: A114-50 )
- Acetonitrile LC-MS grade (Thermo Fisher Scientific, catalog number: A955-4 )
- Isopropanol (Thermo Fisher Scientific, catalog number: A461-4 )
- SPLASH LipidoMIX Internal Standard (Avanti Polar Lipids, catalog number: 330707 )
- Ice
- LC-MS Analysis
- Acquity UPLC BEH C18 Column, 130Å, 1.7 µm, 2.1 mm x 100 mm (Waters, catalog number: 186002352 )
- Acquity UPLC BEH C18 VanGuard Pre-column, 130Å, 1.7 µm, 2.1 mm x 5 mm (Waters, catalog number: 186003975 )
- Acquity UPLC CSH C18 Column, 130Å, 1.7 µm, 2.1 mm x 100 mm (Waters, catalog number: 186005297 )
- Acquity UPLC CSH C18 VanGuard Pre-column, 130Å, 1.7 µm, 2.1 mm x 5 mm (Waters, catalog number: 186005303 )
- Solvent B (100% LC-MS grade methanol)
- Solvent A (see Recipes)
- Solvent C (see Recipes)
- Solvent D (see Recipes)
Equipment
- Surgery Scissors (Fisherbrand, catalog number: 08-935 )
- Surgery Forceps (BrainTree Scientific, catalog number: FC03-41 )
- Thermo Q-Exactive Quadrupole Orbitrap Mass Spectrometer (Thermo Scientific) coupled to a Vanquish Horizon UHPLC (Thermo Scientific)
- Nitrogen Stream Sample Concentrator (Techne, catalog number: FSC400D ), equipped with 127 mm needles (Techne, catalog number: F7210 )
- Vortexer (VWR, catalog number: 10153-838 )
- Micro Centrifuge (Beckman Coulter, model: Microfuge 20 , catalog number: B31599 )
- Benchtop Centrifuge (Thermo, model: IEC Centra CL2 )
- -20 °C Freezer
- 4 °C Fridge
- Standard Fume Hood
- Pipettes (P1000, P200)
- Sterile Cell Culture Hood
- Cell Incubator (37 °C, 5% CO2)
- Countess II Cell Counter (AMQAX1000)
Software
- Thermo Scientific XCalibur 4.1
- MAVEN (v. 682) (http://genomics-pubs.princeton.edu/mzroll/index.php) (Melamud et al., 2010; Clasquin et al., 2012)
- MZMine2 (http://mzmine.github.io/) (Pluskal et al., 2010)
- MSConvertGUI (ProteoWizard, http://proteowizard.sourceforge.net/download.html) (Kessner et al., 2008)
- LipiDex (https://github.com/coongroup/LipiDex) (Hutchins et al., 2018)
Procedure
- Isolation of Bone Marrow Cells and Differentiation into Macrophages (Figure 1)
See additional resource for helpful demonstration of bone marrow isolation (Ying et al., 2013).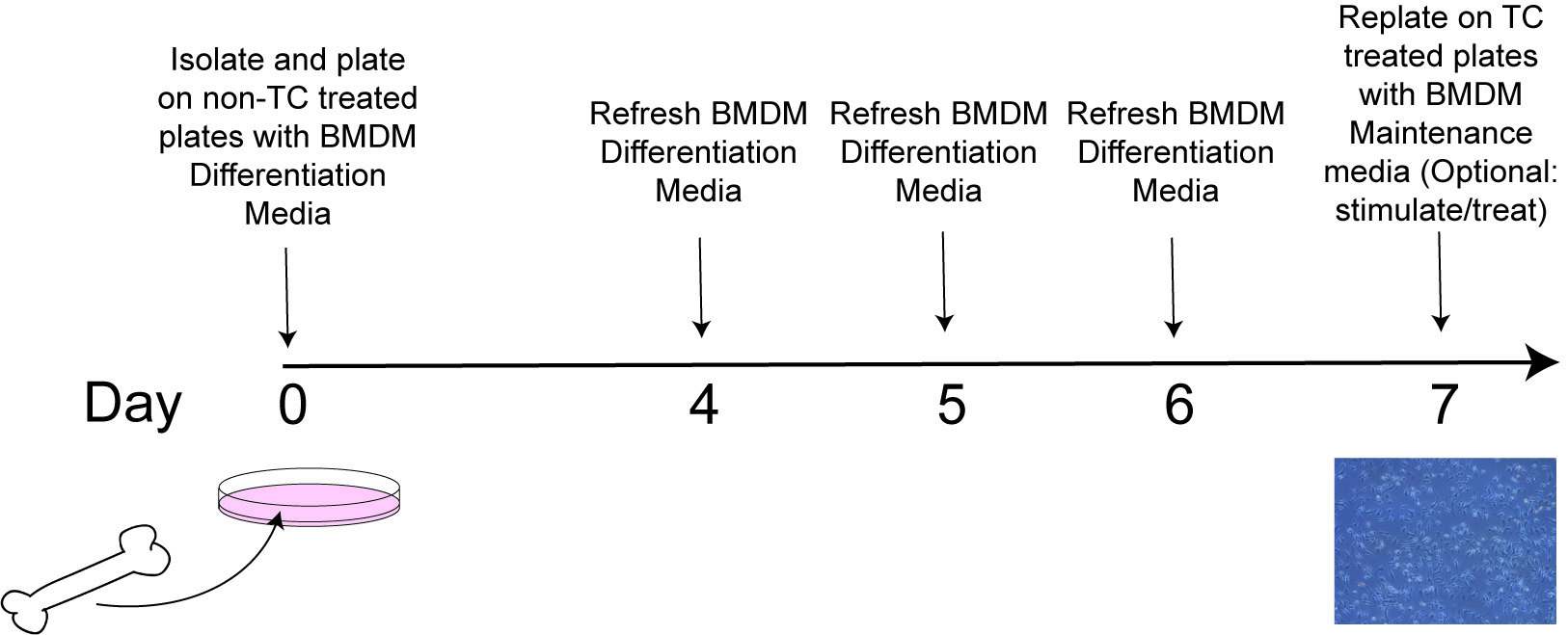
Figure 1. Timing of BMDM Differentiation Protocol. TC treated: tissue culture treated; BMDM: bone marrow derived macrophages.- Euthanize mice by rapid cervical dislocation.
- Thoroughly spray mouse with 70% ethanol and bring into sterile hood.
- Using aseptic technique, isolate femur and tibia from each leg of mouse.
- Using a sharp scissors soaked in ethanol, sever bone proximal to each joint.
- Using a 27½ G needle attached to a 10 ml syringe, flush bones with cold, sterile BMDM Differentiation Media into a 10 cm dish, until bone cavity appears white. Use as much media is required to fully flush the bone.
- Pipette cell suspension thoroughly to fully resuspend bone marrow cells.
- For removal of any muscle or bone fragments, filter cells through 70 µm filter, into a sterile 50 ml conical centrifuge tube.
- Centrifuge cells for 3 min at 500 x g, 4 °C.
- Discard supernatant and resuspend cell pellet in 100 ml of BMDM Differentiation Media.
- Plate 10 ml of cell suspension onto each Petri-dish (non tissue culture treated plates are important here as macrophages will attach too strongly to treated plates)
- Place in cell culture incubator for 3 days.
- Four days post plating aspirate media from cells and replace with 10 ml fresh BMDM Differentiation Media. Repeat on day 5 and day 6.
- Seven days post plating aspirate media from cells and add 4 ml of Accutase per plate. Place back in incubator for approximately 5 min.
- Using pipette, gently dislodge cells from plate and combine all cells in a 50 ml conical tube.
- Spin down (1,000 x g, 5 min) and resuspend into 20 ml BMDM Maintenance media.
- Count cells by staining 1:1 with 0.4% Trypan Blue.
- Resuspend cells to 2.5 x 105 cells per ml in BMDM Maintenance Media and plate on TC treated cell culture plates (2 ml per well of 6-well plate). Three replicates (e.g., 3 wells of a 6-well plate) are recommended for each condition. Additional wells should be designated for protein or cell count normalization.
- Cell will adhere to plate with in a matter of a couple hours. After adherence, cells are ready for stimulation and/or extraction. To stimulate macrophages, treat with 50 ng/ml of LPS and 10 ng/ml IFN-γ, or other stimuli of interest, for desired duration.
- Isolation of Polar Metabolites and Lipids (Figure 2)
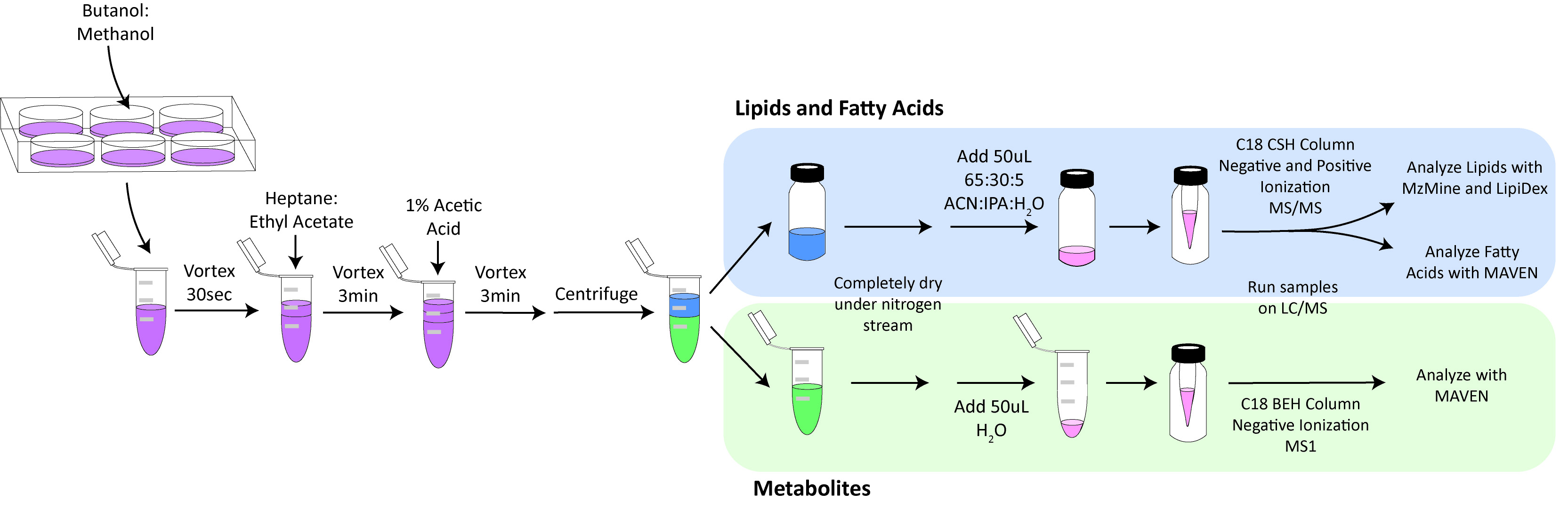
Figure 2. Work flow for extraction of lipids and small polar metabolites. ACN: acetonitrile, IPA: isopropanol.- Before beginning, make the following solutions:
3:1 1-butanol:methanol (LC-MS grade), with 1% SPLASH LipidoMIX Internal Standard (SPLASH)
3:1 heptane:ethyl acetate (LC-MS grade)
1% acetic acid (LC-MS grade)
65:30:5 Acetonitrile:Isopropanol:Water (LC-MS grade, ACN:IPA:H2O) - Chill 3:1 butanol:methanol (+SPLASH) and 3:1 heptane:ethyl acetate in the -20 °C and 1% acetic acid in the 4 °C for at least an hour prior to beginning.
- Aspirate media from cells.
- Wash cells twice with room temperature PBS.
- Add 300 µl of -20 °C 3:1 butanol:methanol (+SPLASH) to each well, place plates immediately on ice.
- While keeping cell plate on ice, scrape cells using cell scraper, and transfer solution from each well to a separate 1.5 ml plastic microcentrifuge tube. Place samples on ice.
Note: Also perform procedural blanks (N > 2) by performing the same Steps B1-B5 with an empty tube/plate. - Vortex each tube for 1 min, place back on ice.
- In the fume hood, add 300 µl -20 °C 3:1 heptane:ethyl acetate to each sample.
- Vortex all tubes for 3 min, place back on ice.
- In the fume hood, add 300 µl 1% acetic acid to each sample.
- Vortex all tubes for 3 min, place back on ice.
- Centrifuge all tubes (20,000 x g, 10 min, 4 °C)–two separate layers should form.
- Using a pipette, transfer a set volume (~400 µl) of the top organic layer to a glass vial (this is the lipid and fatty acid fraction) (perform in fume hood)
- Using a pipette, transfer a set volume (~300 µl) of the bottom aqueous layer to a 1.5 ml plastic tube (this is the polar metabolite fraction) (perform in fume hood)
- Completely dry down all samples by nitrogen stream using a N2 sample concentrator in the fume hood. Samples typically dry within 2h at room temperature. Retrieve samples immediately after drying.
- Resuspend the metabolite fraction in LC-MS grade water (50 µl) and the lipid fraction in 65:30:5 ACN:IPA:H2O (50 µl).
- Transfer metabolite fraction to polypropelyne LC-MS vials and the lipid fraction to glass LC-MS vial, cap and place in LC-MS autosampler for analysis.
- Before beginning, make the following solutions:
- Liquid-chromatography Mass spectrometry
- LC-MS vials should be placed in sample trays in randomized order. Sample runs should start with injection of an equal volume of at least three appropriate blanks: LC-MS H2O for polar metabolites, 65:30:5 ACN:IPA:H2O for lipids. Blanks should additionally be run throughout, between at least every 10 samples.
- Analyses of both the polar metabolite and lipid fractions were performed on a Thermo Q-Exactive mass spectrometer coupled to a Vanquish Horizon UHPLC.
- Polar metabolite samples were separated on a 100 x 2.1 mm 1.7uM Acquity UPLC BEH C18 Column (Waters) coupled to a 130Å, 1.7 µm, 2.1 mm x 5 mm Acquity UPLC BEH C18 VanGuard Pre-column (Waters). A gradient of solvent A (97:3 H2O:methanol, 10 mM TBA, 9 mM acetate, pH 8.2) and solvent B (100% methanol) was used at a 0.2ml/min flow rate. The gradient is: 0 min, 5% B; 2.5 min, 5% B; 17 min, 95% B; 21 min, 95% B; 21.5 min, 5% B (Figure 3A). Five µl of each metabolite sample was injected for analysis. Data was collected on a full scan negative mode. Detailed MS parameters are laid out in in Table 1.
- Lipid samples were separated on a 100 x 2.1 mm 1.7uM Acquity UPLC CSH C18 Column (Waters) coupled to a 130Å, 1.7 µm, 2.1 mm X 5 mm Acquity UPLC CSH C18 VanGuard Pre-column. A gradient of solvent C (70:30 ACN:H2O, 10 mM NH4Ac) and solvent D (90:10 IPA:ACN, 10 mM NH4Ac) was used at a 0.4 ml/min flow rate. The gradient is: 0 min, 2% D; 2 min, 2% D; 5 min, 30% D; 19 min, 85% D; 20 min 99% D; 27 min, 99% D; 28 min, 2% D; 32 min, 2%D (Figure 3B). Five µl of each lipid sample was injected for analysis. Samples were run twice, once in negative mode and once in positive mode both using data dependent MS2 (ddMS2) analysis. Detailed MS parameters are laid out in in Table 1.
Table 1. Mass Spectrometry Parameters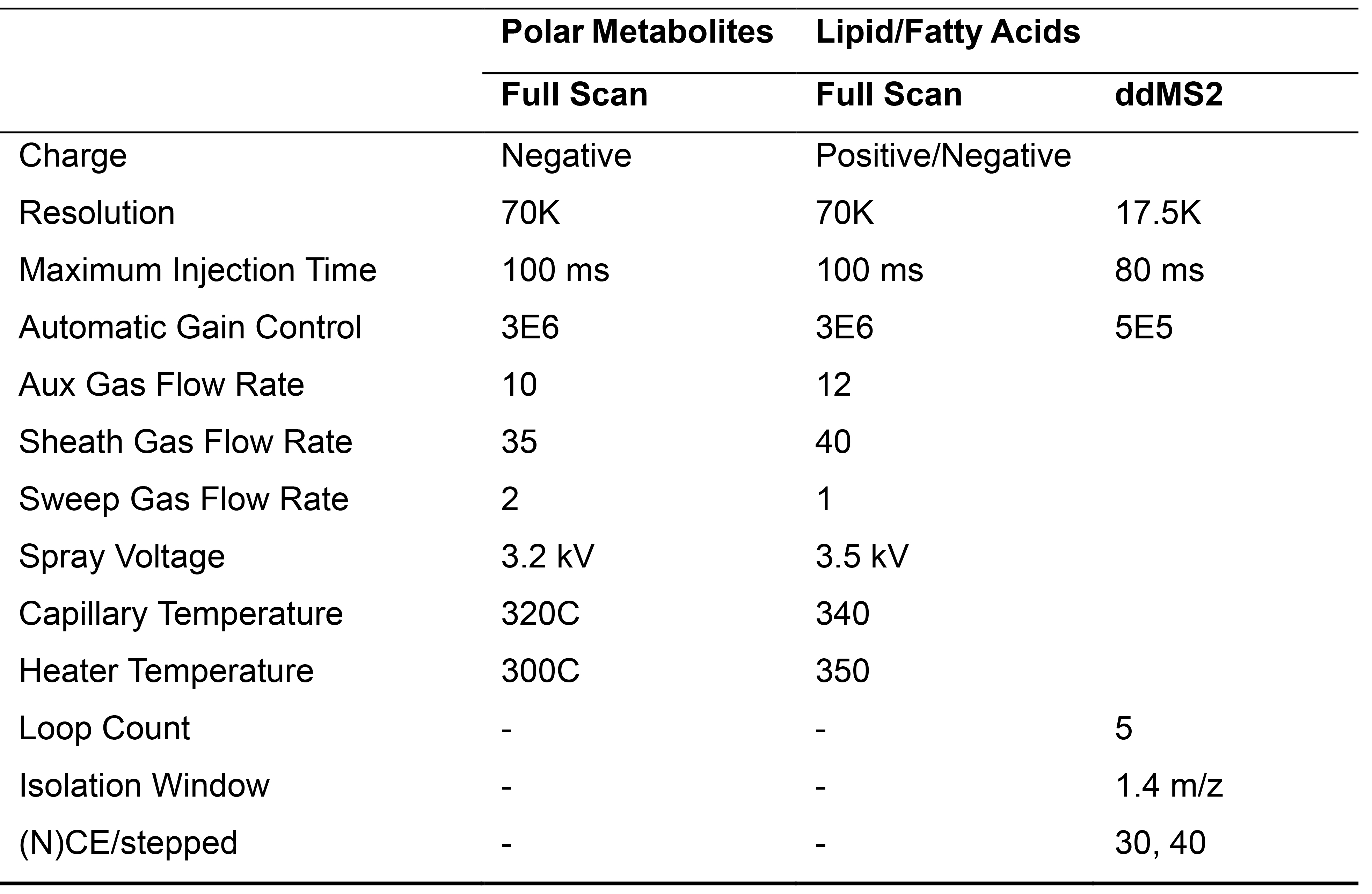
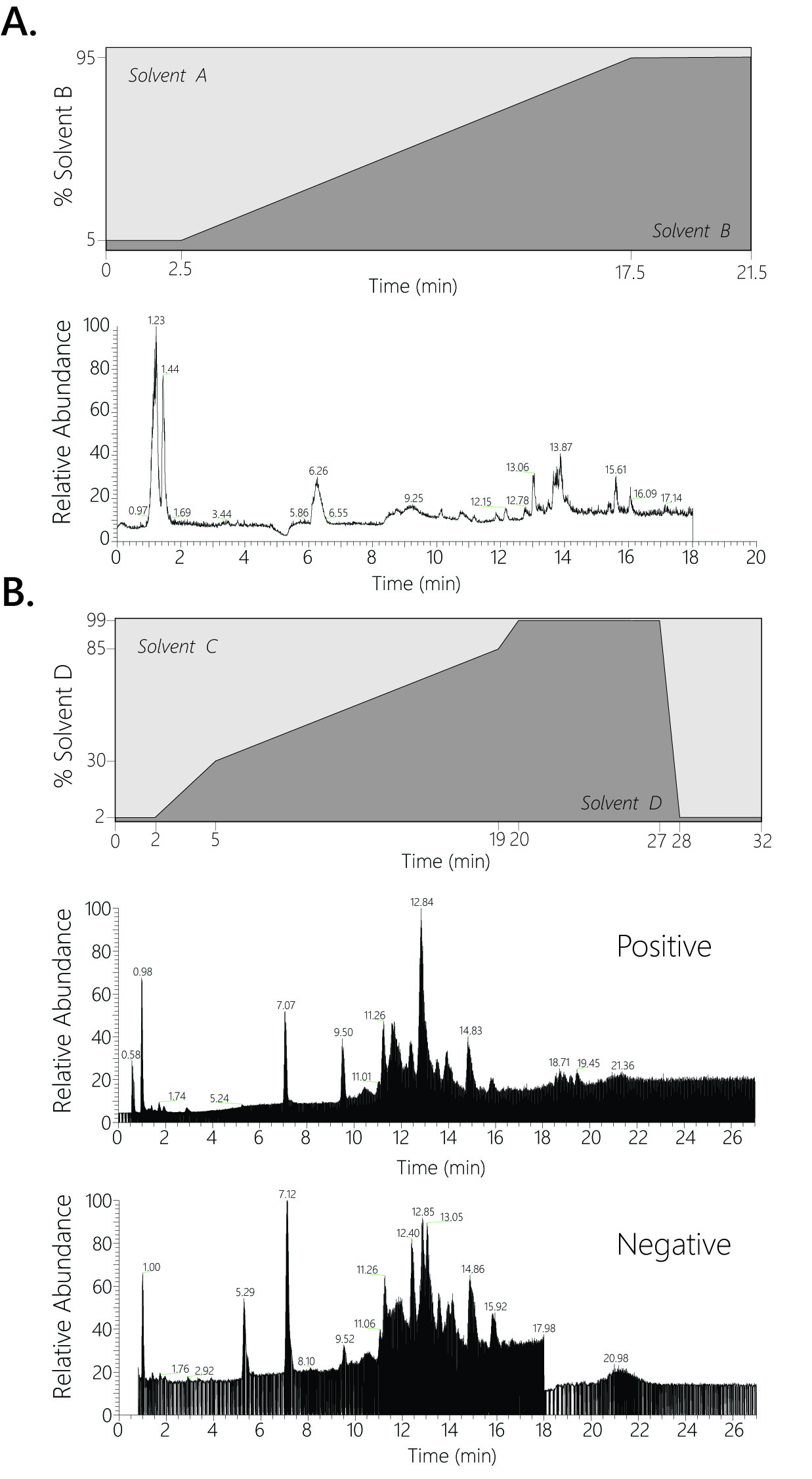
Figure 3. Solvent gradients and elution profiles. A. Solvent gradient and example elution profile for LC-MS polar metabolite method. B. Solvent gradient and example elution profile for LC-MS lipid analysis, profiles from both negative and positive ionization runs are shown.
Data analysis
MS1 data is used for both polar metabolite and free fatty acid quantification using MAVEN software (Melamud et al., 2010; Clasquin et al., 2012). Lipid data is identified and quantified based on full scan–ddMS2 using LipiDex Software (Hutchins et al., 2018). Both methods are described below. Representative data obtained from analysis of metabolite and lipids profiles of unstimulated (M0) and LPS+IFNγ stimulated BMDM (M1) is shown in Figure 4.
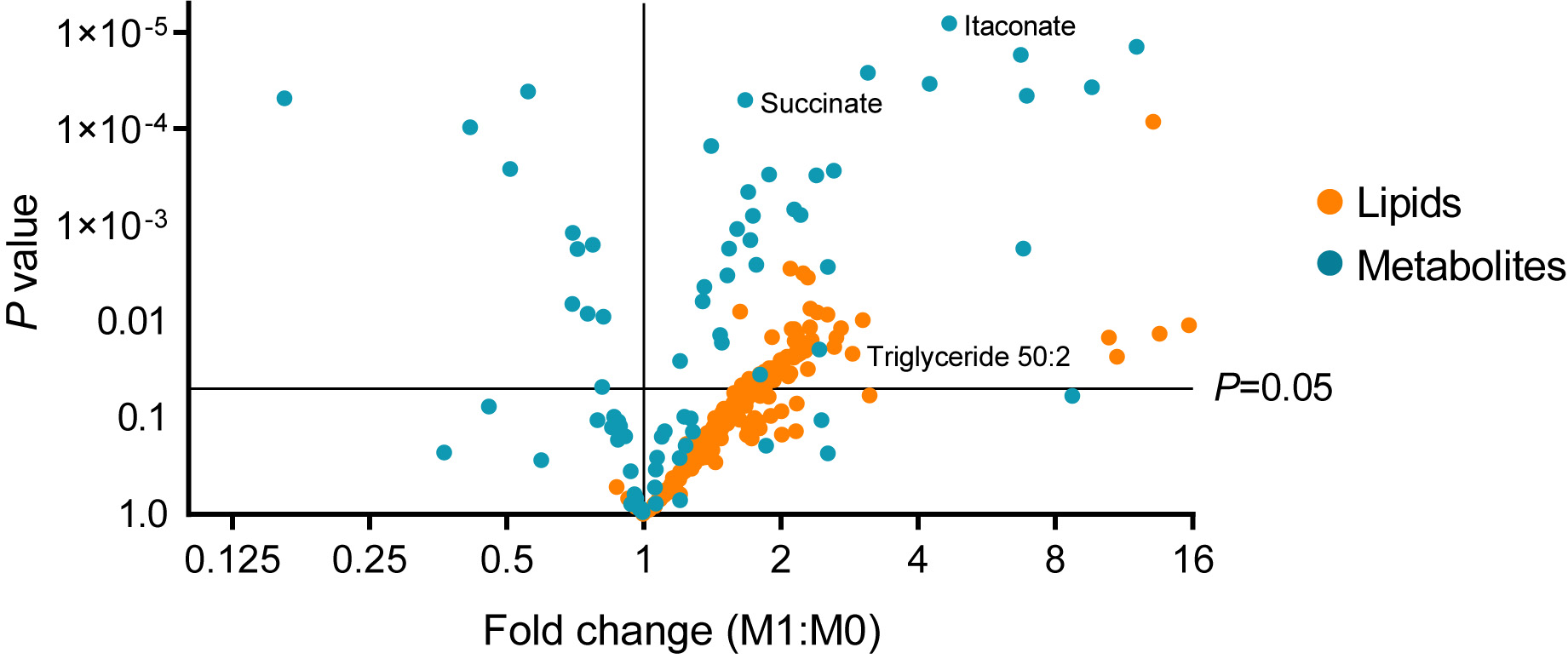
Figure 4. Representative metabolite and lipid data. A volcano plot showing fold changes in identified metabolites and lipids in LPS+IFNγ stimulated (M1) BMDM as compared to unstimulated (M0) BMDM. Highlighted are itaconate and succinate, two immunoregulatory metabolites known to be dynamically upregulated in M1 macrophages (Jha et al., 2015; Cordes et al., 2016; Lampropoulou et al., 2016; Mills et al., 2016). Also highlighted is triglyceride 50:2. Triglyceride accumulation has also been shown to occur in M1 macrophages (Köberlin et al., 2014; Lee et al., 2017; Feingold et al., 2012).
Polar Metabolites and Fatty Acids
Polar metabolites and fatty acids are quantified in a targeted fashion by matching the expected exact mass and retention time. The list of retention times and M/z is determined from analysis of metabolite standards using the same LC-MS method for samples as outlined above, and is in the supplementary information (supplemental file “Polar Metabolites RT List .xlsx”). We suggest that particularly with metabolites of interest, users run standards of their own. To do so, dilute standards to a range of concentrations (e.g., 1 μM, 10 μM, 100 μM, 1,000 μM, for inject 1 μl injection volume) in the appropriate sample solvents, and run on LC-MS using the corresponding method. For untargeted metabolomics analysis, see additional resources (Smith et al., 2005 and 2006; Kind et al., 2018; Wang et al., 2019). Additionally, this method will only describe what is necessary for relative quantification of metabolites. For measurement of absolute concentration of metabolites see additional resources (Bennett et al., 2008). We use MAVEN software for integration of peaks (Melamud et al., 2010; Clasquin et al., 2012). This software and more detailed guides on its use are available here.
- Convert Thermo generated .raw files to .mzXML files using MSConvertGUI (Holman et al., 2014; Adusumilli et al., 2017).
- Open .mzXML files in MAVEN. When properly loaded samples IDs will appear in the samples window to the left.
- Make sure “Compounds Widget” is turned on. Load compound list by clicking on compound tab on left and selecting “Load Custom Compound List”. Load the .csv file with known m/z and retention times generated from metabolite standards or as provided in supplemental material. Adjust the ppm window on top right of Maven window to match that of the sensitivity of your instrument.
- With compound list loaded, click on name of compound of interest and the peaks corresponding to that metabolite in each sample will appear in the main window. The red line corresponds to the observed retention time according to the imported compound list (Figure 5). To save the data from selected peak double click on the peak–the name of the compound and corresponding parameters will be displayed in a list within the software. Save data only from those peaks whose retention time matches that on the compound list, has a relatively smooth profile, and whose signal is above desired cut off (e.g., 5E4 and at least 2x times the blank). See Figure 5 for example.
Note: More detailed information about software functions and parameter setting can be found in MAVEN using manual. - When all desired peaks have been saved, export this data from Maven by selecting “Export Peaks to SpreadSheet (.csv) above the list of saved peaks.
- Blank the data by subtracting the average signal from the blanks from the signal from each sample. Subsequently normalize data to cell count or protein.
Note: Only compare relative abundance from samples run within the same sequence, as changes in instrument conditions over time may cause difference in signal response. If absolute quantitation of metabolite concentration is desired, use of isotopic internal standards is recommended.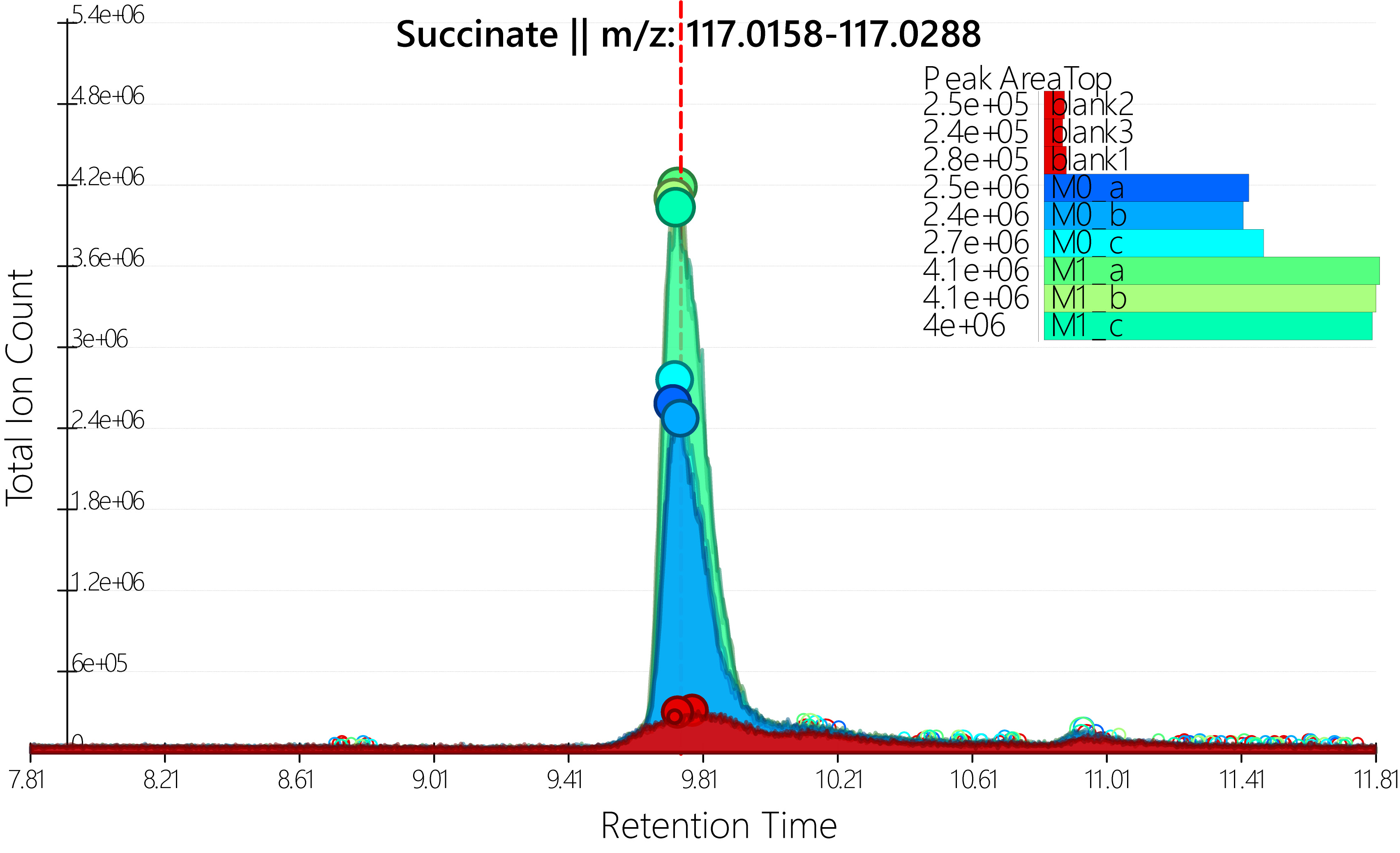
Figure 5. Example of a good LC-MS peak. Succinate peaks from unstimulated (M0) and LPS+IFNγ stimulated (M1) BMDM, as displayed by Maven software.
Lipids
Lipid identification and quantification is performed using the open access LipiDex software (Hutchins et al., 2018). Based on full scan-ddMS2 data, LipiDex identifies lipids. The software is coupled to MZmine2 to quantify lipid species. Below is our workflow for this analysis (Pluskal et al., 2010).
- Convert Thermo generated .raw files to .mgf files (we use MSConvertGUI software [Holman et al., 2014; Adusumilli et al., 2017]).
- Perform Spectrum Search in LipiDex
- Open LipiDex and go to Spectrum Searcher.
- Upload .mgf files.
- Enable the appropriate libraries.
- Specify desired parameters for search (e.g., MS1 search tolerance: 0.01, MS2 search tolerance: 0.01).
- Click “Spectra Search”–this will produce result .csv files with identifications.
- Generate chromatographic feature tables using MZmine2. Do separately for positive and negative ionization runs. Follow step-by-step instructions as provided here.
- Perform Peak Finding in LipiDex
- Open LipiDex and go to Peak Finder.
- Click the MZMine2 option.
- Load the positive and negative polarity peak tables generated by MZMine2.
- Upload the MS/MS .csv result files, making sure to identify the files with the appropriate polarity (positive or negative).
- Set desired filtering parameters, for instance, using the following:
MS2 filtering parameters: Min. Lipid Spectral Purity (%): 75, Min. MS2 Search Dot Product: 500, Min. MS2 Search Rev. Dot Product: 700.
Feature Association Parameters: FWHM Window Multiplier: 2.0, Max. Mass Difference (ppm): 15.
Result Filtering Parameters: Check Adduct/Dimer Filtering and In-source Fragment Filtering. Check Max. RT M.A.D Factor with 3.5. Feature found in n Files: 2. - Click Identify Chromatographic Peaks–this will produce .csv files containing identified lipid species and their quantification.
- Blank data using the procedural blanks that were run parallel to samples, and normalize data to cell count or protein content.
- Quality Control: The SPLASH LipidoMIX Internal Standard is a mixture of isotopic labeled lipids, representing major lipid classes. SPLASH (1%) was added to the initial extraction buffer (3:1 1-butanol:methanol). The presence of this internal standard in all analyzed lipid samples allows for determination of retention time drift across samples, and potential difference in sample recovery. The retention time drift within a sequence is typically < 0.1min, and the variation of internal standards is < 20%. If the variation is significant, data can be normalized to internal standard of the same lipid class or nearest retention time (if there is no standard of the same class). See supplemental file “SPLASH LipidoMix Stds Compound list .xlsx” for list of compounds in SPLASH LipidoMIX and their respective retention times using the outlined method.
Notes
- Keep and measure out all LC-MS grade solvents in clean designated glassware. Do not use this glassware for other purposes and do not wash with tap water or soap. Residual detergent and other material will cause background. When necessary, rinse with HPLC grade solvents only.
- Plastic used during making of extraction solvents and the extraction of lipids can cause background, particularly for fatty acids. Always perform a procedural blank to appropriately account for this background signal. Alternative use of glass vials and pipettes in place of plastic microcentrifuge tubes and pipette tips can help reduce background if a considerable amount is observed.
Recipes
- BMDM Differentiation Media
RPMI 1640 without Glutamine
10% FBS
20% L929 Conditioned Media
10 U/ml penicillin/streptomycin
25 mM HEPES
2 mM glutamine - BMDM Maintenance Media
RPMI 1640 without Glutamine
10% dFBS
10 U/ml penicillin/streptomycin
25 mM HEPES
2 mM glutamine
20 ng/ml m-CSF - L929 Culture Media
DMEM High Glucose
10 U/ml penicillin/streptomycin
10% FBS - L929 Conditioned Media
- Culture L929 cells in L929 culture media until fully confluent
- Trypsinize cells, spin down at 1,000 x g for 5 min
- Aspirate trypsin, resuspend cell pellet in L929 culture media
- Count cells and resuspend to 8 x 103 cells/ml
- Add 130 ml of this suspension to a 175 cm2 flask and place in incubator
- 7 days later collect all the spent media
- Centrifuge at 1,000 x g, 4 °C, for 5 min. Sterile filter using a 0.45 μm filter into a sterile glass bottle
- Aliquot to 50 ml plastic centrifuge tubes, and freeze at -20 °C until desired use (use within 6 months)
- Solvent A
97:3 water:methanol (LC-MS grade)
10 mM tributylamine
Acetic acid (LC-MS grade), pH to 8.2
Note: This takes a great deal of vigorous mixing to fully dissolve the tributylamine and reach a stable pH. - Solvent C
70:30 acetonitrile:water (LC-MS grade)
10 mM Ammonium Acetate (LC-MS grade)
250 µl/L Acetic Acid (LC-MS grade) - Solvent D
90:10 isopropanol:acetonitrile (LC-MS grade)
10 mM Ammonium Acetate (LC-MS grade)
250 µl/L Acetic Acid (LC-MS grade)
Acknowledgments
The methods for bone marrow derived macrophages isolation, metabolites and lipids extraction, LC-MS analysis and data analysis are adapted and optimized based on previous works, each referenced in the respective section.
Competing interests
The authors declare no competing interests.
Ethics
Animal care and experimental procedures were carried out in accordance with a protocol approved by University of Wisconsin-Madison Institutional Animal Care and Use Committee.
References
- Adusumilli, R. and Mallick, P. (2017). Data conversion with proteowizard msConvert. Methods Mol Biol 1550: 339-368.
- Bennett, B. D., Yuan, J., Kimball, E. H. and Rabinowitz, J. D. (2008). Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc 3(8): 1299-1311.
- Clasquin, M. F., Melamud, E. and Rabinowitz, J. D. (2012). LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr Protoc Bioinformatics Chapter 14: Unit14 11.
- Cordes, T., Wallace, M., Michelucci, A., Divakaruni, A. S., Sapcariu, S. C., Sousa, C., Koseki, H., Cabrales, P., Murphy, A. N., Hiller, K. and Metallo, C. M. (2016). Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J Biol Chem 291(27): 14274-14284.
- Feingold, K. R., Shigenaga, J. K., Kazemi, M. R., McDonald, C. M., Patzek, S. M., Cross, A. S., Moser, A. and Grunfeld, C. (2012). Mechanisms of triglyceride accumulation in activated macrophages. J Leukoc Biol 92(4): 829-839.
- Fiehn, O. (2002). Metabolomics--the link between genotypes and phenotypes. Plant Mol Biol 48(1-2): 155-171.
- Galván-Peña, S. and O’Neill, L. A. J. (2014). Metabolic reprogramming in macrophage polarization. Front Immunol 5: 420.
- Gika, H. G., Theodoridis, G. A., Plumb, R. S. and Wilson, I. D. (2014). Current practice of liquid chromatography-mass spectrometry in metabolomics and metabonomics. J Pharm Biomed Anal 87: 12-25.
- Holman, J. D., Tabb, D. L. and Mallick, P. (2014). Employing proteowizard to convert raw mass spectrometry data. Curr Protoc Bioinformatics 46: 13 24 11-19.
- Hutchins, P. D., Russell, J. D. and Coon, J. J. (2018). Lipidex: an integrated software package for high-confidence lipid identification. Cell Syst 6(5): 621-625 e625.
- Jha, A. K., Huang, S. C., Sergushichev, A., Lampropoulou, V., Ivanova, Y., Loginicheva, E., Chmielewski, K., Stewart, K. M., Ashall, J., Everts, B., Pearce, E. J., Driggers, E. M. and Artyomov, M. N. (2015). Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42(3): 419-430.
- Kelly, B. and O'Neill, L. A. (2015). Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res 25(7): 771-784.
- Kessner, D., Chambers, M., Burke, R., Agus, D. and Mallick, P. (2008). ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24(21): 2534-2536.
- Kind, T., Tsugawa, H., Cajka, T., Ma, Y., Lai, Z., Mehta, S. S., Wohlgemuth, G., Barupal, D. K., Showalter, M. R., Arita, M. and Fiehn, O. (2018). Identification of small molecules using accurate mass MS/MS search. Mass Spectrom Rev 37(4): 513-532.
- Köberlin, M. S., Heinz, L. X. and Superti-Furga, G. (2016). Functional crosstalk between membrane lipids and TLR biology. Curr Opin Cell Biol 39: 28-36.
- Lampropoulou, V., Sergushichev, A., Bambouskova, M., Nair, S., Vincent, E. E., Loginicheva, E., Cervantes-Barragan, L., Ma, X., Huang, S. C., Griss, T., Weinheimer, C. J., Khader, S., Randolph, G. J., Pearce, E. J., Jones, R. G., Diwan, A., Diamond, M. S. and Artyomov, M. N. (2016). Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab 24(1): 158-166.
- Lee, J. W., Mok, H. J., Lee, D. Y., Park, S. C., Kim, G. S., Lee, S. E., Lee, Y. S., Kim, K. P. and Kim, H. D. (2017). UPLC-QqQ/MS-Based Lipidomics Approach To Characterize Lipid Alterations in Inflammatory Macrophages. J Proteome Res 16(4): 1460-1469.
- Liu, P. S., Wang, H., Li, X., Chao, T., Teav, T., Christen, S., Di Conza, G., Cheng, W. C., Chou, C. H., Vavakova, M., Muret, C., Debackere, K., Mazzone, M., Huang, H. D., Fendt, S. M., Ivanisevic, J. and Ho, P. C. (2017). α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol 18(9): 985-994.
- Löfgren, L., Forsberg, G. B. and Ståhlman, M. (2016). The BUME method: a new rapid and simple chloroform-free method for total lipid extraction of animal tissue. Sci Rep 6: 27688.
- Löfgren, L., Stahlman, M., Forsberg, G. B., Saarinen, S., Nilsson, R. and Hansson, G. I. (2012). The BUME method: a novel automated chloroform-free 96-well total lipid extraction method for blood plasma. J Lipid Res 53(8): 1690-1700.
- Martin, S. L., Seim, G. L., Wawire, S., Chapleau, G. M., Young, S. L. and Dickin, K. L. (2017). Translating formative research findings into a behaviour change strategy to promote antenatal calcium and iron and folic acid supplementation in western Kenya. Matern Child Nutr 13(1).
- Melamud, E., Vastag, L. and Rabinowitz, J. D. (2010). Metabolomic analysis and visualization engine for LC-MS data. Anal Chem 82(23): 9818-9826.
- Mills, E. L. and O'Neill, L. A. (2016). Reprogramming mitochondrial metabolism in macrophages as an anti-inflammatory signal. Eur J Immunol 46(1): 13-21.
- Mills, E. L., Kelly, B., Logan, A., Costa, A. S. H., Varma, M., Bryant, C. E., Tourlomousis, P., Dabritz, J. H. M., Gottlieb, E., Latorre, I., Corr, S. C., McManus, G., Ryan, D., Jacobs, H. T., Szibor, M., Xavier, R. J., Braun, T., Frezza, C., Murphy, M. P. and O'Neill, L. A. (2016). Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167(2): 457-470 e413.
- O'Neill, L. A. and Pearce, E. J. (2016). Immunometabolism governs dendritic cell and macrophage function. J Exp Med 213(1): 15-23.
- Pluskal, T., Castillo, S., Villar-Briones, A. and Orešič, M. (2010). MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11: 395.
- Smith, C. A., O'Maille, G., Want, E. J., Qin, C., Trauger, S. A., Brandon, T. R., Custodio, D. E., Abagyan, R. and Siuzdak, G. (2005). METLIN: a metabolite mass spectral database. Ther Drug Monit 27(6): 747-751.
- Smith, C. A., Want, E. J., O'Maille, G., Abagyan, R. and Siuzdak, G. (2006). XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78(3): 779-787.
- Tannahill, G. M., Curtis, A. M., Adamik, J., Palsson-McDermott, E. M., McGettrick, A. F., Goel, G., Frezza, C., Bernard, N. J., Kelly, B., Foley, N. H., Zheng, L., Gardet, A., Tong, Z., Jany, S. S., Corr, S. C., Haneklaus, M., Caffrey, B. E., Pierce, K., Walmsley, S., Beasley, F. C., Cummins, E., Nizet, V., Whyte, M., Taylor, C. T., Lin, H., Masters, S. L., Gottlieb, E., Kelly, V. P., Clish, C., Auron, P. E., Xavier, R. J. and O'Neill, L. A. (2013). Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496(7444): 238-242.
- Theodoridis, G. A., Gika, H. G., Want, E. J. and Wilson, I. D. (2012). Liquid chromatography-mass spectrometry based global metabolite profiling: a review. Anal Chim Acta 711: 7-16.
- Van den Bossche, J., O'Neill, L. A. and Menon, D. (2017). Macrophage immunometabolism: where are we (going)? Trends Immunol 38(6): 395-406.
- Wang, L., Xing, X., Chen, L., Yang, L., Su, X., Rabitz, H., Lu, W. and Rabinowitz, J. D. (2019). Peak annotation and verification engine for untargeted LC-MS metabolomics. Anal Chem 91(3): 1838-1846.
- Wenk, M. R. (2005). The emerging field of lipidomics. Nat Rev Drug Discov 4(7): 594-610.
- Williams, N. C. and O'Neill, L. A. J. (2018). A role for the krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front Immunol 9: 141.
- Ying, W., Cheruku, P. S., Bazer, F. W., Safe, S. H. and Zhou, B. (2013). Investigation of macrophage polarization using bone marrow derived macrophages. J Vis Exp(76): 50323.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Seim, G. L., John, S. V. and Fan, J. (2020). Metabolomic and Lipidomic Analysis of Bone Marrow Derived Macrophages. Bio-protocol 10(14): e3693. DOI: 10.21769/BioProtoc.3693.
Category
Immunology > Immune cell isolation > Macrophage
Systems Biology > Metabolomics > Biofluid
Cell Biology > Cell metabolism > Lipid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link




.jpg)







