- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Differential Fractionation of Erythrocytes Infected by Plasmodium berghei
Published: Vol 10, Iss 11, Jun 5, 2020 DOI: 10.21769/BioProtoc.3647 Views: 4485
Reviewed by: Alexandros AlexandratosDhiman Sankar PalKathrin Sutter

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2107 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2184 Views
Abstract
The study of host/pathogen interactions at the cellular level during Plasmodium intra-erythrocytic cycle requires differential extraction techniques aiming to analyze the different compartments of the infected cell. Various protocols have been proposed in the literature to study specific compartments and/or membranes in the infected erythrocyte. The task remains delicate despite the use of enzymes or detergents theoretically capable of degrading specific membranes inside the infected cell.
The remit of this protocol is to propose a method to isolate the erythrocyte cytosol and ghosts from the other compartments of the infected cell via a percoll gradient. Also, the lysis of the erythrocyte membrane is done using equinatoxin II, which has proven to be more effective at erythrocyte lysis regardless of the cell infection status, compared to the commonly used streptolysin. The parasitophorous vacuole (PV) content is collected after saponin lysis, before recovering membrane and parasite cytosol proteins by Triton X-100 lysis. The lysates thus obtained are analyzed by Western blot to assess the accuracy of the various extraction steps. This protocol allows the separation of the host compartment from the parasite compartments (PV and parasite), leading to potential studies of host proteins as well as parasite proteins exported to the host cell.
Background
The protocol presented herein is an improvement of a protocol that we initially published in Scientific Reports. The initial goal of this protocol was to perform the characterization of a P. berghei protein exported by the parasite to its host cell. This allowed the confirmation of immunofluorescence assays showing the export of the protein–and its blockage by Brefeldin A–and the analysis of its interactome upon export in the infected erythrocyte (Gnangnon et al., 2019).
With the aim to provide an improved protocol herein, we refined the technique reported previously by including a percoll gradient step to separate the host cell materials from the parasite ones. After enriching the infected erythrocytes via a Nycodenz® layer, we use Equinatoxin II (EQT–Anderluh et al., 1996) to lyse infected erythrocytes, instead of Streptolysin (SLO) as proposed previously (Nyalwidhe and Lingelbach, 2006; Heiber and Spielmann, 2014; Külzer et al., 2015). We made this choice since a few studies have shown that EQT lyses all erythrocytes independently from their infection status, whereas SLO lyses preferentially uninfected erythrocytes (Jackson et al., 2007; Schön et al., 2008). A percoll gradient is then used to separate the erythrocyte components from the parasite ones. The preparation of this gradient is performed similarly to the one used for P. falciparum (Heiber and Spielmann, 2014).
The parasites embedded in their parasitophorous vacuole (PV) are further treated using saponin, which specifically lyses the PV membrane (Külzer et al., 2015). The final lysis step involves Triton X-100 to recover membrane and parasite cytosol proteins (Heiber and Spielmann, 2014).
Other intermediate lysis steps can also be set up to analyze the parasite compartments more in depth (Ghosh et al., 2017). The different lysates are analyzed by Western blot. The choice of the antibodies used to detect protein markers of each purified compartment was based on previous studies and on antisera availability from different laboratories (see Table 1) (Knapp et al., 1990; Kina et al., 2000; Blisnick et al., 2006; Hliscs et al., 2013; Külzer et al., 2015; Meibalan et al., 2015; Ghosh et al., 2017). The techniques used here are optimized to the study of P. berghei–infected erythrocytes (the antibodies listed in Table 1 cross-react with P. berghei antigens despite being initially raised against proteins from other Plasmodium species) and may require some modifications when studying other Plasmodium species. In the case of P. falciparum, the reader can refer to suggestions made by Külzer et al. (2015).
The protocol presented here will enable the analysis of the membrane and cytosol of erythrocytes infected by Plasmodium (and maybe other erythrocyte-infecting pathogens like Babesia, following protocol optimization). Further refinements could allow a more precise and specific analysis of all the different compartments of the infected cell at once, which will favor the study of host/pathogen interactions over the parasite intra-erythrocytic development (see the “Notes” section at the very end of this manuscript). This is of critical importance since this phase of the development cycle of the parasite is responsible for the symptoms of malaria.
Materials and Reagents
Note: All chemicals are stored in dedicated rooms and safety cabinets. They are manipulated in accordance with product safety data sheets (SDS) and manufacturer instructions were followed for shelf-life (see below for storage temperatures–RT = Room Temperature).
Consumables:
- 1 ml syringes (Terumo, catalog number: SS+01T1 )
- 26 G ½” needles (Terumo, catalog number: AN-2613R1 )
- Transfer pipettes (rod volume 3 ml) (Biosigma, catalog number: 390514 )
- Microcentrifuge tubes (1.5 ml SafeLock) (Eppendorf, catalog number: 7953691744 )
- Polypropylene 15 ml centrifugation tubes (Sarstedt, catalog number: 62.554.502 )
- Parasites and Mice
- Plasmodium berghei ANKA parasites (kind gift from Dr. O. Silvie, CIMI, Paris, stored in liquid nitrogen, diluted in room temperature RPMI post-thawing prior to being i.p.-injected to mice)
- CD-1 mice (25-30 g male CD-1® IGS, Charles River)
Mice are i.p. infected with 106 infected red blood cells and parasitemia is monitored using thin blood smears. For the procedure detailed below, parasitemia of infected mice should be between 5% and 10%. This is usually obtained between day 5 and 10 post infection. - Sodic Heparin (25,000 UI/5 ml, Sanofi, stored at RT)
- iRBC (infected Red Blood Cell) sequential lysis
- Tris Base (Euromedex, catalog number: 200923-A , stored at RT)
- KCl (Sigma-Aldrich, catalog number: 60128 , stored at RT)
- CaNa2·EDTA (Sigma-Aldrich, catalog number: ED2SC , stored at RT)
- NaCl (Euromedex, catalog number: 1112 , stored at RT)
- MgSO4 (Sigma-Aldrich, catalog number: M7506 , stored at RT)
- Triton X-100 (Euromedex, catalog number: EU2000-B , stored at RT)
- D-Sorbitol (Sigma-Aldrich, catalog number: S1876 , stored at RT)
- Nycodenz® powder (Eurobio Ingen, catalog number: 1002424 , stored at RT)
- 10x PBS (Euromedex, catalog number: ET330-A , stored at RT)
- cOmpleteTM EDTA-free anti-protease cocktail (Roche, catalog number: 5056489001 , stored at 4 °C)
- Percoll solution (GE Healthcare, catalog number: 17-0891-01 , stored at 4 °C)
- RPMI medium 1640 w/HEPES (Thermo Fisher Scientific, catalog number: 12017599 , stored at 4 °C)
- Equinatoxin II enzyme (kind gift from Prof.Gregor Anderluh, National Institute of Chemistry, Ljubljana, Slovenia, aliquots are stored in PBS at -80 °C)
- Saponin lysis solution (Sigma Aldrich, catalog number: S7900 , powder stored at RT)
- 60% Nycodenz solution (see Recipes for storage conditions)
Nycodenz buffered medium
Nycodenz stock solution
60% Nycodenz solution (freshly prepared) - Equinatoxin II (EQT) lysis buffer (see Recipes)
- Saponin lysis solution (see Recipes)
Saponin 10x stock solution
1x Saponin lysis solution (freshly prepared) - Triton lysis buffer (see Recipes)
- Percoll gradient layers (see Recipes)
90% Percoll stock solution
80% Percoll layer
70% Percoll layer
60% Percoll layer
40% Percoll layer
- SDS-Page and Western blot analysis
- Tris Base (Euromedex, catalog number: 200923-A , stored at RT)
- SDS 20% solution (Sigma-Aldrich, catalog number: 0 5030 , stored at RT)
- Glycine (Euromedex, catalog number: 26-128-6405 , stored at RT)
- Glycerol (Euromedex, catalog number: EU3550 , stored at RT)
- Tween-20 (Sigma-Aldrich, catalog number: 8221840500 , stored at RT)
- Polyacrylamide gels (Bio-Rad Mini-PROTEAN® TGXTM Precast Gels, catalog number: 4561043 , stored at 4 °C
- PageRuler Prestained Protein Ladder (Thermo Scientific, catalog number: 26619 , stored at -20 °C)
- Low fat milk powder (Régilait, skimmed milk 0.8% fat, stored at RT)
- Western blot detection kit (SuperSignalTM West Dura Extended Duration Substrate, Thermo Fisher Scientific, catalog number: 34075 , stored at 4 °C)
- Antibodies (primary and secondary antibodies are listed in Table 1)
- 3x Sample buffer (see Recipes)
- Running buffer (see Recipes)
- Transfer buffer (see Recipes)
- Blocking solution (see Recipes)
- Western blot washing buffer (see Recipes)
Table 1. List of primary and secondary antibodies used in this study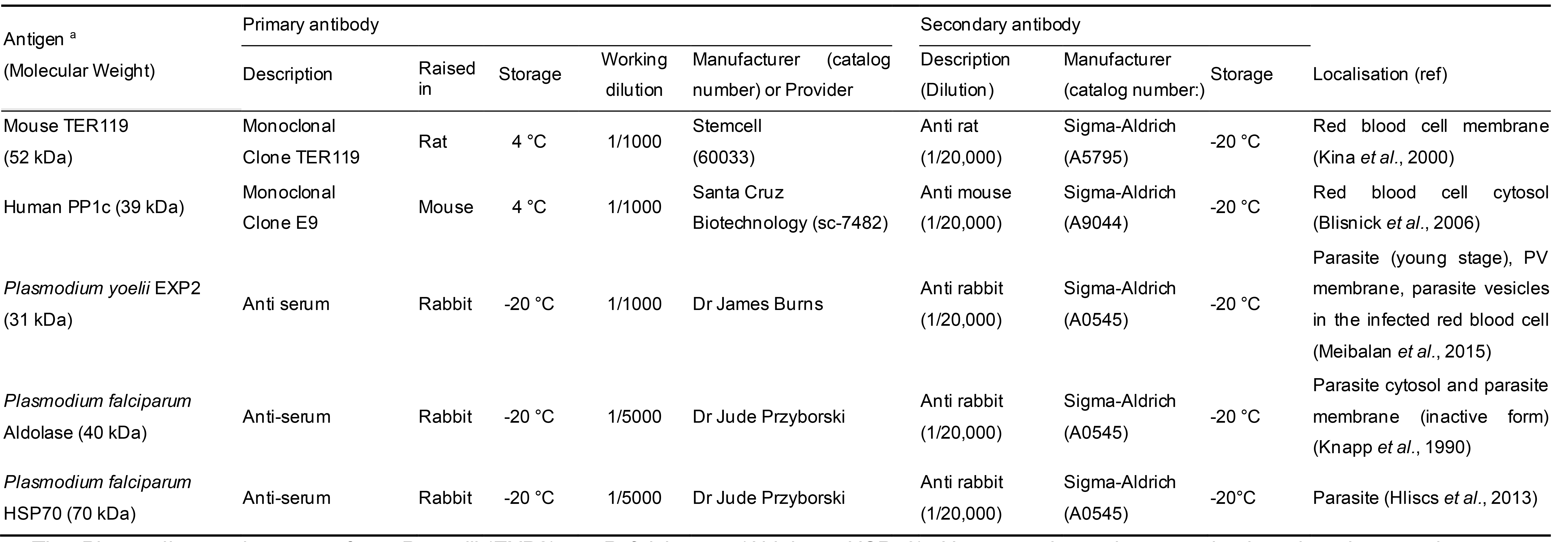
aThe Plasmodium antigens are from P. yoelii (EXP2), or P. falciparum (Aldolase, HSP70). However, the antiserum raised against these antigens cross react with P. berghei EXP2, Aldolase and HSP70 respectively. The antibody targeting human PP1c also recognizes mouse PP1c.
Equipment
- Laboratory rotating wheel (Stuart, model: SB3 )
- 50 ml Nalgene Oak Ridge High-Speed Centrifuge Tubes with sealing cap, PSF (Thermo Fisher Scientific, catalog number: 3137-0050 )
- Refrigerated centrifuge for microtubes (Eppendorf, model: 5427R )
- Centrifuge for 15 ml tubes (Eppendorf, model: 5810R )
- Superspeed centrifuge (Sorvall Lynx 4000 centrifuge with fixed angle rotor F20-12x50 LEX, ThermoScientific)
- Sonication device (Bioruptor plus, Diagenode, catalog number: B01020001 )
- Block heater (Stuart, model: SBH200D )
- Gel electrophoresis and transfer chamber (Bio-Rad Miniprotean Tetra Cell System)
- Filter paper for Western blot transfer (Whatman 3MM CHR, GE Healthcare, catalog number 3030-917 )
- Nitrocellulose membrane (Amersham protran Premium 0.45 Nitrocellulose, Thermo Fisher Scientific, catalog number: 15269804 )
- Vacuum sealer (KitchenGynti)
- Rocker platform (Labnet reciprocal, 3D or rocker shaker)
- Molecular Imager (Bio-Rad ChemiDoc XRS+)
Software
- Image Lab 5.0 (Bio-Rad)
Procedure
Note: This procedure contains 3 sections (A, B and C). Sections A and B must be carried out without interruption (total duration ~1 day). Samples obtained at the end of Section B can be frozen before going to Section C.
- Collection and isolation of P. berghei-infected red blood cells
- Prepare two 15 ml polypropylene tubes with 5 ml of 60% Nycodenz solution (see Recipe 1) and let them stand at room temperature (1-2 h).
- Let the 80%, 70%, 60% and 40% percoll solutions stand at room temperature (1-2 h).
- Heart bleed two CO2-euthanized (see the “Ethics” section for protocol approval) P. berghei-infected mice (parasitemia between 5-10 %) using 26 G ½” needles with attached 1 ml syringes containing 0.15 ml heparin.
- Gently layer 1 ml of heparinized blood on the 60% Nycodenz® (one mouse per Nycodenz® layer; the volume of heparinized blood may vary from 800 µl to 1.2 ml).
- Centrifuge at 450 x g for 20 min at room temperature with no brake and minimal acceleration.
- Carefully collect the brown layer (illustrated in Figure 1) consisting of P. berghei-infected red blood cells at the interface between the upper layer and the Nycodenz layer with a transfer pipette into new 15 ml tubes.
- Add 10 ml of 1x PBS (kept at RT) in each tube.
- Centrifuge at 450 x g for 10 min at room temperature.
- Discard the supernatant.

Figure 1. Graphical abstract of the isolation and differential fractionation of P. berghei-infected erythrocytes. The outcome after Nycodenz® and percoll gradients, and fractions to be collected (infected Red Blood Cells and ①, ② and ③) are illustrated. At the end of each step of the procedure, each fraction is indicated in gray boxes. Western blot analysis of these factions is summarized in Figure 2.
- Sequential lysis of P. berghei-infected red blood cells
Note that Steps B1-B8 are done at RT (pay close attention to acceleration and braking conditions) and that Steps B9-B16 must be done on ice (centrifugations at 4 °C, maximal acceleration and brake).- Prepare EQT lysis buffer (see Recipe 2).
- Add 1 ml of EQT lysis buffer on each pellet of infected erythrocytes.
- Incubate for 15 min on a rotating wheel at room temperature (20-25 rpm) (lysis can be monitored using thin blood smears).
- Prepare the Percoll gradient in a 50 ml Nalgene Oak Ridge High-Speed Centrifuge Tube: add gently successively: 8 ml of 80% Percoll solution, 6 ml of 70% Percoll solution, 6 ml of 60% Percoll solution and 6 ml of 40% Percoll solution (carefully place each layer one above the other). Use the gradient immediately.
- Pool the two tubes of EQT-lysed infected red blood cells (total volume 2-3 ml).
- Pipette slowly the EQT-lysed infected red blood cells on the top of the Percoll gradient and seal the tube.
- Centrifuge immediately at 10,000 x g for 30 min at room temperature, with minimal acceleration and no brake.
- Using a transfer pipette, collect carefully each layer illustrated on Figure 1 into new 1.5 ml tubes. Collect the fraction 1 (supernatant) without reaching the interface with the layer 2 and proceed to Step B9. Collect fractions 2 and 3 and proceed to Step B10 (see Figure 1 for layer/fraction numbers).
- Centrifuge fraction 1 at 18,000 x g for 30 min at 4 °C. Collect the pellet (pellet P1). Collect the supernatant into a new 1.5 ml tube and repeat the centrifugation. Collect the supernatant (supernatant SN1).
Notes:- The supernatant SN1 collected after Equinatoxin lysis and Percoll gradient contains the red blood cell cytosol as attested by the detection of the host PP1. This supernatant must be collected carefully without disturbing the pellet, and centrifuged twice for 30 min at 18,000 x g. It is important to note that the Equinatoxin lysis is performed during 15 min, as shorter incubation may provide incomplete lysis of infected erythrocytes, and longer incubation may increase the leakage of parasite content as illustrated by discrete parasite HSP70 staining observed in some cases.
- The pellet P1 obtained after SN1 first centrifugation contains red blood cell ghosts (TER119 positive signal). This pellet should be red. The presence of a black pellet at the bottom of the red one indicates a contamination with parasites. The EXP2 signal observed in Western blot analysis of P1 may correspond to the presence of parasite vesicles in the red blood cell, as described for P. yoelii-infected cells, that would be pelleted along with ghosts during centrifugation (Meibalan et al., 2015).
- Centrifuge fractions 2 and 3 at 18,000 x g for 30 min at 4 °C and wash three times with 1 ml of 1x PBS to remove the percoll. Note that following the first two centrifugations the fractions may not pellet correctly. Discard all supernatants.
Note: The fractions 2 and 3 obtained after the percoll gradient may correspond to red blood cells infected with different stages of the parasite (as observed in Heiber and Spielmann, 2014), even if this is not clear when these fractions are analyzed on smears. Note that we have always observed the same profile of EXP2, HSP70 and Aldolase expression in these two fractions, and that in P. berghei-infected mice, only ring to trophozoite stages are observed in the circulating blood (Janse et al., 2004). - Prepare the 1x Saponin lysis solution (see Recipe 3).
- Add 1 ml of 1x Saponin lysis solution on pellets 2 and 3 (Step B10) and incubate 10 min on ice.
- Centrifuge Saponin-lysed fractions 2 and 3 at 18,000 x g for 30 min at 4 °C. Collect the pellets (to be processed in Step B15). Collect the supernatants into new 1.5 ml tubes and repeat the centrifugation. Collect the supernatants on ice (supernatant SN2s and SN3s).
Note: The supernatants SN2s and SN3s collected after saponin lysis correspond to the PV content. Note that it is important to centrifuge these fractions twice for at least 30 min at 18,000 x g to avoid contamination with PV and/or parasite membranes. This can be checked by the absence of EXP2 detection in Western blot analysis. However, the HSP70 signal observed in these supernatants confirms previous results showing that the use of saponin to lyse the PV membrane does not prevent leakage from the parasite compartment (Burghaus and Lingelbach, 2001; Nyalwidhe and Lingelbach, 2006). In the absence of a more specific detergent or enzyme capable of lysing specifically the PV membrane, the study of this compartment that plays a key role in the parasite growth and communication with its host cell remains particularly complex (reviewed in Matz et al., 2020). - Prepare the Triton lysis solution (see Recipe 4).
- Add 200 µl of Triton lysis solution on pellets 2 and 3 from Step B13 and incubate 5 min on ice.
- Centrifuge Triton-lysed fractions 2 and 3 at 18,000 x g for 30 min at 4 °C. Collect the pellets (Pellet P2t and P3t). Collect the supernatants into new 1.5 ml tubes and repeat the centrifugation. Collect the new supernatants on ice (supernatant SN2t and SN3t).
Notes:- The supernatants SN2t and SN3t collected after Triton X-100 lysis correspond to the parasite cytosol, as attested by the detection of parasite Aldolase, together with HSP70. The detection of EXP2 signal in these fractions may correspond to EXP2 protein embedded in the PV membrane (and released during Triton X-100 lysis) as well as parasite cytosolic EXP2 which has been observed in young stages of the parasite (Meibalan et al., 2015).
- The pellets P2t and P3t correspond to the PV and parasite membranes. The detection of Aldolase in these pellets can be explained by the presence of an inactive form associated to the parasite membrane (Knapp et al., 1990).
- Add the required amount of Sample buffer to each fraction: add 100 µl of 1x Sample buffer to pellets P1, P2t and P3t, and 100 µl of 3x Sample buffer to 200 µl of supernatants SN1, SN2s, SN3s, SN2t and SN3t.
Note: All the fractions SN1, P1, SN2s, SN3s, SN2t, SN3t, P2t and P3t may be stored at -20 °C in Sample buffer prior Western blot analysis). All the collected fractions may also be used in mass spectrometry analysis. In case an immunoprecipitation (IP) must be performed before mass spectrometry or Western blot, do not add Sample buffer and proceed to the IP using the fractions.
- Western blot analysis of protein extracts
- SDS-PAGE
- Place three polyacrylamide gels into the electrophoresis chamber filled with running buffer.
- Sonicate the samples in a sonication device at 4 °C (10 cycles, 10 s each).
- Incubate the samples at 95 °C for 3 min in a block heater.
- Load the samples on three polyacrylamide gels 30 µl (except for SN1: use 15 µl) of extracts per lane, 3 identical gels). Load the prestained protein ladder (7 µl).
- Run the gels at 110 V for 1 h 30 min.
- Western blot analysis
- Prepare the blot using sponge, filter papers, nitrocellulose membrane and the SDS gel, and perform protein transfer in a transfer chamber filled with transfer buffer (80 V for 1 h 30 min in a 4 °C room).
- Post transfer rinse the three nitrocellulose membranes in 1x PBS (transfer can be confirmed by staining the nitrocellulose membranes using Ponceau red).
- Prepare 100 ml of blocking solution.
- Incubate the membranes in 10 ml blocking solution for 1 h at RT on a rocking platform.
- Place the membranes into transparent sheets sealed on three sides (cut to the size of the membranes). Add 1 ml of blocking solution containing primary antibodies (see Table 1 for antibody dilutions and specificity; note that anti-PfHSP70, anti-PyEXP2 and anti PfAldolase antisera can be incubated together as they have all been raised in rabbit and MW of native proteins are different) and seal the last side of the transparent sheets. Incubate overnight at 4 °C on a rotating wheel.
- Wash the membranes five times in Western blot washing buffer for 15 min at RT on a rocking platform.
- Dilute the secondary antibodies in blocking solution (see Table 1 for antibody dilutions and specificity) and incubate the membranes at RT for 30 min on a rocking platform.
- Wash the membranes five times (15 min each) in Western blot washing buffer at RT on a rocking platform.
- For Western blot detection, incubate the membranes with detection reagent (sufficient quantity to cover the membrane) according to the manufacturer instructions. A graphical abstract of Western blot analysis of the different fractions and representative Western blots are shown in Figure 2.
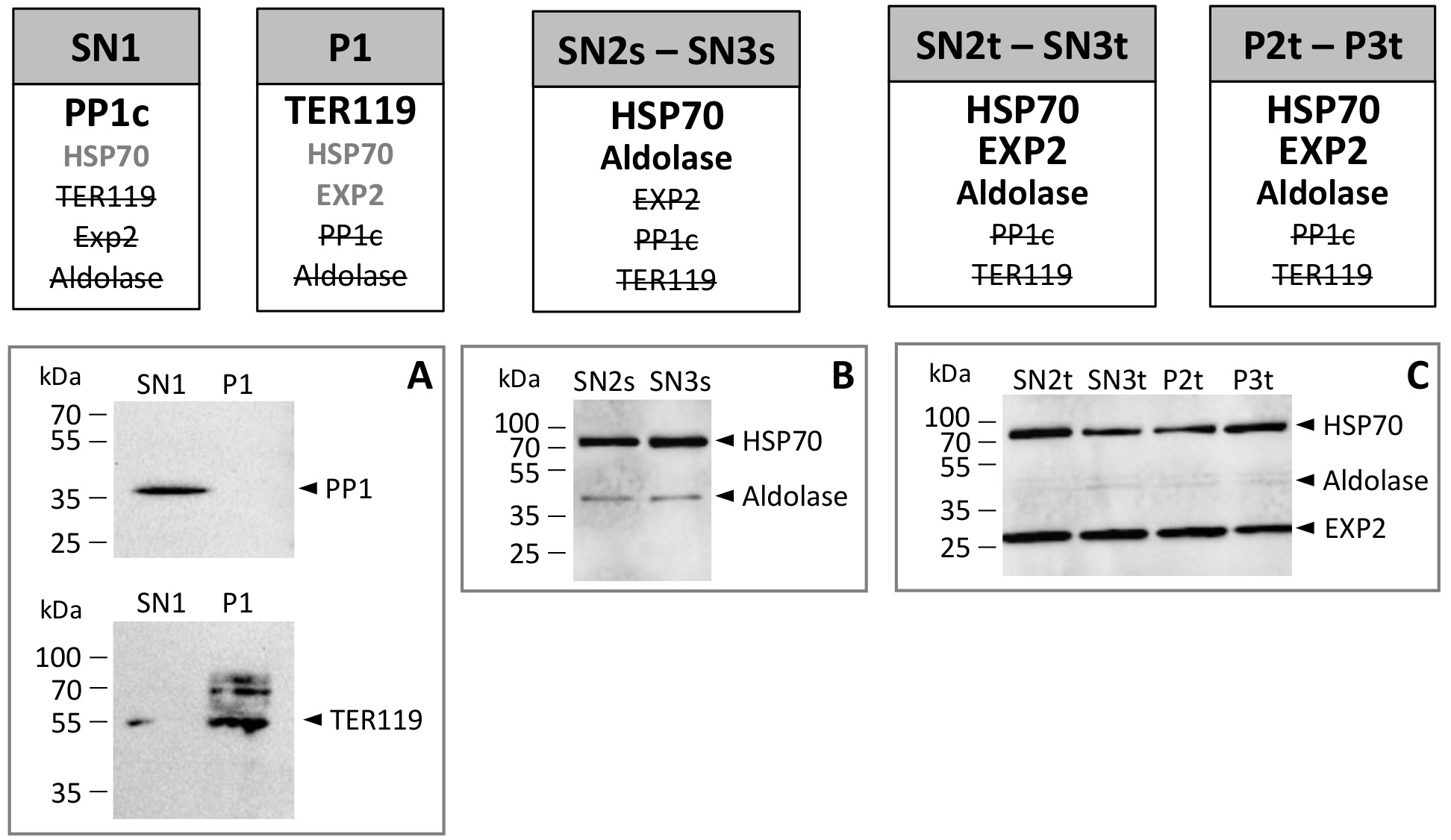
Figure 2. Summary of Western blot analysis of the collected fractions. Fraction names are indicated in gray boxes and refer to fractions illustrated in Figure 1. Summary of Western blot analysis is shown below these gray boxes. In bold black are the major signals observed, in gray are the other antigens detected (low signal and/or not obtained systematically), and in strikethrough characters are the not detected antigens. Host PP1 and TER119 are detected using anti-human PP1c and anti-mouse TER119 respectively (note that the bands above the one labeled as ‘TER119’ may be due to complexes resulting from the ability of this protein to interact with other erythrocyte surface proteins (Kina et al., 2000). Parasite EXP2, HSP70 and Aldolase are detected using anti-P. yoelii EXP2, anti-P. falciparum HSP70 and anti-P. falciparum Aldolase respectively (see Table 1). Panels A, B and C show representative Western blot analysis of host-specific (A) or parasite-specific (B, C) proteins. Note that these Western blots are not quantitative and that the intensity differences of Aldolase signal between B and C may correspond to different quantities of total extracts loaded.
- SDS-PAGE
Notes
Since our initial research goal was to study parasite proteins exported to the host cell, we focused on improving the isolation of the host cell material from the parasite elements in the infected erythrocyte. This implies that we did not refine the sequential lysis of the PV/parasite compartments (the use of antibodies against EXP2, Aldolase and HSP70 is not discriminant between parasite cytosol and PV/parasite membranes). Elegant techniques involving the use of biotin derivatives have however allowed the isolation the PV and the erythrocyte membrane, prior to their study by OMICs approaches (Nyalwidhe and Lingelbach, 2006; Ravenhill et al., 2019). The combination of our protocol with these techniques creates perspectives for more sophisticated ways of deciphering the ins and outs of host/pathogen interactions over the intra-erythrocytic cycle of Plasmodium on both the host and the parasite sides.
Recipes
- 60% Nycodenz solution
Nycodenz buffered medium
5 mM Tris HCl, pH 7.5
3 mM KCl
0.3 mM CaNa2·EDTA
Nycodenz stock solution- Dissolve 27.6 g solid Nycodenz in 60 ml buffered medium and make up to 100 ml with that medium [density (20 °C) 1.15 g/ml]
- Autoclave and store at 4 °C
Add 2 ml of 1x PBS to 3 ml of Nycodenz stock solution - Equinatoxin II (EQT) lysis buffer (to be freshly prepared)
20 mM Tris pH 7.4
130 mM NaCl
1 mM MgSO4
6 µg/ml EQT - Saponin lysis solution
Saponin 10x stock solution
Dissolve 0.7 g Saponin in 100 ml 1x PBS; store at 4 °C
1x Saponin lysis solution (to be freshly prepared)
Mix 1 vol of Saponin stock solution with 9 vol 1x PBS - Triton lysis buffer (to be freshly prepared)
50 mM Tris pH 7.4
100 mM NaCl
1% Triton X-100
0.5 mM EDTA
Roche cOmpleteTM EDTA-free anti-protease cocktail
0.2 U/ml DNase, store on ice until use - Percoll gradient layers
90% Percoll stock solution
Mix 45 ml Percoll with 5 ml 10x PBS
80% Percoll layer
Mix 0.5 g D-sorbitol with 8.9 ml 90% Percoll stock solution and 1.1 ml RPMI
70% Percoll layer
Mix 0.5 g D-sorbitol with 7.8 ml 90% Percoll stock solution and 2.2 ml RPMI
60% Percoll layer
Mix 0.5 g D-sorbitol with 6.7 ml 90% Percoll stock solution and 3.3 ml RPMI
40% Percoll layer
Mix 0.5 g D-sorbitol with 4.4 ml 90% Percoll stock solution and 5.6 ml RPMI
Filter all solutions (0.22 µm) and store at 4 °C - 3x Sample buffer (long storage at -20 °C–thaw at RT for use)
150 mM Tris pH 6.5
7.5% SDS
15% β-Mercaptoethanol
30% glycerol
0.25% bromophenol blue - Running buffer (stored for a couple of weeks at 4 °C)
3.03 g Tris Base
14.41 g Glycine
5 ml SDS 20%
Add H2O to 1 L - Transfer buffer (stored for a couple of weeks at 4 °C)
2.42 g Tris Base
11.26 g Glycine
200 ml EtOH
Add H2O to 1 L - Blocking solution (freshly prepared prior to blocking)
5% Low fat milk in 1x PBS - Western blot washing buffer (stored for a couple of weeks at 4 °C)
0.4% Tween-20 in 1x PBS
Acknowledgments
This work was supported by Université de Lille, CNRS, Inserm and Institut Pasteur de Lille. This work is based on a protocol initially published in Scientific Reports (Gnangnon et al., 2019). The authors want to thank Dr. Jamal Khalife for critical reading of the manuscript.
Competing interests
The authors declare no competing interests.
Ethics
Experiments were carried out in accordance with the principles of the European Community Council recommendations (86/609/EEC) for animal experimentations. All animal procedures were approved and supervised by the local Ethics Animal Committee (CEEA-75 Comité d’Ethique en Expérimentation Animale, Nord - Pas de Calais, France). The ethical approval number for protocols and procedures on mice is APAFIS#18905-2019020111166978v2.
References
- Anderluh, G., Pungercar, J., Strukelj, B., Macek, P. and Gubensek, F. (1996). Cloning, sequencing, and expression of equinatoxin II. Biochem Biophys Res Commun 220(2): 437-442.
- Blisnick, T., Vincensini, L., Fall, G. and Braun-Breton, C. (2006). Protein phosphatase 1, a Plasmodium falciparum essential enzyme, is exported to the host cell and implicated in the release of infectious merozoites. Cell Microbiol 8(4): 591-601.
- Burghaus, P. A. and Lingelbach, K. (2001). Luciferase, when fused to an N-terminal signal peptide, is secreted from transfected Plasmodium falciparum and transported to the cytosol of infected erythrocytes. J Biol Chem 276(29): 26838-26845.
- Ghosh, S., Kennedy, K., Sanders, P., Matthews, K., Ralph, S. A., Counihan, N. A. and de Koning-Ward, T. F. (2017). The Plasmodium rhoptry associated protein complex is important for parasitophorous vacuole membrane structure and intraerythrocytic parasite growth. Cell Microbiol 19(8).
- Gnangnon, B., Freville, A., Cailliau, K., Leroy, C., De Witte, C., Tulasne, D., Martoriarti, A., Jung, V., Guerrera, I. C., Marion, S., Khalife, J. and Pierrot, C. (2019). Plasmodium pseudo-Tyrosine Kinase-like binds PP1 and SERA5 and is exported to host erythrocytes. Sci Rep 9(1): 8120.
- Heiber, A. and Spielmann, T. (2014). Preparation of parasite protein extracts and Western blot analysis. Bio-protocol 4(11): e1136.
- Hliscs, M., Nahar, C., Frischknecht, F. and Matuschewski, K. (2013). Expression profiling of Plasmodium berghei HSP70 genes for generation of bright red fluorescent parasites. PLoS One 8(8): e72771.
- Jackson, K. E., Spielmann, T., Hanssen, E., Adisa, A., Separovic, F., Dixon, M. W., Trenholme, K. R., Hawthorne, P. L., Gardiner, D. L., Gilberger, T. and Tilley, L. (2007). Selective permeabilization of the host cell membrane of Plasmodium falciparum-infected red blood cells with streptolysin O and equinatoxin II. Biochem J 403(1): 167-175.
- Janse, C. J., Ramesar, J. and Waters, A. P. (2004). Plasmodium berghei : general parasitological methods. Leiden, The Netherlands.
- Kina, T., Ikuta, K., Takayama, E., Wada, K., Majumdar, A. S., Weissman, I. L. and Katsura, Y. (2000). The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br J Haematol 109(2): 280-287.
- Knapp, B., Hundt, E. and Kupper, H. A. (1990). Plasmodium falciparum aldolase: gene structure and localization. Mol Biochem Parasitol 40(1): 1-12.
- Külzer, S., Bittl, V. and Przyborski, J. M. (2015). Fractionation of Plasmodium-infected human red blood cells to study protein trafficking. Methods Mol Biol 1270: 71-80.
- Matz, J. M., Beck, J. R. and Blackman, M. J. (2020). The parasitophorous vacuole of the blood-stage malaria parasite. Nat Rev Microbiol. DOI: 10.1038/s41579-019-0321-3.
- Meibalan, E., Comunale, M. A., Lopez, A. M., Bergman, L. W., Mehta, A., Vaidya, A. B. and Burns, J. M., Jr. (2015). Host erythrocyte environment influences the localization of exported protein 2, an essential component of the Plasmodium translocon. Eukaryot Cell 14(4): 371-384.
- Nyalwidhe, J. and Lingelbach, K. (2006). Proteases and chaperones are the most abundant proteins in the parasitophorous vacuole of Plasmodium falciparum-infected erythrocytes. Proteomics 6(5): 1563-1573.
- Ravenhill, B. J., Kanjee, U., Ahouidi, A., Nobre, L., Williamson, J., Goldberg, J. M., Antrobus, R., Dieye, T., Duraisingh, M. T. and Weekes, M. P. (2019). Quantitative comparative analysis of human erythrocyte surface proteins between individuals from two genetically distinct populations. Commun Biol 2: 350.
- Schön, P., Garcia-Saez, A. J., Malovrh, P., Bacia, K., Anderluh, G. and Schwille, P. (2008). Equinatoxin II permeabilizing activity depends on the presence of sphingomyelin and lipid phase coexistence. Biophys J 95(2): 691-698.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Gnangnon, B., Peucelle, V. and Pierrot, C. (2020). Differential Fractionation of Erythrocytes Infected by Plasmodium berghei. Bio-protocol 10(11): e3647. DOI: 10.21769/BioProtoc.3647.
Category
Microbiology > Microbial biochemistry > Protein > Isolation and purification
Microbiology > Microbe-host interactions > In vitro model > Cell line
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










