- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Split Nano Luciferase-based Assay to Measure Assembly of Japanese Encephalitis Virus
Published: Vol 10, Iss 9, May 5, 2020 DOI: 10.21769/BioProtoc.3606 Views: 4321
Reviewed by: David PaulAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Selective Isolation of Retroviruses from Extracellular Vesicles by Intact Virion Immunoprecipitation
Tyler Milston Renner [...] Marc-André Langlois
Sep 5, 2018 12159 Views

Fluorescence Microscopy Assay to Measure HIV-1 Capsid Uncoating Kinetics in vitro
Chantal L. Márquez [...] Till Böcking
Jul 5, 2019 6844 Views

Preparation of a Bacteriophage T4-based Prokaryotic-eukaryotic Hybrid Viral Vector for Delivery of Large Cargos of Genes and Proteins into Human Cells
Jingen Zhu [...] Venigalla B. Rao
Apr 5, 2020 5629 Views
Abstract
Cells infected with flavivirus release various forms of infectious and non-infectious particles as products and by-products. Comprehensive profiling of the released particles by density gradient centrifugation is informative for understanding viral particle assembly. However, it is difficult to detect low-abundance minor particles in such analyses. We developed a method for viral particle analysis that integrates a high-sensitivity split luciferase system and density gradient centrifugation. This protocol enables high-resolution profiling of particles produced by cells expressing Japanese encephalitis virus factors.
Keywords: FlavivirusBackground
The flavivirus, a group of arboviruses including dengue virus, West Nile virus, tick-borne encephalitis virus, Zika virus, yellow fever virus, and Japanese encephalitis virus (JEV), is associated with substantial morbidity and mortality in human populations (Chambers et al., 1990). Flaviviruses have a single ORF genome encoding a long polypeptide, which is post-translationally cleaved into three structural (C, prM and E) and seven non-structural (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) proteins by the host signal peptidase and NS2B-NS3 viral protease (Chambers et al., 1990).
Viral genomic RNA synthesized by the NS5 RNA-dependent RNA polymerase associates with C protein to form the nucleocapsid, which in turn is packaged in the envelope composed of host cell-derived lipid membrane, and viral prM and E transmembrane proteins to form the virion. It has been found that a reduced form of flaviviral genomic RNA, designated as subgenomic replicon, which lacks all structural genes, can replicate itself in host cells (Suzuki et al., 2014), and that the subgenomic RNA is incorporated into the virus-like particle to form single-round infectious particles (SRIPs) when C, prM and E proteins are supplied in trans (Suzuki et al., 2014; Matsuda et al., 2018).
Besides major infectious particles, virus-infected cells often produce minor particles, some of which are hard to detect with immunochemical methods, due to their small amounts. To study typical and atypical particles released from flavivirus-infected cells, we modified a SRIP system (Matsuda et al., 2018) to develop a JEV HiBiT-SRIP assay system. In this system, the C protein is labeled with a highly sensitive HiBiT peptide tag, to enable high-resolution sedimentation profiling of particles produced by the cells expressing viral structural and nonstructural factors (Ishida et al., 2019). Cells transfected with a set of plasmids (referred to as SRIPs-producer cells) produce SRIPs and non-infectious subviral particles containing C-HiBiT, which can be separated by sucrose density gradient centrifugation. SRIPs in sedimentation fractions can be detected by measurement of cellular NanoLuc levels after inoculation of other cells (referred to as target cells) with all the fractions, which allows SRIP-borne NanoLuc expression (Figure 1). It is possible to apply the principle of JEV HiBiT-SRIP assay to other viruses.
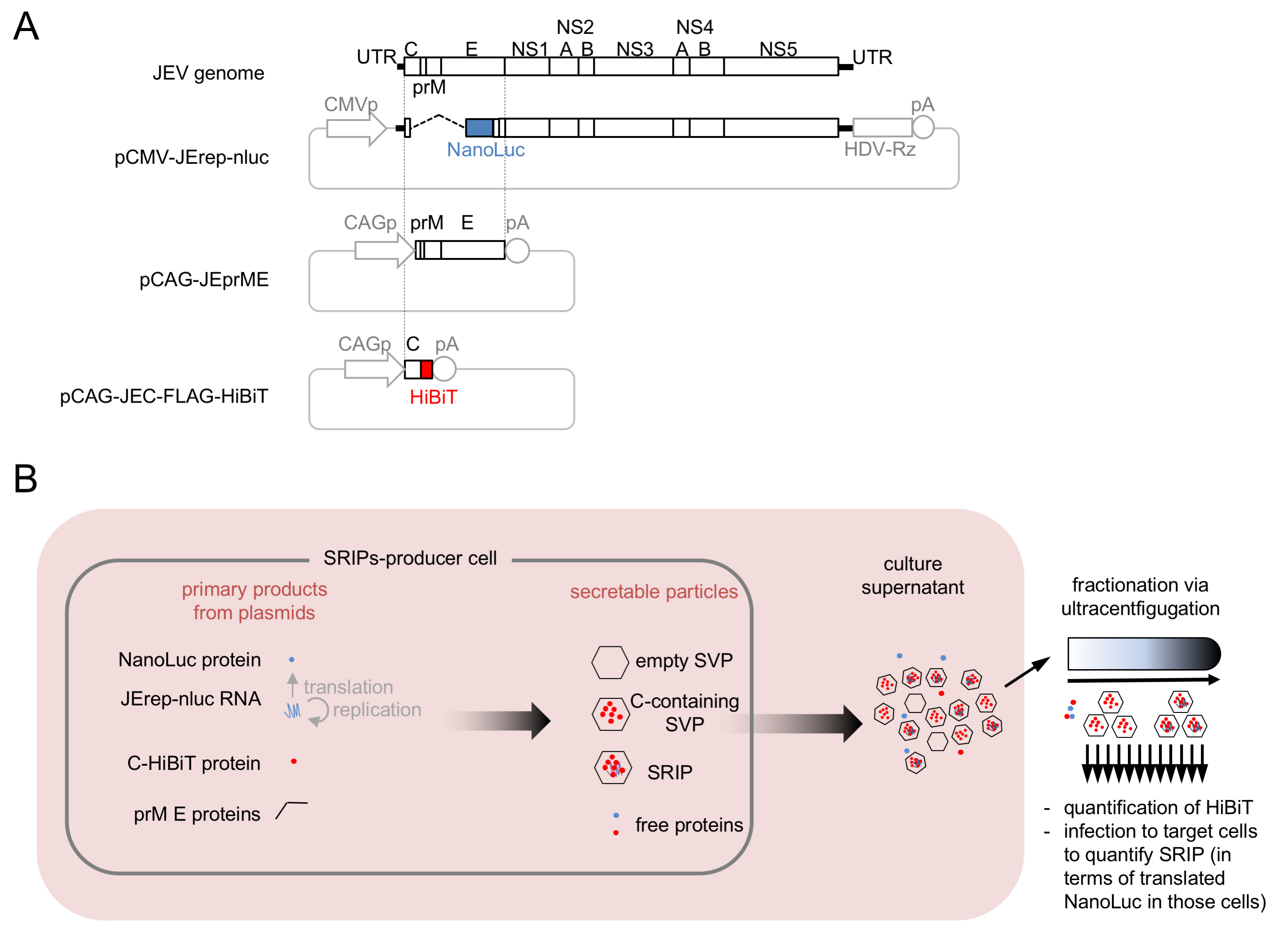
Figure 1. Overview of HiBiT-SRIP system. A. HiBiT-SRIP system plasmids. HDV-Rz: self-cleaving HDV Ribozyme; pA: SV40 polyadenylation signal; CMVp: CMV promoter; CAGp: CAG promoter. B. Schematic presentation of HiBiT-SRIP experiments. JErep-nluc: subgenomic replicon with a gene for NanoLuc as the reporter.
Materials and Reagents
- 1.5 ml tube (Watson, catalog number: 131-815C )
- 2 ml tube (Watson, catalog number: 132-620C )
- 6-well cell culture plate (Violamo, catalog number: 2-8588-01 )
- 96-well cell culture plate (Violamo, catalog number: 2-8588-05 )
- 14 x 95 mm open-top polyclear centrifuge tubes (Seton Scientific, catalog number: 7031 )
- White 384-well immuno plates (Thermo, catalog number: 460372 )
- 0.22 μm filter (Merck Millipore, catalog number: UFC30GV00 )
- 293T cells (ATCC, catalog number: CRL-3216 )
- Huh7 cells (JCRB, catalog number: JCRB0403 )
- Plasmid pCAG-JEC-FLAG-HiBiT which encodes HiBiT-tagged wild-type JEV C protein (Ishida et al., 2019; available from authors upon request) or its variant encoding mutant forms of C protein
- Plasmid pCAG-prME which encodes JEV prM and E proteins (Suzuki et al., 2014; available from authors upon request)
- Plasmid pCMV-JErep-nluc which expresses a subgenomic replicon RNA of JEV, which contains a NanoLuc reporter gene (Matsuda et al., 2018, Ishida et al., 2019; available from authors upon request)
- Nano Glo HiBiT lytic detection system (Promega, catalog number: N3040 )
- Nano-Glo luciferase assay system (Promega, catalog number: N1120 )
- Dulbecco’s modified Eagle’s media (Nacalai, catalog number: 08458-16 )
- FBS (fetal bovine serum) (Thermo, catalog number: 10270-106 )
- PBS (phosphate-buffered saline without calcium and magnesium) (Nacalai, catalog number: 14249-24 )
- Benzylpenicillin potassium (Fujifilm, catalog number: 021-07732 )
- Streptomycin sulfate (Tokyo Chemical Industry, catalog number: S0585 )
- PEI MAX (Polyscience, catalog number: 24765-1 ), a transfection reagent
- Opti-MEM (Thermo, catalog number: 31985062 ), reduced serum medium which allows to keep high transfection efficiency with less cytotoxicity
- Sucrose, centrifugal density-gradient grade (Nacalai, catalog number: 30406-25 )
- Tris (Tris (hydroxymethyl) aminomethane) (Nacalai, catalog number: 35406-91 )
- NaCl (Nacalai, catalog number: 31320-05 )
- Triton X-100 (Nacalai, catalog number: 35501-15 )
- KCl (Nacalai, catalog number: 28513-85 )
- Na2HPO4 (Nacalai, catalog number: 31726-05 )
- KH2PO4 (Nacalai, catalog number: 28720-65 )
- PEI solution (1 mg/ml) (see Recipes)
- 100x penicillin G + streptomycin stock solution (see Recipes)
- Culture media (see Recipes)
- 10x PBS (pH 7.4) (see Recipes)
- 10% sucrose/PBS (see Recipes)
- 45% sucrose/PBS (see Recipes)
- 10-45% sucrose linear gradient/PBS (see Recipes)
- Lysis buffer (see Recipes)
Equipment
- Humidified incubator (PHCBI, model: MCO-170AICUVD-PJ , 37 °C, 5% CO2)
- Ultracentrifuge (Hitachi, model: CP80NX )
- Swing bucket rotor (Hitachi, model: P40ST )
- Gradient mixer (BIOCOMP, Gradient Mate)
- Piston gradient fractionator (BIOCOMP, catalog number: 153 )
- Microplate luminometer (Thermo, Varioskan LUX Multimode Microplate Reader)
- -30 °C freezer (PHCBI, catalog number: MDF-MU539D )
Procedure
Flow of following procedures is presented in Figure 2.
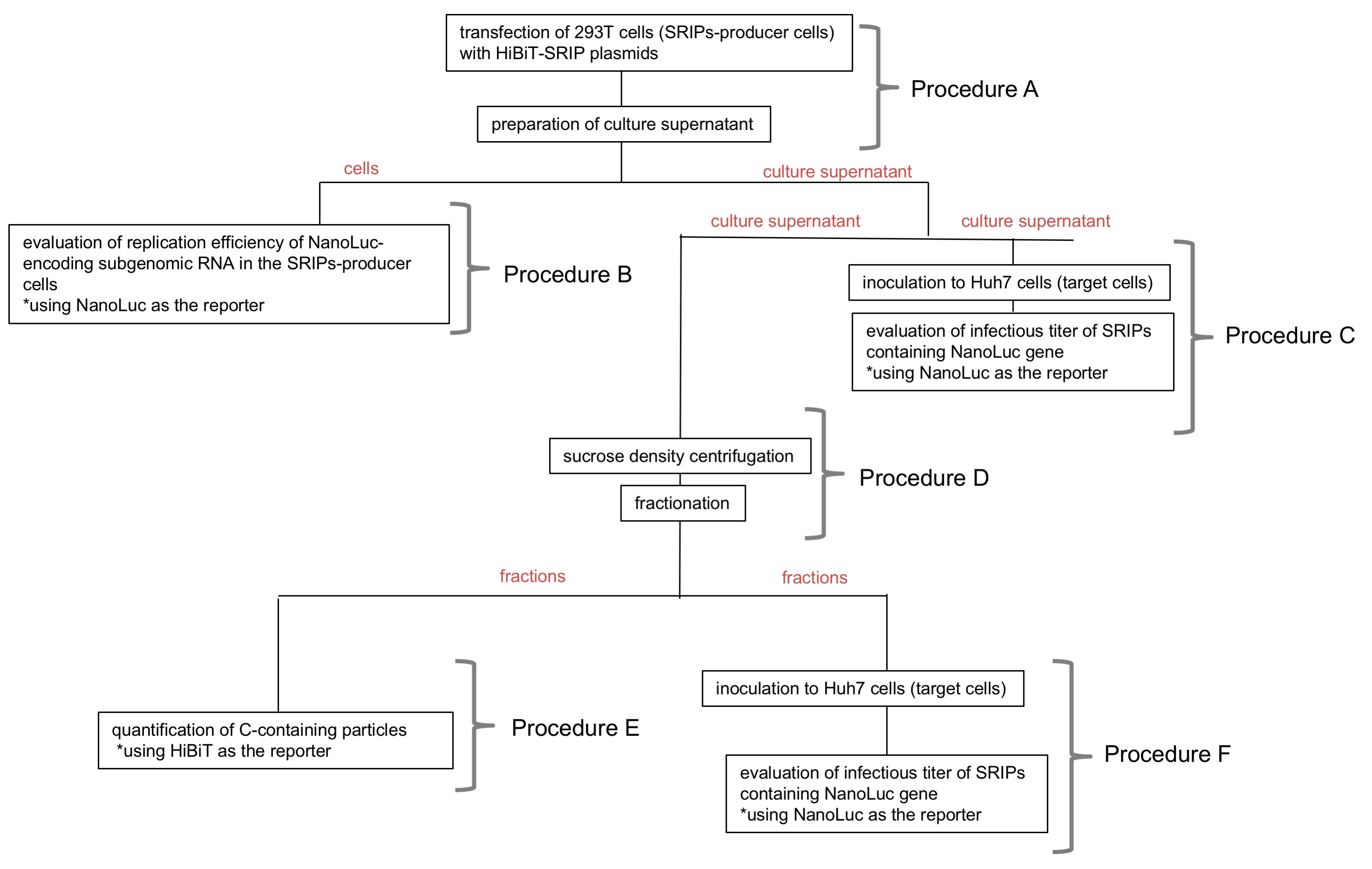
Figure 2. Flow of procedures
- Production of HiBiT-containing particles
- Seed 293T cells in 6-well plates, at density of ~50,000 cells/well in 2 ml growth medium.
- Place the plates in a CO2 incubator for 24 h.
- Mix 0.2 µg pCAG-JEC-HiBiT, 0.4 µg pCMV-JErep-nluc and 0.2 µg pCAG-JEprME in 200 µl Opti-MEM, mix well with a vortex mixer.
(Optional) Set control experiments using an empty vector in place of each plasmid.
(Optional) Set experiments using plasmids encoding mutant forms of viral elements of interest. - Add 2.4 µl PEI solution (1 mg/ml) to the mixture and mix well with a vortex mixer.
- Incubate the DNA-PEI mixture for 15 min at room temperature.
- Apply the DNA-PEI mixture to cell-containing wells (~200 μl/well).
- Place the plates in a CO2 incubator.
- Replenish growth media 6 h post-transfection.
Note: Add media slowly to the side of the well to avoid cell detachment. - Replenish growth media 48 h post-transfection.
Note: Add media slowly to the side of the well to avoid cell detachment. - Transfer the culture supernatant 72 h post-transfection to 2 ml tubes.
- Centrifuge the tube at 20,000 x g, 4 °C for 10 min.
- Transfer supernatant to fresh tubes and proceed to Procedure B or Procedure C.
- Evaluation of replication efficiency of the subgenomic replicon RNA in SRIPs-producer cells (a typical result is presented in Figure 2A)
- Suspend cells in each well in 1 ml PBS and transfer to a 1.5 ml tube.
- Centrifuge the tubes at 2,400 x g, 4 °C for 5 min.
- Aspirate the supernatant, add 50 μl lysis buffer and mix by pipetting.
- Centrifuge the tubes at 20,000 x g, 4 °C for 10 min.
- Transfer 5 μl of the clear lysate in each tube to 384-well plate.
- Add 5 μl of NanoLuc substrate diluted 1:600 with lysis buffer.
- Incubate for 10 min at room temperature.
- Measure the luminescence using a microplate reader.
- Titration of SRIPs in culture supernatant (a typical result is presented in Figure 2C)
- Seed Huh7 cells in 96-well plates, at density of ~10,000 cells/well and incubate in a CO2 incubator overnight.
- Inoculate 100 μl of culture supernatant (prepared with Procedure B) into each well.
- Incubate for 72 h.
- Aspirate growth media and add 50 μl lysis buffer to each well and mix by pipetting.
- Transfer 5 μl of the lysate in each well to a 384-well plate.
- Add 5 µl of NanoLuc substrate diluted 1:600 with lysis buffer.
- Incubate for 10 min at room temperature.
- Measure the luminescence using a microplate reader.
- Sucrose density gradient centrifugation of the culture supernatant
- Layer 300 μl of the culture supernatant containing HiBiT-labeled particles on continuous 10-45% sucrose gradient/PBS in Open-Top Polyclear centrifuge tubes (SETON, 14 x 95 mm).
- Centrifuge the gradients at 100,000 x g for 3 h at 4 °C with a pre-chilled swing bucket rotor.
- Fractionate the gradients from top to bottom (total < 40 fraction), with a piston fractionator (2.3 mm/fraction, 0.3 mm/s) to a 96-well plate.
- Titration of C-HiBiT-containing particles in sedimentation fractions (a typical result is presented in Figure 2B)
- Transfer 5 μl of the fraction in each well to 384-well plate.
- Add 5 μl of HiBiT reagent to each well.
- Incubate for 10 min at room temperature.
- Measure the luminescence using a microplate reader. Typical results out of 3 > experiments are indicated in the figures.
- Titration of SRIPs in sedimentation fractions (a typical result is presented in Figure 2D)
- Seed Huh7 cells in 96-well plates, at density of ~10,000 cells/well and incubate in a CO2 incubator overnight.
- Inoculate 5 μl of sedimentation fractions to each well.
- Incubate for 72 h.
- Aspirate growth media and add 50 μl lysis buffer to each well and mix by pipetting.
- Transfer 5 μl of the lysate in each well to 384-well plate.
- Add 5 µl of NanoLuc substrate diluted 600 fold with lysis buffer.
- Incubate for 10 min at room temperature.
- Measure the luminescence using a microplate reader (Figure 3).
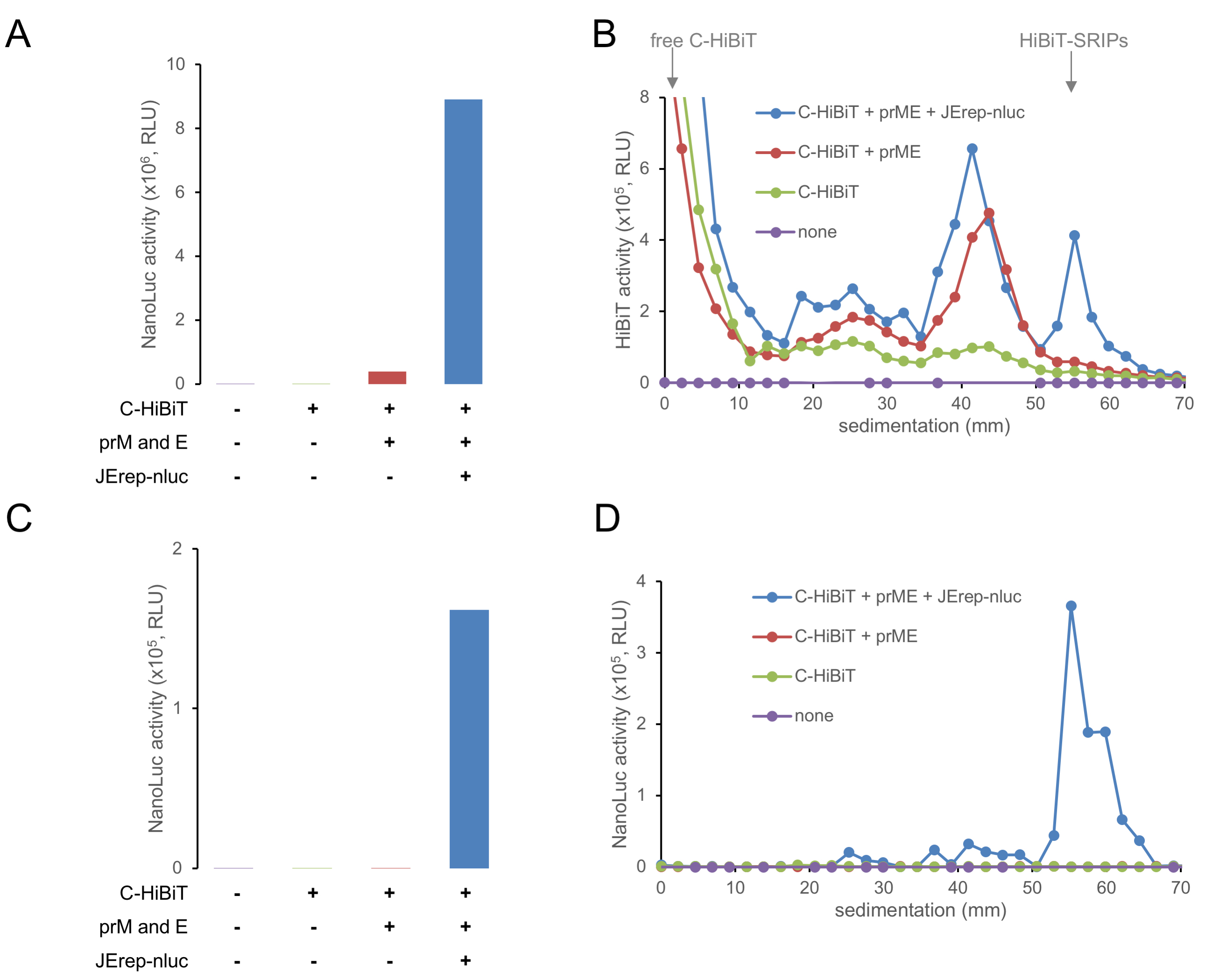
Figure 3. Typical results. (A) NanoLuc activity of producer cell lysate to evaluate autonomous replication of the subgenomic RNA (B) HiBiT activities of fractions of producer cell culture supernatant, fractionated via sucrose density gradient centrifugation. (C) NanoLuc activity of target cells inoculated with producer cell supernatant, which reflects the infectious titer of SRIPs in the supernatant. (D) NanoLuc activities of target cells inoculated with sucrose density gradient centrifugation fractions of producer cell supernatant, which reflect the infectious titer of SRIPs in the fractions.
Notes
- Strong luminescence emitted by NanoLuc/HiBiT system sometimes leaks to adjacent wells in 384-well plates. Therefore, the recommendation is to load samples into every other well. In addition, sometimes it is better to dilute lysates or fractions which produce strong signals to minimize leakage of luminescence to neighboring wells (Procedures B, D, E and F).
- Be aware that it is difficult to estimate relative amounts of C-HiBiT in top fractions when titrating HiBiT-SRIPs in sedimentation fractions (Procedure B), because free NanoLuc proteins that leak from cells in the fractions also contribute to signal production.
Recipes
- PEI solution (1 mg/ml)
PEI MAX 100 mg
ddH2O up to 100 ml
Sterilize via a 0.22 μm filter, aliquot to 15 ml tubes and keep a tube as a working stock in a refrigerator and the rest of the tubes in a -30 °C freezer - 100x penicillin G + streptomycin stock solution
PBS 100 ml
Benzylpenicillin potassium 0.626 g
Streptomycin sulfate 1 g
Sterilize via a 0.22 μm filter and store in a refrigerator - Culture media
Dulbecco’s modified Eagle’s medium 500 ml
FBS, heat-inactivated by incubation at 56 °C for 45 min 50 ml
100x penicillin G + streptomycin stock solution 5 ml - 10x PBS (pH 7.4)
NaCl 80 g
KCl 2.0 g
Na2HPO4 14.4 g
KH2PO4 2.4 g
ddH2O up to 1 L - 10% sucrose/PBS
Sucrose 100 g
10x PBS 100 ml
ddH2O up to 1 L
Sterilize via a 0.22 μm filter and store at room temperature - 45% sucrose/PBS
Sucrose 450 g
10x PBS 100 ml
ddH2O up to 1 L
Sterilize via a 0.22 μm filter and store at room temperature - 10-45% sucrose linear gradient/PBS
Pour appropriate volume of 10% sucrose/PBS to 14 x 95 mm open-top polyclear centrifuge tubes, underlay 45% sucrose/PBS from bottom of the tubes using a needle attached syringe. Form linear gradients using gradient former with following parameters:
Time: 2 min 5 s
Angle: 82°
Speed: 16 rpm
After forming the gradients, chill them by standing several h at 4 °C - Lysis buffer
1 M Tris pH 7.5 20 ml
5 M NaCl 30 ml
Triton X-100 10.7 g
ddH2O up to 1 L
Acknowledgments
We thank Jiahui Ong for proofreading the manuscript. We thank Mami Matsuda for the helpful advice and suggestions to develop HiBiT-SRIP system.
Competing interests
The authors have no competing interests directly relevant to the content of this article.
References
- Chambers, T. J., Hahn, C. S., Galler, R. and Rice, C. M. (1990). Flavivirus genome organization, expression, and replication. Annu Rev Microbiol 44: 649-688.
- Ishida, K., Goto, S., Ishimura, M., Amanuma, M., Hara, Y., Suzuki, R., Katoh, K. and Morita, E. (2019). Functional correlation between subcellular localizations of japanese encephalitis virus capsid protein and virus production. J Virol 93(19).
- Matsuda, M., Yamanaka, A., Yato, K., Yoshii, K., Watashi, K., Aizaki, H., Konishi, E., Takasaki, T., Kato, T., Muramatsu, M., Wakita, T. and Suzuki, R. (2018). High-throughput neutralization assay for multiple flaviviruses based on single-round infectious particles using dengue virus type 1 reporter replicon. Sci Rep 8(1): 16624.
- Suzuki, R., Ishikawa, T., Konishi, E., Matsuda, M., Watashi, K., Aizaki, H., Takasaki, T. and Wakita, T. (2014). Production of single-round infectious chimeric flaviviruses with DNA-based Japanese encephalitis virus replicon. J Gen Virol 95(Pt 1): 60-65.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Goto, S., Ishida, K., Suzuki, R. and Morita, E. (2020). Split Nano Luciferase-based Assay to Measure Assembly of Japanese Encephalitis Virus. Bio-protocol 10(9): e3606. DOI: 10.21769/BioProtoc.3606.
Category
Molecular Biology > Nanoparticle > Plan-derived nanoparticles
Molecular Biology > Protein > Protein-protein interaction
Biochemistry > Protein > Self-assembly
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








