- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Site-specific DNA Mapping of Protein Binding Orientation Using Azidophenacyl Bromide (APB)
Published: Vol 10, Iss 12, Jun 20, 2020 DOI: 10.21769/BioProtoc.3649 Views: 4857
Reviewed by: Gal HaimovichSalma MerchantHAILONG ZHANG

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Combining Gel Retardation and Footprinting to Determine Protein-DNA Interactions of Specific and/or Less Stable Complexes
Meng-Lun Hsieh [...] Deborah M. Hinton
Dec 5, 2020 3604 Views

Rapid Genome Engineering of Pseudomonas Assisted by Fluorescent Markers and Tractable Curing of Plasmids
Daniel C. Volke [...] Pablo I. Nikel
Feb 20, 2021 7434 Views

Characterize the Interaction of the DNA Helicase PriA with the Stalled DNA Replication Fork Using Atomic Force Microscopy
Yaqing Wang [...] Yuri L. Lyubchenko
Mar 5, 2021 4876 Views
Abstract
The orientation of a DNA-binding protein bound on DNA is determinative in directing the assembly of other associated proteins in the complex for enzymatic action. As an example, in a replisome, the orientation of the DNA helicase at the replication fork directs the assembly of the other associated replisome proteins. We have recently determined the orientation of Saccharalobus solfataricus (Sso) Minichromosome maintenance (MCM) helicase at a DNA fork utilizing a site-specific DNA cleavage and mapping assay. Here, we describe a detailed protocol for site-specific DNA footprinting using 4-azidophenacyl bromide (APB). This method provides a straightforward, biochemical method to reveal the DNA binding orientation of SsoMCM helicase and can be applied to other DNA binding proteins.
Keywords: MCM helicaseBackground
DNA replication is the process in which the duplex genomic strands separate into two template strands, the leading and lagging strands. This function is executed by a ring-shaped hexameric helicase in all Domains of life. Like other ring-shaped hexameric helicases, MCM consists of two domains; an N-terminal domain (NTD) and a C-terminal domain (CTD). In theory, either of these domains could be oriented towards the replication fork during translocation and be consistent with the known 3′-5′ translocation directionality. The MCM helicase loads onto DNA origins as a double hexamer with NTDs facing each other. The orientation of the helicase during translocation determines whether the two hexamers dissociate away from each other or bypass one another during active unwinding. Our recent paper shows that the Saccharolobus solfataricus (SsoMCM) unwinds DNA with NTD leading the way (Perera and Trakselis, 2019).
In order to directly determine the translocation orientation, we utilized a combination of site-specific DNA footprinting, single turnover unwinding, and translocation assays. Here, we provide detailed protocols of site-specific DNA footprinting assays with 4-azidophenacyl bromide (APB) to analyze the translocation orientation of SsoMCM (Pendergrast et al., 1992, Kassabov and Bartholomew, 2004, Nodelman et al., 2017).
APB is a heterobifunctional photoactivatable crosslinking agent. Its bromide functional group reacts by S-alkylation with reduced thiols (i.e., cysteines) to form stable thioether products (Figure 1). After binding of the functionalized protein to DNA and then exposure to UV light, a reactive singlet nitrene forms that can crosslink to either protein or DNA (in close proximity) through multiple insertion or addition mechanisms (Figure 1). The resulting crosslinked protein-DNA complex can be cleaved at the crosslinked nucleotide(s) under induced alkali/heat treatment. The lengths of the resulting DNA fragments can be used to determine the orientation distribution of the SsoMCM helicase on DNA. This is a straightforward biochemical method that reveals unique positions of SsoMCM helicases on DNA. It has an important advantage over traditional footprinting in that it can determine protein binding orientation on DNA instead of just binding site size.
Useful applications of this method can be employed at instances where the 3D structure of the protein is known or can be predicted, but the 3D structure of the protein-DNA complex is unknown (Pendergrast et al., 1992). It is especially useful in determining the orientation of proteins that translocate along DNA or for those that bind to specific DNA sequences. Alternatively, the orientation of DNA-binding proteins can also be determined by other biochemical methods that employ localized hydroxyl radical Fenton footprinting reactions utilizing 1-(p-Bromoacetamidobenzyl) ethylenediamine N,N,N (Fe-BABE), similarly (Owens et al., 1998).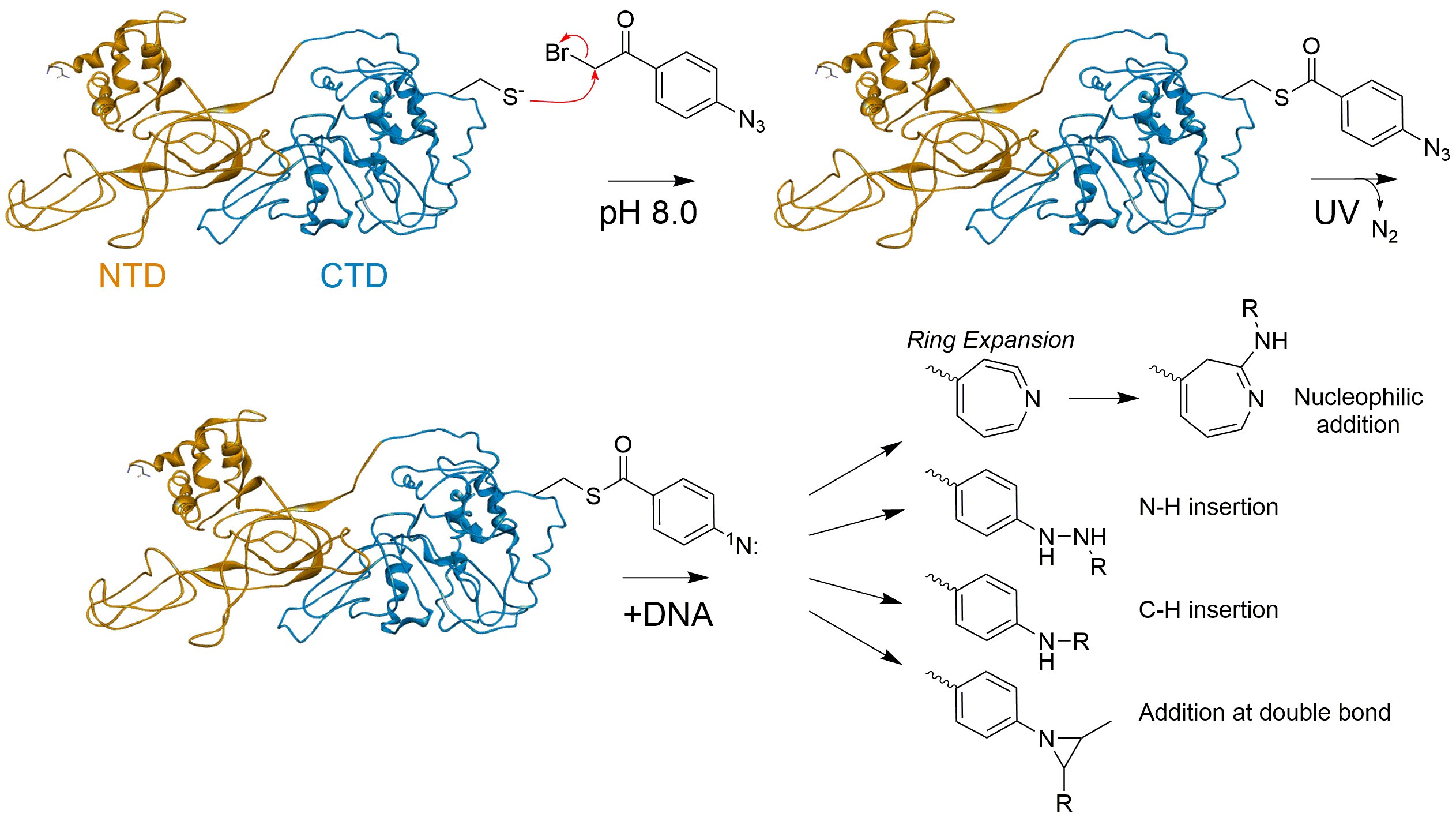
Figure 1. Conjugation of APB to free Cys on SsoMCM and UV induced crosslinking reaction mechanism to DNA
Materials and Reagents
- Cover slips (Fisher Scientific, catalog number: 12-546 )
- Glass Petri dish (Corning, catalog number: 3160100 )
- Kimwipe (Kimberly-Clark, catalog number: 06-666 )
- p-Azidophenacyl bromide (APB) (Sigma-Aldrich, catalog number: 57018-46-9 , storage: 4 °C or (Fisher Scientific, catalog number: 50-520-767 )
- Dimethylformamide (DMF) (EMD Millipore Corporation, catalog number: DX1730-6 )
- Tris Base (Fisher Scientific, catalog number: 77-86-1 )
- Glacial acetic acid (Mallinckrodt Baker, Inc., catalog number: UN 2789 )
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-3 )
- Sodium hydroxide (NaOH) (Mallinckrodt Baker, Inc., catalog number: 7708-10)
- Hydrochloric acid (HCl) (Fisher Scientific, catalog number: A144-212 )
- Magnesium chloride (MgCl2) (Spectrum Chemical, catalog number: M1035 )
- Glycerol (Fisher Scientific, catalog number: BP-229-4 )
- Sodium acetate (NaOAc) (Fisher Scientific, catalog number: S210-500 )
- Potassium acetate (KOAc) (EM Science Industries, catalog number: PX1330-1 )
- Magnesium acetate (MgOAc) (Sigma-Aldrich, catalog number: M-0631 )
- Ammonium acetate (NH4OAc) (Mallinckrodt Baker, Inc., catalog number: 0596-01 )
- Bovine serum albumin (BSA) (Fisher Scientific, catalog number: BP1600-100 )
- Dithiotheritol (DTT) (Fisher Scientific, catalog number: BP172-5 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E5134 )
- Sodium dodecyl sulfate (SDS) (Fisher Scientific, catalog number: 151-21-3 )
- Salmon sperm DNA (Invitrogen, catalog number: 15632-011 , storage temperature: -20 °C)
- Ethanol (Fisher Scientific, catalog number: A962-4 )
- Poly-L-lysine (Sigma-Aldrich, catalog number: P4832 )
- Orange G dye (EMD Millipore Corporation, catalog number: 312-12 )
- SsoMCM protein
SsoMCM protein was purified as previously described (McGeoch et al., 2005; Graham et al., 2011). Any DNA binding protein with a suitable single cysteine residue can be utilized with this approach. It will help if that cysteine residue is solvent accessible from structural data. If a single cysteine is not available, then site-directed mutagenesis can be used to add a cysteine at the appropriate position to test. - Fluorescent (5′ or 3′) labelled (Cy3 or Cy5) DNA (purchased from IDT (Coralville, IA) or Sigma-Aldrich (St. Louis, MO)) (Synthesis scale–0.025 μmole, Purification: Desalt, Format: Dry).
Note: PAGE purification of DNA can be performed depending on the purity of the DNA. - DNA Sequences for the 3’-long arm fork substrates
- DNA 165-3 (5′ Cy3)
5′ 3TCCCACCCAACCCGACCGGCATCTAGTCTGGTAGCGTGAGCGAACGGACC - DNA 171
5′ CTAACTGCGCCGGTCGGGTTGGGTGGGA
- DNA 165-3 (5′ Cy3)
- 1x complex buffer (CB) (see Recipes)
- Annealing buffer (see Recipes)
- Post irradiation buffer (see Recipes)
- Glycerol loading buffer (see Recipes)
- 20% Native PAGE (see Recipes)
Equipment
- Pipettes
- -80 °C freezer
- Benchtop 3UV transilluminator (UVP, model: LMS-20E , catalog number: P/N 95-0220-01)
- Typhoon FLA 9000 imager (GE Healthsciences)
- PCR Instrument (BioRad laboratories (Hercules, CA)
- Benchtop microcentrifuge (Eppendorf, model: 5424R )
- Mini-PROTEAN Tetra Vertical Electrophoresis Cell for Mini Precast Gels (Bio-Rad Laboratories)
- LightSafe micro centrifuge tubes (Sigma-Aldrich, catalog number: Z688312 )
- Dark room (Recommended)
Software
- Image Quant (v.5.0) (GE Healthsciences, (Pittsburgh, PA))
Procedure
- SsoMCM Protein labelling with APB
- In the dark, dissolve APB in 100% DMF at a concentration of 40 mM.
Notes:- Dark conditions can be provided by using LightSafe micro centrifuge tubes, covering Eppendorf tubes in foil or turning off the lights.
- Use LightSafe micro centrifuge tubes when preparing APB solutions.
- Use clear micro centrifuge tubes covered in foil in subsequent reactions for easy visualization of a pellet and/or to achieve complete discarding of a supernatant.
- Use LightSafe micro centrifuge tubes when preparing APB solutions.
- When measuring APB, quantities as small as ~2 mg can be measured under dark conditions.
- Use freshly resuspended APB for each new experiment. Discard the unused APB.
- Dark conditions can be provided by using LightSafe micro centrifuge tubes, covering Eppendorf tubes in foil or turning off the lights.
- Dilute the APB to 4 mM in 20 mM Tris pH 7.5, 75 mM NaCl, 10% glycerol and 10% DMF.
- Add the diluted APB to a sample of the protein (~10 µM monomer) containing a single reduced cysteine (in 20 mM Tris [pH 7.5], 75 mM NaCl, 10% glycerol), to achieve a final concentration of 0.4 mM APB and 1% DMF.
Note: Cysteines must be reduced before conjugation with APB, followed by gel filtration or dialysis to remove the reducing agent. Alternatively, tris(2-carboxyethyl)phosphine (TCEP) can be used without subsequent removal. - Proceed labelling for 2-3 h at room temperature.
- In the dark, dissolve APB in 100% DMF at a concentration of 40 mM.
- Preparing silanized coversli
- Dilute poly-L-lysine solution with distilled water for 1:10.
- Place the coverslips (~15) in a Petri dish.
- Pour the solution (~20 ml) on the coverslips (make sure the coverslips sink in).
- Leave it in the solution for 15-30 min.
- Rinse the coverslips with water three times at Room Temperature (RT).
- Air dry the coverslips placing a kimwipe on top of the Petri dish (avoid touching the coverslips).
- Coverslips can be prepared ahead of time and can be stored at RT for several weeks.
- Preparing annealed fork DNA
- Prepare stock solutions (100 μM) of each complementary DNA strand in water (DNA 165-3 and DNA 171).
- Mix the DNA in equal 1:1 (5 μM) concentration in water or annealing buffer.
Note: EDTA (chelates divalent metal ions) and salt in the annealing buffer facilitate duplex stability. But annealing DNA in water has also yielded in a complete duplex. - Heat at 95 °C for 5 min and cool at a rate of 1 °C/min to room temperature in a PCR instrument.
- Preparing the crosslinked protein-DNA complex
- Incubate APB labelled SsoMCM with fluorescent (Cy3) fork DNA (150 nM) for 10-20 min in 1x CB buffer in 50 µl volumes (maintaining stochiometric ~1:1 MCM6:DNA ratio).
Note: Fluorescently labelled DNA is used here, but 32P labelled DNA can also be utilized. - Transfer the sample into silanized coverslips. Pipette 50 µl of the sample into the middle of the coverslip.
- UV irradiate for 15 s (302 nm UV-B).
- Transfer the sample into an Eppendorf tube.
- Add 150 µl of post irradiation buffer.
- Vortex at RT and place at 70 °C for 20 min.
- Incubate APB labelled SsoMCM with fluorescent (Cy3) fork DNA (150 nM) for 10-20 min in 1x CB buffer in 50 µl volumes (maintaining stochiometric ~1:1 MCM6:DNA ratio).
- Separate the crosslinked protein-DNA complex
- Next, add 1 µl of 10 mg/ ml Salmon sperm DNA, 30 µl of 3.0 M NaOAc, 750 µl of ice cold 100% ethanol, vortex, and leave on ice for 1-2 h at -80 °C.
- Spin down the samples in microfuge at 4 °C, 16,000 x g for 30 min.
- Discard the supernatant and wash the pellet twice with ice cold 70% ethanol.
Note: In most of the steps, the DNA pellets are invisible. Discard the supernatant completely and resuspend in the subsequent buffer. - Spin down the samples in microfuge at 4 °C, 16,000 x g for 30 min.
- Remove ethanol and air dry the pellets by inverting on bench for 1 h.
- Resuspend the pellet in 100 µl: 20 mM NH4OAc, 2% SDS, 0.1 mM EDTA pH 8.0 by vortexing.
- Spin down the samples in microfuge at room temperature, 16,000 x g for 10 min.
- Transfer the supernatant into fresh tubes and place in heat block at 90 °C for 2 min.
- Then, add 1 µl of 2 M NaOH, vortex briefly, and incubate at 90 °C for 20 min.
- After incubation, pulse spin the samples, add 101 µl 20 mM Tris-HCl pH 8.0, 1 µl of 2 M HCl, 1 µl of 2 M MgCl2, 480 µl 100% ethanol, vortex, and place at -80 °C for 1-2 h.
- Pellet the samples in microfuge at 4 °C at 16,000 x g for 30 min, wash two times with ice cold 70% ethanol, and air dry on bench for 1 h.
- Resuspend the DNA pellet with 5 µl of 40% glycerol loading buffer containing Orange G dye for gel loading.
- Electrophorese the samples on a 20% TBE-PAGE (native PAGE) in 1x TBE (Tris base, Boric acid, EDTA) at constant 30 mA for 45 min, and visualize on a Typhoon FLA 9000 imager (GE Healthsciences).
Data analysis
- Quantifying the density of DNA bands on the gel (Figure 2A) is performed by ImageQuant software.
- Calculation of the footprinting percentages (Figure 2B) is performed by quantifying the relative density (minus background) for the labelled strand, divided at the midpoint on the ssDNA arm according to the following equation:
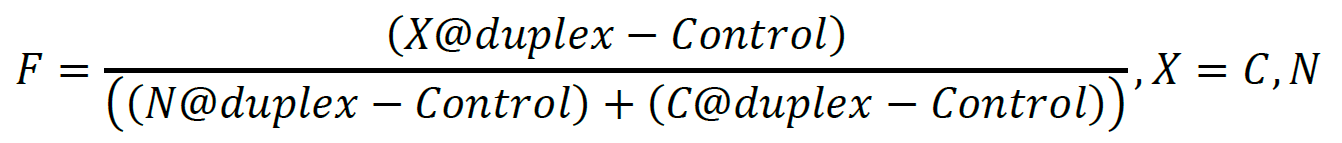
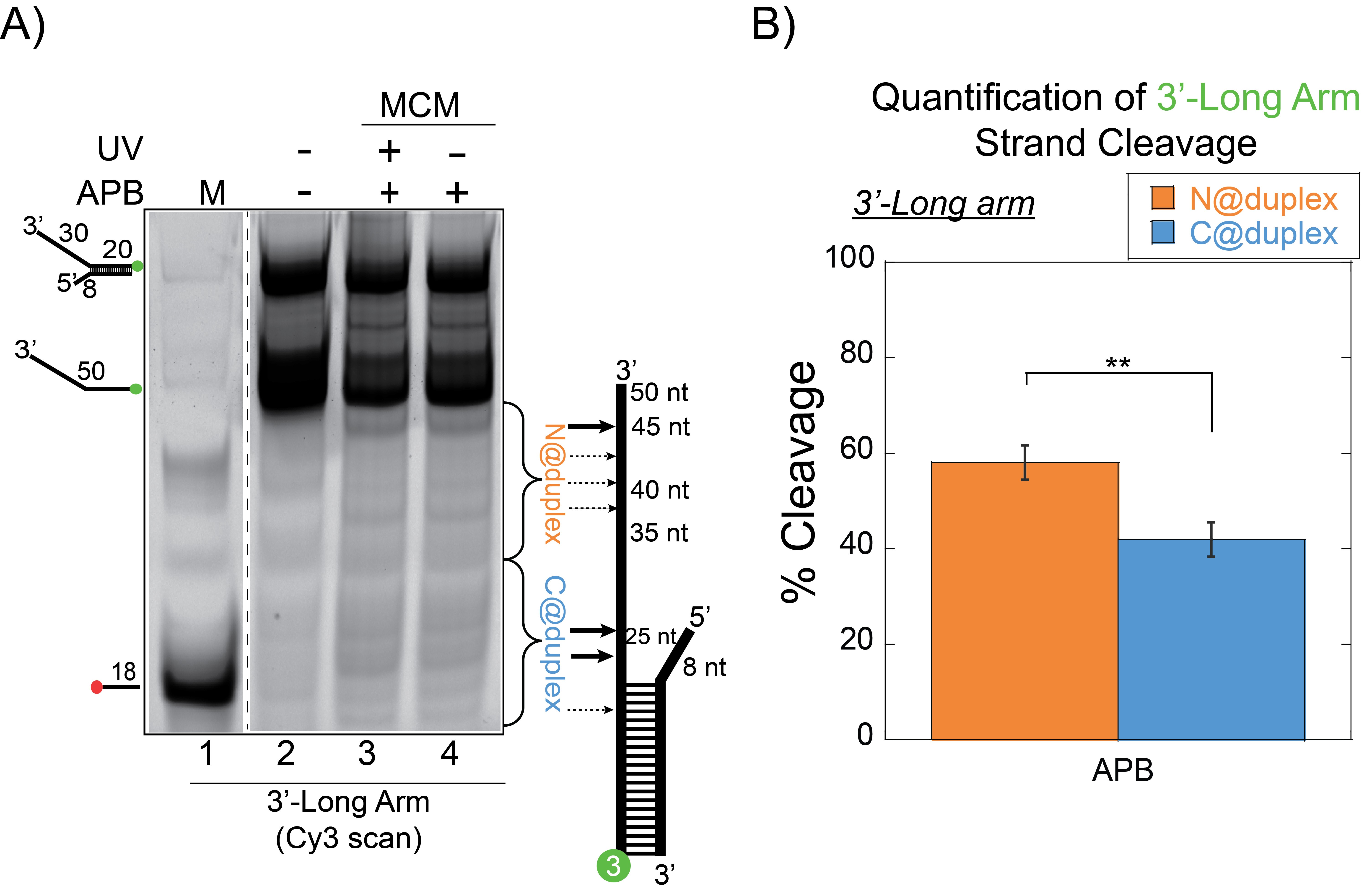
Figure 2. SsoMCM orientation mapping onto 3′-(DNA171/165-3) long arm fork DNA substrate. A. APB orientation mapping of the 3′-encircled strand labelled at the 5′ duplex end with Cy3 on a 3′-long arm fork DNA substrate with a 20 base duplex. SsoMCM was labelled at C682 with APB specifically. B. Quantification of the relative amount of DNA cleavage for bases 20-35 or 36-50 from the 5′-end indicate the relative orientation for placing the N @duplex (orange) or C@duplex (blue), respectively closer to the duplex junction. DNA markers (M) indicate 18, 50 bases and fork DNA. Error bars represent standard error from 3-5 independent experiments. P-value is defined as ** < 0.01. - A standard two-tailed equal variance Student’s t-test is used to determine significant differences of C@duplex versus N@duplex. P-values are reported for each experimental condition. Detailed explanation on data analysis can be found in the original research paper Figures 1C and 1F, 2B and 2D, 3C and 2E (Perera and Trakselis, 2019).
Notes
- Control experiments are performed with APB-MCM-DNA in the absence of UV light and/or with DNA alone in the absence of UV and APB. After activation by UV light, the crosslinking of APB to DNA bases is generally non-specific, yet we detected significant crosslinking and subsequent ssDNA cleavage even in the absence of direct UV light.
- Similarly, 1-(p-Bromoacetamidobenzyl) ethylenediamine N,N,N (Fe-BABE) (Dojindo, Rockville, Maryland) can also be used in orientation mapping studies (Figure 1B and 1E in Perera and Trakselis, 2019). FeBABE utilizes a localized hydroxyl radical Fenton footprinting reaction. Rapid reactions generating high yields, mild protein conjugation and cleavage reaction conditions, non-sequence specific DNA cleavage are some advantages of utilizing FeBABE (Ishihama, 2000).
Recipes
- 1x CB (complex buffer)
20 mM TrisOAc
25 mM KOAc
10 mM MgOAc
0.1 mg/ml BSA
1 mM DTT - Annealing buffer
10 mM Tris pH 7.5
50 mM NaCl
1 mM EDTA - Post irradiation buffer
20 mM Tris-HCl pH 8.0
0.1% SDS
50 mM NaCl - Glycerol loading buffer
40% glycerol
10 mM EDTA (pH 8.0)
7.5% Orange G dye - 20% Native PAGE
1x TBE
20% acrylamide
0.05% APS
4 μl TEMED
Acknowledgments
This work was supported by the National Science Foundation Division of Molecular and Cellular Biosciences [NSF1613534 to M.A.T.] and supported by Baylor University. We thank Gregory Bowman for introducing us to the feasibility of this technique for orientation mapping (Nodelman et al., 2017).
Competing interests
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Graham, B. W., Schauer, G. D., Leuba, S. H. and Trakselis, M. A. (2011). Steric exclusion and wrapping of the excluded DNA strand occurs along discrete external binding paths during MCM helicase unwinding. Nucleic Acids Res 39: 6585-6595.
- Ishihama, A. (2000). Molecular anatomy of RNA polymerase using protein-conjugated metal probes with nuclease and protease activities. Chem Commun 13: 1091-1094.
- Kassabov, S. R. and Bartholomew, B. (2004). Site-directed histone-DNA contact mapping for analysis of nucleosome dynamics. Methods Enzymol 375: 193-210.
- McGeoch, A. T., Trakselis, M. A., Laskey, R. A. and Bell, S. D. (2005). Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol 12:756-762.
- Nodelman, I. M., Bleichert, F., Patel, A., Ren, R., Horvath, K. C., Berger, J. M. and Bowman, G. D. (2017). Interdomain communication of the Chd1 chromatin remodeler across the DNA Gyres of the nucleosome. Mol Cell 65(3): 447-459 e446.
- Owens, J. T., Miyake, R., Murakami, K., Chmura, A. J., Fujita, N., Ishihama, A. and Meares, C. F. (1998). Mapping the σ70 subunit contact sites on Escherichia coli RNA polymerase with a σ70-conjugated chemical protease. PNAS 95(11): 6021-6026.
- Pendergrast, P. S., Chen, Y., Ebright, Y. W. and Ebright, R. H. (1992). Determination of the orientation of a DNA binding motif in a protein-DNA complex by photocrosslinking. Proc Natl Acad Sci U S A 89(21): 10287-10291.
- Perera, H. M. and Trakselis, M. A. (2019). Amidst multiple binding orientations on fork DNA, Saccharolobus MCM helicase proceeds N-first for unwinding. Elife 8: e46096.
Article Information
Copyright
Perera and Trakselis. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Perera, H. M. and Trakselis, M. A. (2020). Site-specific DNA Mapping of Protein Binding Orientation Using Azidophenacyl Bromide (APB). Bio-protocol 10(12): e3649. DOI: 10.21769/BioProtoc.3649.
- Perera, H. M. and Trakselis, M. A. (2019). Amidst multiple binding orientations on fork DNA, Saccharolobus MCM helicase proceeds N-first for unwinding. Elife 8: e46096.
Category
Microbiology > Microbial biochemistry > DNA
Molecular Biology > Protein > Protein-DNA binding
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









