- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of Ureides Content in Plant Tissues
Published: Vol 10, Iss 11, Jun 5, 2020 DOI: 10.21769/BioProtoc.3642 Views: 5271
Reviewed by: Amey RedkarSolmaz IraniMarisa Rosa

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Seed Collection in Temperate Trees—Clean, Fast, and Effective Extraction of Populus Seeds for Laboratory Use and Long-term Storage
Naima Bhutta [...] Katharina Bräutigam
Feb 5, 2024 2036 Views

Vegetative Propagation of Cannabis sativa and Resin Obtained From its Female Inflorescences
Sebastián D´Ippolito [...] Silvana L. Colman
Feb 20, 2025 1735 Views

A New Approach to Detect and Semi-quantify All Molecular Species and Classes of Anionic Phospholipids Simultaneously in Plant Samples
Manon Genva [...] Laetitia Fouillen
Apr 20, 2025 1718 Views
Abstract
The ureides allantoin and allantoate are the main organic nitrogen compounds transported in several legumes, predominantly from N2 fixation. Moreover, recent studies point out a remarkable role for allantoin during several stress responses of plants other than legumes. The goal of this protocol is to determine ureides concentration in different plant tissues. Ureides are extracted from plant material by boiling it in phosphate buffer. The allantoin and allantoate present in the supernatants are subjected to alkaline-acidic hydrolysis to glyoxylate. The glyoxylate is converted into glycoxylic acid phenylhydrazone, that is then oxidized to red-colored 1,5-diphenylformazan. The absorbance of supernatants is measured using a spectrophotometer at 520 nm. Ureides concentration can be inferred by using a glyoxylate calibration curve. Ureide quantification of different tissues of Arabidopsis thaliana and soybean plants were carried out following this protocol.
Keywords: AllantoinBackground
Determination of ureides is important for characterizing N2 fixation and assimilation in legume plants as well as stress and nutritional responses of non-ureidic plants like Arabidopsis (Brychkova et al., 2008; Watanabe et al., 2014; Irani and Todd, 2016 and 2018). The formation of allantoate is also useful for in vivo and in vitro determination of allantoinase activity (Duran and Todd, 2012). Techniques requiring expensive equipment, such as high-performance liquid chromatography (HPLC), are commonly used for ureide quantification while ethanol extraction is mostly employed for ureide recovery from plant tissues. The present protocol is based on flexible spectrometry that uses low volume of cheap reagents. The use of phosphate buffer instead of ethanol for ureide extraction and quantification greatly improved the reproducibility of the measurements, possibly by decreasing the interfering compounds that abound with the ethanol extraction.
Allantoin and allantoate present in plant extracts are converted to glyoxylate by alkaline-acidic hydrolysis (Vogels and Van der Drift, 1970). To carry out this protocol, three tubes (A, B and C) for alkaline and/or acid hydrolysis are prepared for each sample (Figure 1). The glyoxylate is converted into glycoxylic acid phenylhydrazone and is then oxidized by ferricyanide to form red-colored 1,5-diphenylformazan. The absorbance of supernatants is measured using a spectrophotometer at 520 nm. Allantoin content is obtained by subtracting the levels of glyoxylate resultant of allantoate degradation (tube B) from glyoxylate derived of allantoin alkaline-acid hydrolysis (tube C). Likewise, allantoate content can be inferred by the subtraction of basal glyoxylate levels (tube A) from glyoxylate converted from allantoate (tube B).
The described protocol was carried out for determine ureide concentration for different tissues, nutritional and stress conditions in Arabidopsis thaliana (Lescano et al., 2016 and 2020); and in roots and shoots of nodulating and non-nodulating soybean plants (Muñoz et al., 2016).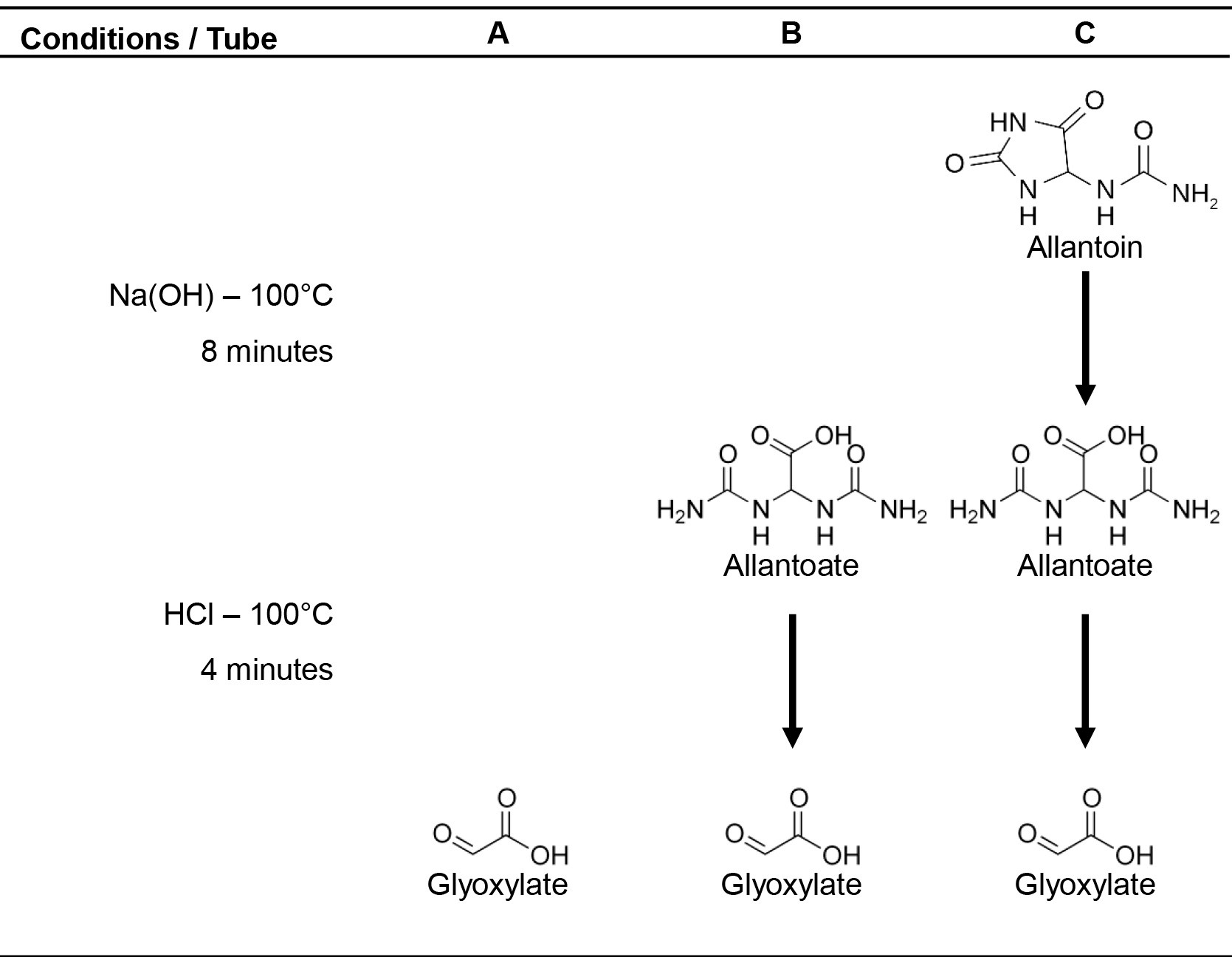
Figure 1. Alkaline-acidic hydrolysis of ureides
Materials and Reagents
- 1.5 ml microcentrifugue tubes (Eppendorf)
- 1.5 ml safe-lock microcentrifugue tubes (Eppendorf)
- 50 ml Falcon tubes
- Chemical resistant gloves
- Seeds (e.g., Arabidopsis)
- Growing medium [e.g., for Arabidopsis 0.5x MS (1% w/v agar) plates or soil: vermiculite (1:1) mix pots]
- Distillated water (dH2O)
- (Optional) Liquid air/N2
- Monopotassium phosphate (Cicarelli® Laboratories, catalog number: 1057211 )
- Dipotassium phosphate (Cicarelli® Laboratories, catalog number: 1015211 )
- Sodium hydroxide (NaOH) (Cicarelli® Laboratories, catalog number: 843211 )
- Hydrochloric acid (HCl) (Biopack, catalog number: 2000963208 )
- Phenylhydrazine (Merck, catalog number: 107251 ). Store at room temperature (RT) in the dark
- Potassium ferricyanide (Anedra, Research AG, catalog number: 7039 )
- Sodium glyoxylate monohydrate (Sigma-Aldrich, Merck, catalog number: G4502 )
- Extraction Buffer (see Recipes)
- Reaction Buffer (see Recipes)
Equipment
- Beaker
- (Optional) Mortar and pestle
- Plant growth chamber/greenhouse
- 60-65 °C incubator
- Analytical balance 0.5/0.0001 g (e.g., Mettler Toledo, model: AL204 )
- Centrifuge at 4°C with microliter rotor (e.g., Thermo Scientific, Sorval Biofuge Primo R)
- Ice bath
- Hot air bath at 100 °C
- Vortex mixer (e.g., Decalab)
- Fume hood
- VIS Spectrophotometer (e.g., Bioamerican Science, model: SP 2000 UV )
- Spectrophotometric plastic or glass VIS/UV-VIS semi-micro 0.75-1.5 ml cuvettes
Procedure
- Preparation of plant materials
- Grow plants in a chamber/greenhouse with the corresponding conditions.
Note: For grow Arabidopsis thaliana, stratify seeds at 4 °C for 2-3 days. Sow seeds on 0.5x MS agar plates or on soil:vermiculite (1:1) mix, and transfer to a growth chamber under a 16 h light/8 h dark photoperiod at 22 °C and a light intensity of 100-150 μmol photons m-2s-1. - Collect about 20 mg-0.5 g of plant samples in 1.5 ml safe-lock microcentrifugue tubes.
Notes:- For Arabidopsis harvest young, senescent and caulinar leaves, buds, flowers, siliques and dry seeds of adult plants (Figures 2A-2F) or collect young seedlings grown in 0.5x MS agar plates (Figure 2G).
- It is recommended to collect plant samples of similar weights between biological replicates.
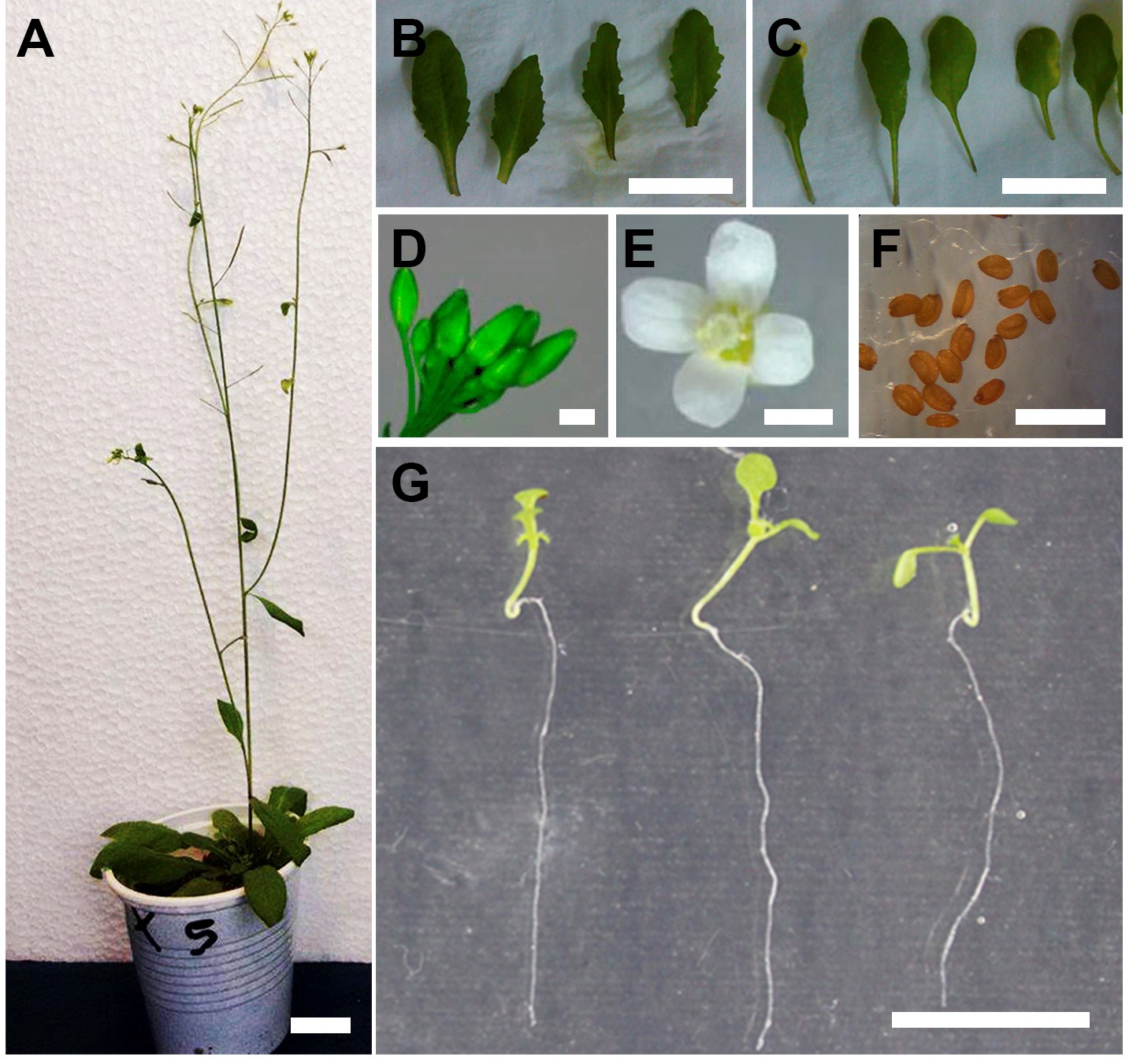
Figure 2. Examples of Arabidopsis thaliana tissues used for ureide quantification. (A) 8-week old plant. Details of (B) young leaves, (C) senescent leaves, (D) buds, (E) flowers and (F) seeds are shown. (G) 7 day-old seedlings grown on 0.5x agar plates. Scale bars = 2 cm (A-C, G), = 1 mm (D-F). - For Arabidopsis harvest young, senescent and caulinar leaves, buds, flowers, siliques and dry seeds of adult plants (Figures 2A-2F) or collect young seedlings grown in 0.5x MS agar plates (Figure 2G).
- Put microcentrifugue tubes with open lids in an oven at 60-65 °C overnight.
- Weight dry plant samples and put them back in the tubes. Samples can be stored at -20 °C.
- Grow plants in a chamber/greenhouse with the corresponding conditions.
- Ureide extraction from plant tissues
- Optional step: ground plant tissues to fine powder with a mortar and a pestle. Transfer the powder to microcentrifugue tubes.
Note: Grinding plant samples to fine powder depends on the type of tissue and plant species. For example, it is not necessary for ureide extraction from Arabidopsis, but could be required for ureide extraction from hard tissues of other plant species, i.e., soybean (Muñoz et al., 2016). - Add 0.5 ml of Extraction Buffer.
- Centrifuge at 17,758 x g for 5 min at 4 °C.
- Incubate in a hot air bath at 100 °C for 20 min. Ensure that all the tubes keep well closed.
- Transfer to ice bath for 1 min. Ensure that all the tubes keep well closed.
- Centrifuge at 17,758 x g for 5 min at 4 °C.
- Transfer the supernatant to a new microcentrifugue tube (not necessary safe-lock tube). Samples can be stored at -20 °C for several months, or at 4-8 °C prior to the next step.
- Optional step: ground plant tissues to fine powder with a mortar and a pestle. Transfer the powder to microcentrifugue tubes.
- Alkaline/Acid Hydrolysis
- Prepare microcentrifugue tubes in which reactions A-C for each sample will be carried out, according the compound/s to be measured (Figure 1). Designate the tubes according which reaction take place in it: A, B and C.
Notes:- Prepare tubes according to the compound/s to be measured as indicated in Table 1. For example, to measure allantoin and allantoate concentrations in sample #1 tubes can be designated 1A, 1B and 1C.
- The use of duplicates for each sample is highly recommended.
Table 1. Tubes preparation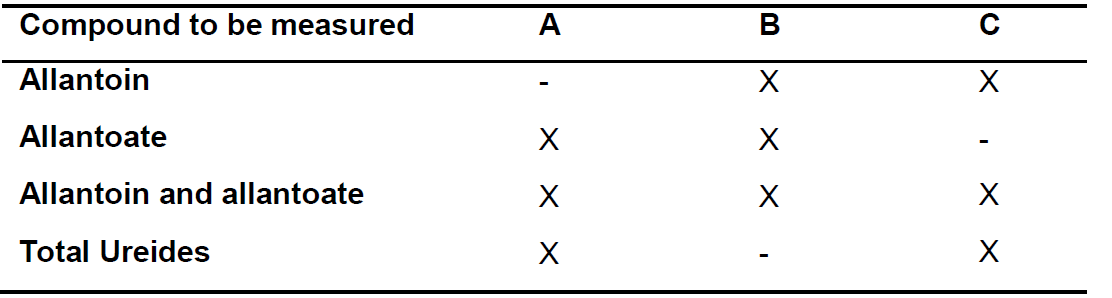
- Prepare tubes according to the compound/s to be measured as indicated in Table 1. For example, to measure allantoin and allantoate concentrations in sample #1 tubes can be designated 1A, 1B and 1C.
- For the glyoxylate standard curve prepare a fresh serial dilution of 0, 2, 4, 6, 8 and 10 nmols sodium glyoxylate monohydrate in 100 µl Extraction Buffer and follow the steps corresponding to glyoxylate quantification (reaction A). It is not necessary to repeat this step once a proper glyoxylate standard curve is obtained.
Note: It is recommended to use triplicates for each concentration of the glyoxylate standard curve.
Follow the corresponding steps for each reaction as indicated in Table 2. Perform the same steps for the corresponding blank tubes, but adding 100 µl Extraction Buffer instead of the sample (Step C3a). In Steps C3d, C3e, C3h and C3i. ensure that all the tubes keep well closed.
Samples can be stored at -20 °C for several months after Step C3j, or maintained at 4-8 °C to continue the protocol immediately.
Note: It is recommended to perform the same reactions simultaneously for the analyzed samples. In the meantime, tubes of other reactions can be stored at 4-8 °C.
Table 2. Ureide hydrolysis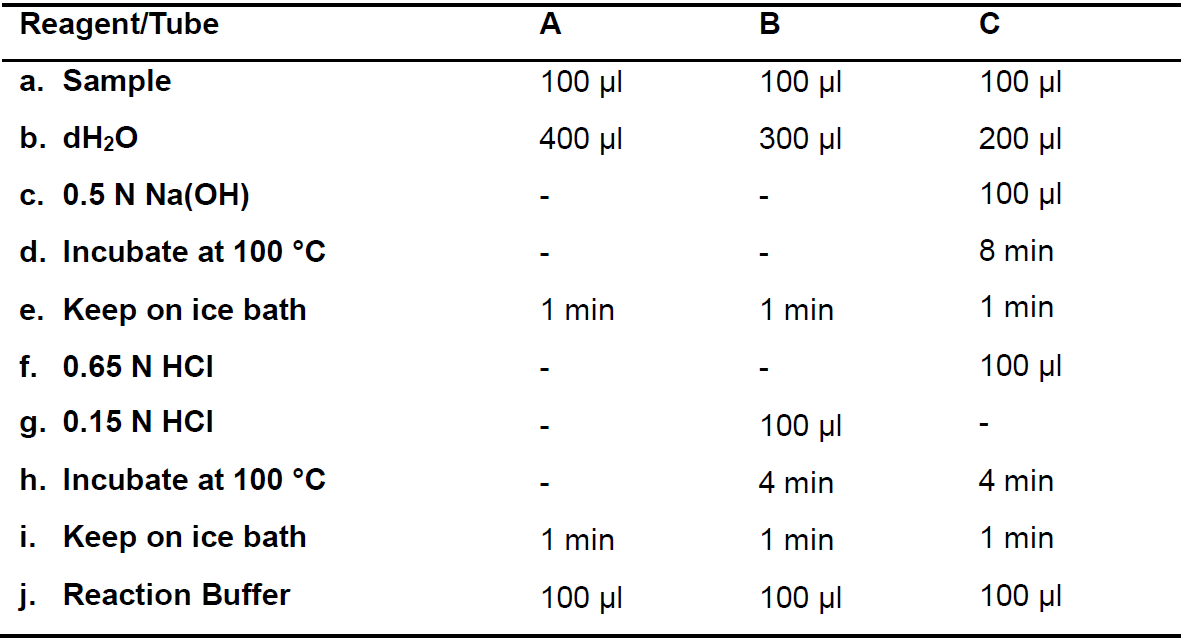
- Prepare microcentrifugue tubes in which reactions A-C for each sample will be carried out, according the compound/s to be measured (Figure 1). Designate the tubes according which reaction take place in it: A, B and C.
- Spectrophotometric measurements
Note: Perform HCl work in a fume hood, with the appropriate personal protective equipment.- Prepare fresh 3mg/ml phenylhydrazine and 16 mg/ml potassium ferricyanide with dH2O in Falcon tubes. Mix the tubes content by inverting and shaking vigorously on a vortex. Store these solutions in the dark at RT.
- Add 100 µl phenylhydrazine 3 mg/ml. Mix on a vortex and incubate for 5 min at RT.
- Add 500 µl 37% HCl and mix by inversion.
- Add 100 µl 16 mg/ml potassium ferricyanide and mix by inversion. A pink to red color should be observed.
- Incubate for 10 min at RT.
- Measure the absorbance of 1,5-diphenylformazan at 520 nm (Abs520) at RT. One value of Abs520 is obtained for each reaction tube, namely Abs520A, Abs520B, Abs520C.
Notes:- If the Abs520 is above the linear range determined with the standard curve, make a dilution of the sample in 18.5% HCl and measure again immediately. 1/5 and 1/10 dilution are recommended (Dilution factors 5 and 10, respectively).
- Make a table to write the obtained data for each sample: Fresh weight, Dry weight, Dilution factor, Abs520A, Abs520B, Abs520C.
- If the Abs520 is above the linear range determined with the standard curve, make a dilution of the sample in 18.5% HCl and measure again immediately. 1/5 and 1/10 dilution are recommended (Dilution factors 5 and 10, respectively).
Data analysis
- Determine values of the intercept and slope of the standard curve. A typical glyoxylate standard curve is shown in Figure 3.
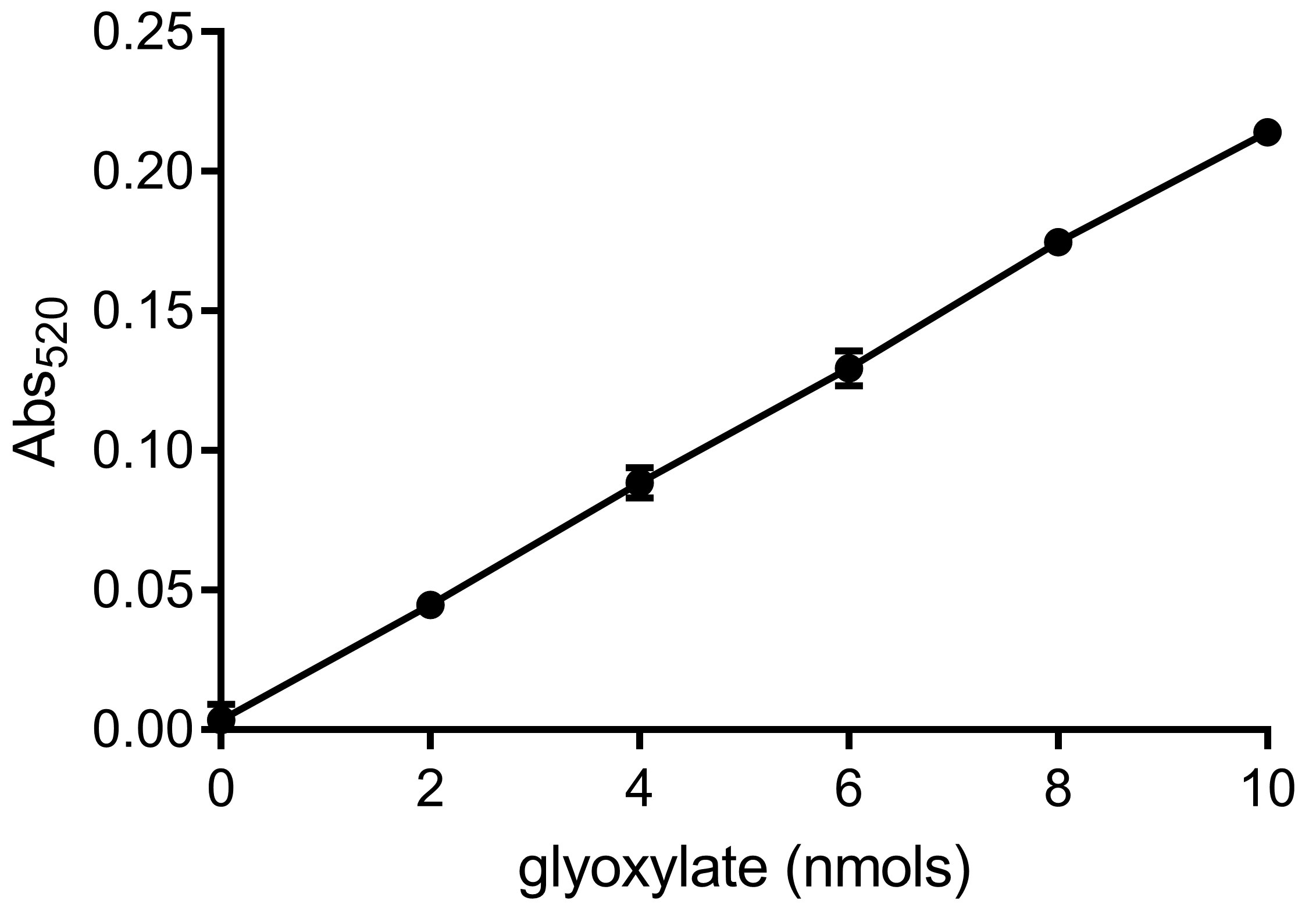
Figure 3. A representative glyoxylate standard curve. The standard curve is generated by performing a serial dilution of sodium glyoxylate monohydrate. Intercept = 0.0006; Slope = 0.0214; R2 = 0.9998 (10 nmol point was not taken in account for calculation). - Calculate glyoxylate content (G) in tubes A, B and C using the formula [Abs520 – intercept]/slope to obtain values GA, GB and GC.
- Ureide content in the sample can be calculated using the following equation:
total ureide content (nmol) = [GC-GA] = [Allantoin content + Allantoate content]
allantoin content (nmol) = [GC-GA]
allantoate content (nmol) = [GB-GA] - Determine ureide concentration of the sample (nmol/mg dry weight) using the formula:
total ureide concentration = [total ureide content (nmol)] × [dilution factor]/[dry weight (mg)]
allantoin concentration = [allantoin content (nmol)] × [dilution factor]/[dry weight (mg)]
allantoate concentration = [total allantoate content (nmol)] × [dilution factor]/[dry weight (mg)] - Report ureide concentrations of the analyzed material/s as nmol/mg dry weight (DW), e.g., allantoin and allantoate concentrations in different Arabidopsis thaliana tissues are shown in Figure 4. The described protocol was also used for determine changes in ureide concentration in shoots of Arabidopsis plants grown under different nutritional and stress conditions (Lescano et al., 2016 and 2020). Allantoin concentration was 18.2 nmol/mg in the roots and 6.3 nmol/mg in the shoots of 2 week-old Arabidopsis roots (Lescano et al., 2016, Figure 8). Allantoin and allantoate concentration from xylem sap of 6 week-old Arabidopsis were 0.27 ± 0.07 nmol/μl and 0.09 ± 0.08 nmol/μl, respectively. This protocol was also carried out for determine ureide concentration in roots and shoots of nodulating and non-nodulating soybean plants (Muñoz et al., 2016).
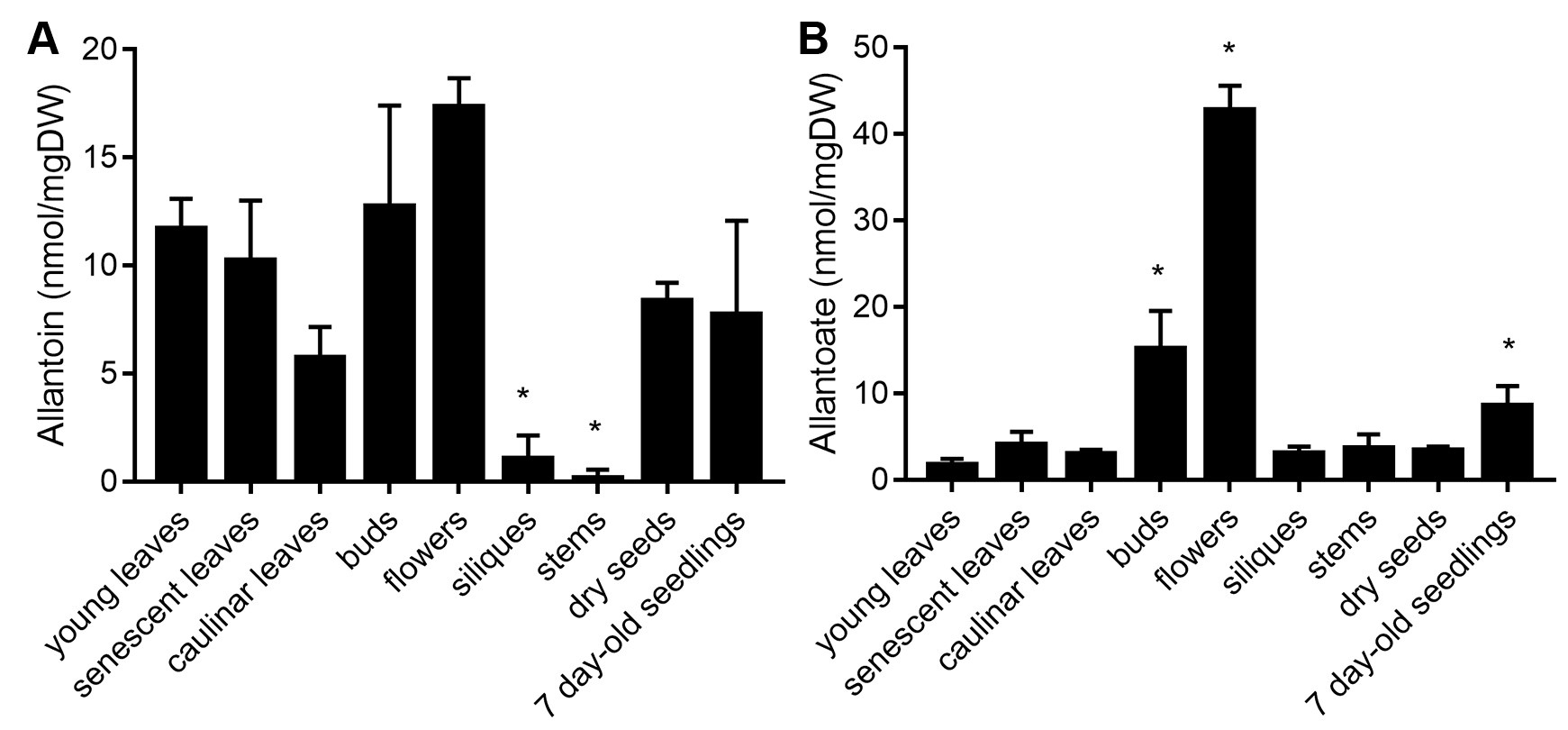
Figure 4. Ureide concentration in different Arabidopsis tissues. Allantoin and allantoate content of different organs and tissues of adult plants and 7 day-old whole seedlings. DW = Dry weight. Asterisks indicates significant differences in comparison to young leaves samples (P < 0.05, Dunn’s test)
Recipes
- Reaction Buffer (0.4 M potassium phosphate buffer)
- Prepare 500 ml 0.4 M monopotassium phosphate and 500 ml 0.4 M dipotassium phosphate by dissolving the corresponding amount of salts in dH2O
- Put 200 ml of 0.4 M dipotassium phosphate in a beaker
- Bring the solution to pH 7 by adding as much as needed of 0.4 M monopotassium phosphate. The resulting solution is 0.4 M potassium phosphate buffer. Store Reaction Buffer at 4-8 °C and stock solutions at -20 °C
- Prepare 500 ml 0.4 M monopotassium phosphate and 500 ml 0.4 M dipotassium phosphate by dissolving the corresponding amount of salts in dH2O
- Extraction Buffer (25 mM potassium phosphate buffer)
- Put 31.25 ml Reaction Buffer in a beaker
- Bring the solution to pH 7 with 0.5 N HCl
- And add dH2O to give a final volume of 500 ml
- Put 31.25 ml Reaction Buffer in a beaker
Acknowledgments
The present procedures are derived from the work of Lescano et al. (2016) and Lescano et al. (2020). This work was supported by the National Fund of Science and Technology (FONCyT, Argentina) [PICT-2009-0114] and of the Secretary of Science and Technology of the National University of Córdoba (SECyT-UNC, Argentina). The author gratefully acknowledges to Biol. María Laura Rojas for the critical revision of this protocol.
Competing interests
There are no conflict of interests.
References
- Brychkova, G., Alikulov, Z., Fluhr, R. and Sagi, M. (2008). A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. Plant J 54(3): 496-509.
- Duran, V. A. and Todd, C. D. (2012). Four allantoinase genes are expressed in nitrogen-fixing soybean. Plant Physiol Biochem 54: 149-155.
- Irani, S. and Todd, C. D. (2016). Ureide metabolism under abiotic stress in Arabidopsis thaliana. J Plant Physiol 199: 87-95.
- Irani, S. and Todd, C. D. (2018). Exogenous allantoin increases Arabidopsis seedlings tolerance to NaCl stress and regulates expression of oxidative stress response genes. J Plant Physiol 221: 43-50.
- Lescano, C. I., Martini, C., González, C. A. and Desimone, M. (2016). Allantoin accumulation mediated by allantoinase downregulation and transport by Ureide Permease 5 confers salt stress tolerance to Arabidopsis plants. Plant Mol Biol 91(4-5): 581-595.
- Lescano, I., Bogino, M. F., Martini, C., Tessi, T. M., González, C. A., Schumacher, K. and Desimone, M. (2019). Arabidopsis thaliana Ureide Permease 5 (AtUPS5) connects cell compartments involved in Ureide metabolism. Plant Physiol: pp.01136.2019.
- Muñoz, N., Qi, X., Li, M. W., Xie, M., Gao, Y., Cheung, M. Y., Wong, F. L. and Lam, H. M. (2016). Improvement in nitrogen fixation capacity could be part of the domestication process in soybean. Heredity (Edinb) 117(2): 84-93.
- Vogels, G. D. and Van der Drift, C. (1970). Differential analyses of glyoxylate derivatives. Anal Biochem 33(1): 143-157.
- Watanabe, S., Matsumoto, M., Hakomori, Y., Takagi, H., Shimada, H. and Sakamoto, A. (2014). The purine metabolite allantoin enhances abiotic stress tolerance through synergistic activation of abscisic acid metabolism. Plant Cell Environ 37(4): 1022-1036.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Lescano, I. (2020). Determination of Ureides Content in Plant Tissues. Bio-protocol 10(11): e3642. DOI: 10.21769/BioProtoc.3642.
- Lescano, I., Bogino, M. F., Martini, C., Tessi, T. M., González, C. A., Schumacher, K. and Desimone, M. (2019). Arabidopsis thaliana Ureide Permease 5 (AtUPS5) connects cell compartments involved in Ureide metabolism. Plant Physiol: pp.01136.2019.
Category
Plant Science > Plant biochemistry > Metabolite
Plant Science > Plant physiology > Metabolism
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










